400:
861:
221:
1168:-ATPase that sodium levels will begin to increase within the cell which ultimately increases the concentration of intracellular calcium via the sodium-calcium exchanger. This increased presence of calcium is what allows for the force of contraction to be increased. In the case of patients where the heart is not pumping hard enough to provide what is needed for the body, use of digoxin helps to temporarily overcome this.
1283:-ATPase has two paralogs, ATPα (ATPα1) and JYalpha (ATPα2), resulting from an ancient duplication in insects. In Drosophila, ATPα1 is ubiquitously and highly expressed, whereas ATPα2 is most highly expressed in male testes and is essential for male fertility. Insects have at least one copy of both genes, and occasionally duplications. Low expression of ATPα2 has also been noted in other insects. Duplications and
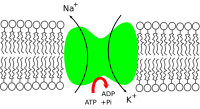
313:). For every ATP molecule that the pump uses, three sodium ions are exported and two potassium ions are imported. Thus, there is a net export of a single positive charge per pump cycle. The net effect is an extracellular concentration of sodium ions which is 5 times the intracellular concentration, and an intracellular concentration of potassium ions which is 30 times the extracellular concentration.
5575:
229:
1164:) which essentially binds "to the extracellular part of enzyme i.e. that binds potassium, when it is in a phosphorylated state, to transfer potassium inside the cell" After this essential binding occurs, a dephosphorylation of the alpha subunit occurs which reduces the effect of cardiac disease. It is via the inhibiting of the
43:
864:
The sodium–potassium pump is found in many cell (plasma) membranes. Powered by ATP, the pump moves sodium and potassium ions in opposite directions, each against its concentration gradient. In a single cycle of the pump, three sodium ions are extruded from and two potassium ions are imported into the
1129:, which may be the mechanism of the long-term inotropic effect of cardiac glycosides such as digoxin. The problem with this hypothesis is that at pharmacological concentrations of digitalis, less than 5% of Na/K-ATPase molecules – specifically the α2 isoform in heart and arterial smooth muscle (
1264:
Several studies have detailed the evolution of cardiotonic steroid resistance of the alpha-subunit gene family of Na/K-ATPase (ATP1A) in vertebrates via amino acid substitutions most often located in the first extracellular loop domain. Amino acid substitutions conferring cardiotonic steroid
1265:
resistance have evolved independently many times in all major groups of tetrapods. ATP1A1 has been duplicated in some groups of frogs and neofunctionlised duplicates carry the same cardiotonic steroid resistance substitutions (Q111R and N122D) found in mice, rats and other muroids.
641:
by comparing the active and estivating states. They concluded that reversible phosphorylation can control the same means of coordinating ATP use by this ion pump with the rates of the ATP generation by catabolic pathways in estivating
359:(ROS), as well as intracellular calcium. In fact, all cells expend a large fraction of the ATP they produce (typically 30% and up to 70% in nerve cells) to maintain their required cytosolic Na and K concentrations. For neurons, the
453:
ions have the same charge, they can still have very different equilibrium potentials for both outside and/or inside concentrations. The sodium-potassium pump moves toward a nonequilibrium state with the relative concentrations of
320:, who was awarded a Nobel Prize for his work in 1997. Its discovery marked an important step forward in the understanding of how ions get into and out of cells, and it has particular significance for excitable cells such as
3138:
Mohammadi, Shabnam; Yang, Lu; Harpak, Arbel; Herrera-Álvarez, Santiago; Rodríguez-Ordoñez, María del Pilar; Peng, Julie; Zhang, Karen; Storz, Jay F.; Dobler, Susanne; Crawford, Andrew J.; Andolfatto, Peter (2021-06-21).
3406:"Structure-function relationships in the sodium-potassium ATPase .alpha. subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme"
1295:. Insects adapted to cardiotonic steroids typically have a number of amino acid substitutions, most often in the first extra-cellular loop of ATPα1, that confer resistance to cardiotonic steroid inhibition.
3305:
Mohammadi, Shabnam; Özdemir, Halil İbrahim; Ozbek, Pemra; Sumbul, Fidan; Stiller, Josefin; Deng, Yuan; Crawford, Andrew J; Rowland, Hannah M; Storz, Jay F; Andolfatto, Peter; Dobler, Susanne (2022-12-06).
3246:
Mohammadi, Shabnam; Herrera-Álvarez, Santiago; Yang, Lu; Rodríguez-Ordoñez, María del Pilar; Zhang, Karen; Storz, Jay F.; Dobler, Susanne; Crawford, Andrew J.; Andolfatto, Peter (2022-08-16).
433:
for details). In addition, there is a short-circuit channel (i.e. a highly K-permeable ion channel) for potassium in the membrane, thus the voltage across the plasma membrane is close to the
3719:
701:
pump. However, upon subsequent ouabain binding, the Src kinase domain is released and then activated. Based on this scenario, NaKtide, a peptide Src inhibitor derived from the
4402:
2501:
Young EA, Fowler CD, Kidd GJ, Chang A, Rudick R, Fisher E, Trapp BD (April 2008). "Imaging correlates of decreased axonal Na/K ATPase in chronic multiple sclerosis lesions".
3125:
Hernández Poveda M (2022) Convergent evolution of neo-functionalized duplications of ATP1A1 in dendrobatid and grass frogs. MS Thesis
Dissertation. Universidad de los Andes
625:
Within the last decade, many independent labs have demonstrated that, in addition to the classical ion transporting, this membrane protein can also relay extracellular
184:
327:
All mammals have four different sodium pump sub-types, or isoforms. Each has unique properties and tissue expression patterns. This enzyme belongs to the family of
1707:
Watanabe D, Wada M (December 2019). "Effects of reduced muscle glycogen on excitation-contraction coupling in rat fast-twitch muscle: a glycogen removal study".
203:
371:. This was first discovered in red blood cells (Schrier, 1966), but has later been evidenced in renal cells, smooth muscles surrounding the blood vessels, and
425:). The sodium–potassium pump mechanism moves 3 sodium ions out and moves 2 potassium ions in, thus, in total, removing one positive charge carrier from the
832:
inhibits sodium–potassium pumps in the cerebellum and this is likely how it corrupts cerebellar computation and body coordination. The distribution of the
3198:"Mutations to the cardiotonic steroid binding site of Na+/K+-ATPase are associated with high level of resistance to gamabufotalin in a natricine snake"
5095:
1664:
Dutka TL, Lamb GD (September 2007). "Na-K pumps in the transverse tubular system of skeletal muscle fibers preferentially use ATP from glycolysis".
421:
In order to maintain the cell membrane potential, cells keep a low concentration of sodium ions and high levels of potassium ions within the cell (
4395:
1227:. ATP1A1 is expressed ubiquitously in vertebrates, and ATP1A3 in neural tissue. ATP1A2 is also known as "alpha(+)". ATP1A4 is specific to mammals.
4625:
1543:
Sanders MJ, Simon LM, Misfeldt DS (March 1983). "Transepithelial transport in cell culture: bioenergetics of Na-, D-glucose-coupled transport".
1140:. However, apart from the population of Na/K-ATPase in the plasma membrane, responsible for ion transport, there is another population in the
2951:
2618:
1478:
1453:
1160:-ATPase may need to be inhibited via pharmacological means. A commonly used inhibitor used in the treatment of cardiac disease is digoxin (a
4594:
4167:
1038:-ATPase can be pharmacologically modified by administering drugs exogenously. Its expression can also be modified through hormones such as
3781:
3363:
Ujvari, Beata; Mun, Hee-chang; Conigrave, Arthur D.; Bray, Alessandra; Osterkamp, Jens; Halling, Petter; Madsen, Thomas (January 2013).
4163:
2818:"Signaling mechanisms that link salt retention to hypertension: endogenous ouabain, the Na pump, the Na/Ca exchanger and TRPC proteins"
4388:
1621:
Glitsch HG, Tappe A (January 1993). "The Na/K pump of cardiac
Purkinje cells is preferentially fuelled by glycolytic ATP production".
601:
pump helps to maintain the right concentrations of ions. Furthermore, when the cell begins to swell, this automatically activates the
2703:
Lin HH, Tang MJ (January 1997). "Thyroid hormone upregulates Na,K-ATPase α and β mRNA in primary cultures of proximal tubule cells".
1000:-ATPase is endogenously negatively regulated by the inositol pyrophosphate 5-InsP7, an intracellular signaling molecule generated by
5294:
2738:
Blaustein MP (May 1977). "Sodium ions, calcium ions, blood pressure regulation, and hypertension: a reassessment and a hypothesis".
3724:
3519:
Yang, L.; Ravikanthachari, N.; Mariño-Pérez, R.; Deshmukh, R.; Wu, M.; Rosenstein, A.; Kunte, K.; Song, H.; Andolfatto, P. (2019).
2476:
3756:
2426:"Simulation of alcohol action upon a detailed Purkinje neuron model and a simpler surrogate model that runs >400 times faster"
647:
630:
196:
5185:
2226:"The Slow Dynamics of Intracellular Sodium Concentration Increase the Time Window of Neuronal Integration: A Simulation Study"
5180:
3734:
964:
163:
139:
5450:
3365:"Isolation Breeds Naivety: Island Living Robs Australian Varanid Lizards of Toad-Toxin Immunity Via Four-Base-Pair Mutation"
1752:"Inhibition of glycogenolysis prolongs action potential repriming period and impairs muscle function in rat skeletal muscle"
2016:"NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells"
363:-ATPase can be responsible for up to 3/4 of the cell's energy expenditure. In many types of tissue, ATP consumption by the
5202:
5565:
3308:"Epistatic Effects Between Amino Acid Insertions and Substitutions Mediate Toxin resistance of Vertebrate Na+,K+-ATPases"
5250:
4567:
3571:"Convergently Evolved Toxic Secondary Metabolites in Plants Drive the Parallel Molecular Evolution of Insect Resistance"
690:
2908:
Pavlovic D (2014). "The role of cardiotonic steroids in the pathogenesis of cardiomyopathy in chronic kidney disease".
5600:
5172:
3657:"Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase"
3765:
491:
5435:
1090:
is quickly returned to its normal concentration by a carrier enzyme in the plasma membrane, and a calcium pump in
5551:
5538:
5525:
5512:
5499:
5486:
5473:
5231:
4772:
4670:
4495:
4491:
4468:
4441:
4428:
4415:
4370:
4130:
4125:
1005:
490:
Export of sodium ions from the cell provides the driving force for several secondary active transporters such as
5445:
2980:
Skou JC (February 1957). "The influence of some cations on an adenosine triphosphatase from peripheral nerves".
157:
5399:
5342:
4786:
4481:
4446:
4419:
3713:
3066:
Moore, David J.; Halliday, Damien C. T.; Rowell, David M.; Robinson, Anthony J.; Keogh, J. Scott (2009-08-23).
2781:
Schoner W, Scheiner-Bobis G (September 2008). "Role of endogenous cardiotonic steroids in sodium homeostasis".
1304:
1195:
62:
3248:"Constraints on the evolution of toxin-resistant Na,K-ATPases have limited dependence on sequence divergence"
144:
5347:
5156:
5119:
4458:
3068:"Positive Darwinian selection results in resistance to cardioactive toxins in true toads (Anura: Bufonidae)"
1275:
1018:-ATPase is also regulated by reversible phosphorylation. Research has shown that in estivating animals, the
651:
646:. The downstream signals through ouabain-triggered protein phosphorylation events include activation of the
356:
1086:
release from the muscle cells' sarcoplasmic reticulum. Immediately after muscle contraction, intracellular
4886:
4526:
4223:
1091:
882:
310:
2175:"Prolonged Intracellular Na Dynamics Govern Electrical Activity in Accessory Olfactory Bulb Mitral Cells"
1586:
Lynch RM, Paul RJ (March 1987). "Compartmentation of carbohydrate metabolism in vascular smooth muscle".
208:
5368:
5287:
5016:
4924:
905:
659:
399:
372:
280:
132:
5440:
3709:
3197:
3749:
3467:
2651:
2561:
2127:
1812:
1284:
1184:
550:
549:
and glucose, which is far more efficient than simple diffusion. Similar processes are located in the
4258:
5404:
5221:
3454:
Zhen, Ying; Aardema, Matthew L.; Medina, Edgar M.; Schumer, Molly; Andolfatto, Peter (2012-09-28).
3141:"Concerted evolution reveals co-adapted amino acid substitutions in frogs that prey on toxic toads"
1248:
1236:
1180:
808:, which has symptoms to indicate that it is a pathology of cerebellar computation. Furthermore, an
467:
426:
317:
160:
2959:
84:
5337:
5236:
4802:
4380:
3598:
3005:
2933:
2890:
2763:
2685:
2526:
1898:
1781:
1732:
1689:
1646:
1568:
1077:
522:
3567:
Petschenka Georg, Vera
Wagschal, Michael von Tschirnhaus, Alexander Donath, Susanne Dobler 2017
852:
pump disfunction has been tied to various diseases, including epilepsy and brain malformations.
772:
mitral cells and probably other neuron types. This suggests that the pump might not simply be a
5595:
4476:
3688:
3637:
3590:
3550:
3501:
3483:
3433:
3425:
3386:
3345:
3327:
3287:
3269:
3225:
3217:
3178:
3160:
3105:
3087:
2997:
2925:
2882:
2847:
2798:
2755:
2720:
2677:
2614:
2589:
2518:
2457:
2406:
2357:
2308:
2257:
2206:
2155:
2096:
2047:
1996:
1947:
1890:
1840:
1773:
1724:
1681:
1638:
1603:
1560:
1525:
1474:
1449:
1423:
1372:
1287:
of ATPα1 have been observed in insects that are adapted to cardiotonic steroid toxins such as
1161:
1054:
931:
416:
408:
344:
151:
4357:
4352:
4337:
4025:
3196:
Mohammadi, Shabnam; Brodie, Edmund D.; Neuman-Lee, Lorin A.; Savitzky, Alan H. (2016-05-01).
2608:
993:
effect, but these were later found to be inaccurate due to additional complicating factors.
915:
ions to the extracellular region. The phosphorylated form of the pump has a low affinity for
840:
pump on myelinated axons in the human brain has been demonstrated to be along the internodal
5383:
5378:
5352:
5280:
4866:
4812:
4794:
4660:
4650:
4604:
4321:
4315:
4310:
4288:
4278:
4268:
4235:
4117:
4112:
4102:
3883:
3823:
3678:
3668:
3629:
3582:
3540:
3532:
3491:
3475:
3417:
3376:
3335:
3319:
3277:
3259:
3209:
3168:
3152:
3095:
3079:
3023:
2989:
2917:
2874:
2837:
2829:
2790:
2747:
2712:
2667:
2659:
2640:
drives sodium-potassium pump degradation by relieving an autoinhibitory domain of PI3K p85α"
2579:
2569:
2510:
2447:
2437:
2396:
2388:
2347:
2298:
2288:
2247:
2237:
2196:
2186:
2145:
2135:
2086:
2078:
2037:
2027:
1986:
1978:
1937:
1929:
1880:
1830:
1820:
1763:
1716:
1673:
1630:
1595:
1552:
1515:
1507:
1413:
1403:
1362:
1352:
1076:
Muscle contraction is dependent on a 100- to 10,000-times-higher-than-resting intracellular
1039:
983:
972:
860:
765:
434:
352:
306:
4679:
4299:
4139:
4134:
2116:"The sodium-potassium pump controls the intrinsic firing of the cerebellar Purkinje neuron"
120:
5430:
5414:
5327:
5213:
5102:
4916:
4616:
4436:
4247:
4073:
3769:
3742:
1343:
1309:
897:
655:
295:
3618:"Molecular adaptation of Chrysochus leaf beetles to toxic compounds in their food plants"
3041:
96:
3617:
3471:
2655:
2565:
2131:
1918:"Na/K-ATPase tethers phospholipase C and IP3 receptor into a calcium-regulatory complex"
1816:
67:
5579:
5468:
5409:
4536:
4171:
4155:
3683:
3656:
3545:
3520:
3496:
3455:
3340:
3307:
3282:
3247:
3173:
3140:
3100:
3067:
2865:
Fuerstenwerth H (2014). "On the differences between ouabain and digitalis glycosides".
2842:
2817:
2672:
2635:
2584:
2545:
2452:
2425:
2401:
2376:
2303:
2276:
2252:
2225:
2201:
2174:
2150:
2115:
2091:
2066:
2042:
2015:
1991:
1966:
1942:
1917:
1520:
1495:
1418:
1391:
1367:
1338:
829:
769:
251:
179:
2716:
1835:
1800:
474:
is 10 mM. On the other hand, in extracellular space, the usual concentration range of
220:
5589:
5373:
5332:
5080:
4690:
4251:
4063:
3381:
3364:
2993:
2689:
2634:
Chin AC, Gao Z, Riley AM, Furkert D, Wittwer C, Dutta A, et al. (October 2020).
2277:"The sodium-potassium pump is an information processing element in brain computation"
1785:
1736:
1292:
518:
422:
328:
48:
3009:
2894:
2751:
2530:
1902:
1650:
1599:
1572:
978:-coupled GPCRs. In contrast, substances causing a decrease in cAMP downregulate the
5322:
5065:
5038:
4967:
4960:
4943:
4938:
4912:
3790:
3777:
3728:
3602:
3405:
2937:
2767:
1693:
1145:
934:
of the pump, reverting it to its previous conformational state, thus releasing the
805:
277:
3569:
Petschenka, G.; Wagschal, V.; von
Tschirnhaus, M.; Donath, A.; Dobler, S. (2017).
3213:
1136:= 32 nM) – are inhibited, not enough to affect the intracellular concentration of
3264:
2878:
2833:
2544:
Smith RS, Florio M, Akula SK, Neil JE, Wang Y, Hill RS, et al. (June 2021).
2352:
2327:
2191:
2140:
5546:
5481:
5317:
5161:
2082:
1288:
1260:
The parallel evolution of resistance to cardiotonic steroids in many vertebrates
777:
773:
5574:
2554:
Proceedings of the
National Academy of Sciences of the United States of America
1805:
Proceedings of the
National Academy of Sciences of the United States of America
1720:
1677:
1496:"Updated energy budgets for neural computation in the neocortex and cerebellum"
1252:, although closely related to ATP1B1, ATP1B2, and ATP1B3, lost its function as
521:, for example, sodium is transported out of the reabsorbing cell on the blood (
5146:
4835:
3156:
2442:
1469:
Voet D, Voet JG (December 2010). "Section 20-3: ATP-Driven Active
Transport".
1392:"The Structure and Function of the Na,K-ATPase Isoforms in Health and Disease"
1357:
1183:
in 1957 while working as assistant professor at the
Department of Physiology,
893:
781:
762:
686:
582:
570:
499:
384:
375:. Recently, glycolysis has also been shown to be of particular importance for
368:
3487:
3429:
3331:
3323:
3273:
3221:
3164:
3091:
2293:
2242:
1408:
1022:-ATPase is in the phosphorylated and low activity form. Dephosphorylation of
47:
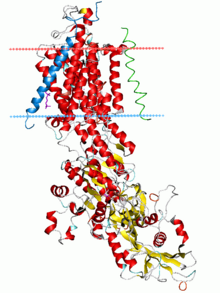
Sodium–potassium pump, E2-Pi state. Calculated hydrocarbon boundaries of the
17:
5520:
5494:
4852:
4411:
3673:
3633:
3570:
3479:
2574:
2032:
1982:
1933:
1825:
901:
738:
538:
248:
245:
3692:
3641:
3594:
3554:
3536:
3505:
3390:
3349:
3291:
3229:
3182:
3109:
3083:
3001:
2929:
2886:
2851:
2802:
2681:
2663:
2593:
2522:
2461:
2410:
2361:
2312:
2261:
2210:
2159:
2100:
2051:
2000:
1951:
1894:
1844:
1777:
1728:
1685:
1556:
1529:
1427:
1376:
407:-ATPase, as well as effects of diffusion of the involved ions maintain the
3655:
Dobler, Susanne; Dalla, Safaa; Wagschal, Vera; Agrawal, Anurag A. (2012).
3521:"Predictability in the evolution of Orthopteran cardenolide insensitivity"
3437:
2724:
1965:
Tian J, Cai T, Yuan Z, Wang H, Liu L, Haas M, et al. (January 2006).
1916:
Yuan Z, Cai T, Tian J, Ivanov AV, Giovannucci DR, Xie Z (September 2005).
1642:
1607:
1564:
1511:
693:, to form a signaling receptor complex. Src is initially inhibited by the
581:
and other organic compounds inside the cell. When this is higher than the
4231:
4030:
4020:
4015:
4000:
3995:
3990:
3980:
3975:
3970:
3960:
3955:
3945:
3940:
3935:
3930:
3911:
3903:
3799:
2794:
2759:
1865:"Suppression of Na/K-ATPase activity during estivation in the land snail
1314:
1141:
1066:
841:
825:
801:
789:
578:
380:
324:, which depend on this pump to respond to stimuli and transmit impulses.
316:
The sodium–potassium pump was discovered in 1957 by the Danish scientist
287:
3421:
462:
for both inside and outside of cell. For instance, the concentration of
228:
108:
5257:
5226:
5004:
4999:
4758:
4748:
4723:
4718:
4453:
4347:
4342:
4010:
4005:
3985:
3965:
3950:
3925:
3920:
3915:
1634:
1188:
1062:
1058:
1053:-ATPase found in the membrane of heart cells is an important target of
1043:
809:
742:
726:
626:
586:
495:
127:
2921:
2514:
2173:
Zylbertal A, Kahan A, Ben-Shaul Y, Yarom Y, Wagner S (December 2015).
1885:
1864:
761:
pump has been shown to control and set the intrinsic activity mode of
351:. It also functions as a signal transducer/integrator to regulate the
5533:
5303:
5134:
4776:
4753:
4743:
4713:
4708:
4703:
4698:
4655:
4645:
4640:
4635:
4630:
4599:
4587:
4582:
4577:
4572:
4560:
4555:
4550:
4499:
4331:
4326:
4305:
4283:
4273:
4210:
4205:
4200:
4195:
4190:
4185:
4180:
4175:
4147:
4107:
4097:
4092:
4087:
4082:
4077:
4047:
4037:
3873:
3853:
3848:
3843:
3838:
3818:
3808:
3773:
2014:
Li Z, Cai T, Tian J, Xie JX, Zhao X, Liu L, et al. (July 2009).
1768:
1751:
1241:
1231:
1224:
1220:
1216:
1212:
1126:
1008:
919:
ions, so they are released; by contrast it has high affinity for the
821:
502:
and other nutrients into the cell by use of the sodium ion gradient.
348:
321:
291:
283:
273:
241:
191:
103:
91:
79:
2392:
1967:"Binding of Src to Na/K-ATPase forms a functional signaling complex"
3586:
3525:
Philosophical
Transactions of the Royal Society of London, Series B
5507:
5190:
5129:
5112:
5107:
4955:
4933:
4899:
4894:
4879:
4857:
4845:
4840:
4830:
4825:
4820:
4545:
4521:
4294:
4218:
4159:
3893:
3888:
3878:
3868:
3863:
3858:
3833:
3828:
3813:
1156:
In certain conditions such as in the case of cardiac disease, the
1070:
1001:
859:
785:
590:
1339:"Sodium Transporters in Human Health and Disease (Figure 2)"
820:
pumps in the cerebellum of a live mouse results in it displaying
5195:
5151:
5139:
5124:
5090:
5085:
5070:
5053:
5048:
5043:
5031:
5026:
5021:
5011:
4987:
4982:
4977:
4972:
4874:
4262:
2822:
Biochimica et
Biophysica Acta (BBA) - Molecular Basis of Disease
941:
The unphosphorylated form of the pump has a higher affinity for
734:
730:
663:
115:
51:
are shown as blue (intracellular) and red (extracellular) planes
5276:
4384:
3738:
2375:
Calderon DP, Fremont R, Kraenzlin F, Khodakhah K (March 2011).
869:
Looking at the process starting from the interior of the cell:
844:, and not within the nodal axolemma as previously thought. The
42:
5245:
5241:
2328:"Paying the price at the pump: dystonia from mutations in a Na
2067:"Interaction of the alpha subunit of Na,K-ATPase with cofilin"
967:. Thus, substances causing an increase in cAMP upregulate the
776:, "housekeeping" molecule for ionic gradients, but could be a
574:
2377:"The neural substrates of rapid-onset Dystonia-Parkinsonism"
1097:
According to the Blaustein-hypothesis, this carrier enzyme (
908:. This process leads to a conformational change in the pump.
1801:"The Na/K pump, Cl ion, and osmotic stabilization of cells"
669:
Protein-protein interactions play a very important role in
517:
gradient that is used by certain carrier processes. In the
5272:
3456:"Parallel Molecular Evolution in an Herbivore Community"
1198:"for the first discovery of an ion-transporting enzyme,
637:-ATPase in foot muscle and hepatopancreas in land snail
629:-binding signalling into the cell through regulation of
2114:
Forrest MD, Wall MJ, Press DA, Feng J (December 2012).
609:
pump because it changes the internal concentrations of
585:
outside of the cell, water flows into the cell through
1101:
exchanger, NCX) uses the Na gradient generated by the
533:
pump, whereas, on the reabsorbing (lumenal) side, the
5563:
1113:
from the intracellular space, hence slowing down the
633:. For instance, a study investigated the function of
1473:(4th ed.). John Wiley & Sons. p. 759.
1144:
which acts as digitalis receptor and stimulates the
1073:
performance by increasing its force of contraction.
677:
pump-mediated signal transduction. For example, the
5459:
5423:
5392:
5361:
5310:
5211:
5170:
4910:
4784:
4771:
4736:
4688:
4669:
4615:
4535:
4514:
4507:
4490:
4467:
4427:
4062:
4046:
3902:
3798:
3789:
569:pumps can result in swelling of the cell. A cell's
202:
190:
178:
173:
150:
138:
126:
114:
102:
90:
78:
73:
61:
56:
32:
3449:
3447:
1750:Jensen R, Nielsen J, Ørtenblad N (February 2020).
1026:-ATPase can recover it to the high activity form.
989:-coupled GPCRs. Note: Early studies indicated the
3404:Price, Elmer M.; Lingrel, Jerry B. (1988-11-01).
2477:"The Neuroscience Reason We Fall Over When Drunk"
2065:Lee K, Jung J, Kim M, Guidotti G (January 2001).
379:-ATPase in skeletal muscles, where inhibition of
573:is the sum of the concentrations of the various
478:is about 3.5-5 mM, whereas the concentration of
27:Enzyme found in the membrane of all animal cells
3661:Proceedings of the National Academy of Sciences
1666:American Journal of Physiology. Cell Physiology
1390:Clausen MV, Hilbers F, Poulsen H (June 2017).
1332:
1330:
945:ions. ATP binds, and the process starts again.
545:gradient as a source of energy to import both
5288:
4396:
3750:
3720:RCSB Protein Data Bank: Sodium–Potassium Pump
1500:Journal of Cerebral Blood Flow and Metabolism
1494:Howarth C, Gleeson P, Attwell D (July 2012).
1004:, which relieves an autoinhibitory domain of
391:-ATPase activity and lower force production.
8:
3042:"ATPase Na+/K+ transporting subunits (ATP1)"
1709:Journal of Muscle Research and Cell Motility
709:pump, was developed as a functional ouabain–
347:, affects transport, and regulates cellular
1858:
1856:
1854:
666:) in different intracellular compartments.
654:(ROS) production, as well as activation of
5295:
5281:
5273:
4781:
4511:
4504:
4403:
4389:
4381:
3795:
3757:
3743:
3735:
982:-ATPase. These include the ligands of the
971:-ATPase. These include the ligands of the
589:. This can cause the cell to swell up and
170:
41:
4146:3.A.3.1.4: H/K transporting, nongastric:
3712:at the U.S. National Library of Medicine
3682:
3672:
3544:
3495:
3380:
3339:
3281:
3263:
3172:
3099:
2841:
2816:Blaustein MP, Hamlyn JM (December 2010).
2671:
2583:
2573:
2451:
2441:
2400:
2351:
2302:
2292:
2251:
2241:
2200:
2190:
2149:
2139:
2090:
2041:
2031:
1990:
1941:
1884:
1834:
1824:
1767:
1519:
1417:
1407:
1366:
1356:
1863:Ramnanan CJ, Storey KB (February 2006).
1439:
1437:
398:
294:cells. It performs several functions in
227:
219:
5570:
2230:Frontiers in Computational Neuroscience
2224:Zylbertal A, Yarom Y, Wagner S (2017).
1326:
1121:pump results in a permanently elevated
2952:"Na/K-ATPase and inhibitors (Digoxin)"
1011:to drive endocytosis and degradation.
911:The conformational change exposes the
650:(MAPK) signal cascades, mitochondrial
29:
3241:
3239:
3133:
3131:
3121:
3119:
3061:
3059:
2783:Nephrology, Dialysis, Transplantation
1194:In 1997, he received one-half of the
7:
1448:. St. Louis, Mo: Elsevier Saunders.
3024:"The Nobel Prize in Chemistry 1997"
2020:The Journal of Biological Chemistry
1873:The Journal of Experimental Biology
1191:. He published his work that year.
873:The pump has a higher affinity for
717:pump-mediated signal transduction.
3030:. Nobel Media AB. 15 October 1997.
2740:The American Journal of Physiology
2636:"The inositol pyrophosphate 5-InsP
1588:The American Journal of Physiology
1082:concentration, which is caused by
904:residue and subsequent release of
900:of the pump at a highly conserved
749:Controlling neuron activity states
25:
3791:F-, V-, and A-type ATPase (3.A.2)
5573:
3382:10.1111/j.1558-5646.2012.01751.x
2867:American Journal of Therapeutics
648:mitogen-activated protein kinase
631:protein tyrosine phosphorylation
621:Functioning as signal transducer
617:to which the pump is sensitive.
3807:H transporting, mitochondrial:
3622:Molecular Biology and Evolution
3312:Molecular Biology and Evolution
2752:10.1152/ajpcell.1977.232.5.C165
2546:"Early role for a Na,K-ATPase (
1600:10.1152/ajpcell.1987.252.3.c328
1094:, causing the muscle to relax.
926:The pump binds 2 extracellular
470:, whereas the concentration of
1545:Journal of Cellular Physiology
1446:Textbook of medical physiology
505:Another important task of the
430:
367:-ATPases have been related to
1:
3616:Labeyrie E, Dobler S (2004).
3214:10.1016/j.toxicon.2016.02.019
2982:Biochimica et Biophysica Acta
2717:10.1016/S0024-3205(96)00661-3
1971:Molecular Biology of the Cell
1922:Molecular Biology of the Cell
1337:Gagnon KB, Delpire E (2021).
685:pump interacts directly with
3265:10.1371/journal.pgen.1010323
2994:10.1016/0006-3002(57)90343-8
2879:10.1097/MJT.0b013e318217a609
2834:10.1016/j.bbadis.2010.02.011
2610:Sodium In Health And Disease
2353:10.1016/j.neuron.2004.07.002
2275:Forrest MD (December 2014).
2192:10.1371/journal.pbio.1002319
2141:10.1371/journal.pone.0051169
691:non-receptor tyrosine kinase
5173:Protein-synthesizing GTPase
1444:Hall JE, Guyton AC (2006).
492:membrane transport proteins
383:breakdown (a substrate for
5617:
4217:3.A.3.5: Cu transporting:
3766:Membrane transport protein
2475:Forrest M (4 April 2015).
1721:10.1007/s10974-019-09524-y
1678:10.1152/ajpcell.00132.2007
1152:Pharmacological regulation
963:-ATPase is upregulated by
414:
309:(i.e. it uses energy from
5451:Michaelis–Menten kinetics
5232:Guanylate-binding protein
4416:acid anhydride hydrolases
4366:
3912:H transporting, lysosomal
3157:10.1016/j.cub.2021.03.089
2910:Nephron Clinical Practice
2613:. CRC Press. p. 15.
2443:10.1186/s12868-015-0162-6
2424:Forrest MD (April 2015).
2083:10.1042/0264-6021:3530377
1799:Armstrong CM (May 2003).
1756:The Journal of Physiology
1358:10.3389/fphys.2020.588664
881:ions, thus after binding
725:pump also interacts with
169:
40:
5343:Diffusion-limited enzyme
4787:Heterotrimeric G protein
4482:Phosphoadenylylsulfatase
3714:Medical Subject Headings
3710:Sodium,+Potassium+ATPase
2294:10.3389/fphys.2014.00472
2243:10.3389/fncom.2017.00085
1409:10.3389/fphys.2017.00371
1305:Sodium-calcium exchanger
1196:Nobel Prize in Chemistry
1179:-ATPase was proposed by
885:, binds 3 intracellular
800:pump causes rapid onset
770:accessory olfactory bulb
4459:Thiamine-triphosphatase
3674:10.1073/pnas.1202111109
3575:The American Naturalist
3480:10.1126/science.1226630
2575:10.1073/pnas.2023333118
2550:) in brain development"
2326:Cannon SC (July 2004).
2281:Frontiers in Physiology
2071:The Biochemical Journal
2033:10.1074/jbc.M109.013821
1983:10.1091/mbc.E05-08-0735
1934:10.1091/mbc.E05-04-0295
1826:10.1073/pnas.0931278100
1396:Frontiers in Physiology
1344:Frontiers in Physiology
1279:, the alpha-subunit of
1276:Drosophila melanogaster
652:reactive oxygen species
557:Controlling cell volume
357:reactive oxygen species
343:-ATPase helps maintain
270:sodium–potassium ATPase
3537:10.1098/rstb.2018.0246
3324:10.1093/molbev/msac258
3084:10.1098/rsbl.2009.0281
2664:10.1126/sciadv.abb8542
1557:10.1002/jcp.1041140303
1256:-ATPase beta subunit.
1092:sarcoplasmic reticulum
1069:drugs used to improve
866:
412:
373:cardiac Purkinje cells
233:
225:
5436:Eadie–Hofstee diagram
5369:Allosteric regulation
5214:Polymerization motors
4925:Rho family of GTPases
3634:10.1093/molbev/msg240
3151:(12): 2530–2538.e10.
1512:10.1038/jcbfm.2012.35
863:
660:inositol triphosphate
513:pump is to provide a
482:is about 135-145 mM.
411:across the membranes.
402:
238:sodium–potassium pump
231:
223:
5446:Lineweaver–Burk plot
1285:neofunctionalization
1185:University of Aarhus
930:ions, which induces
551:renal tubular system
232:Alpha and beta units
5222:dynamin superfamily
4320:Class VI, type 11:
3667:(32): 13040–13045.
3472:2012Sci...337.1634Z
3466:(6102): 1634–1637.
3422:10.1021/bi00422a016
2656:2020SciA....6.8542C
2566:2021PNAS..11823333S
2560:(25): e2023333118.
2503:Annals of Neurology
2381:Nature Neuroscience
2132:2012PLoSO...751169F
1817:2003PNAS..100.6257A
1594:(3 Pt 1): C328-34.
1181:Jens Christian Skou
938:ions into the cell.
427:intracellular space
387:) leads to reduced
318:Jens Christian Skou
5601:Transport proteins
5405:Enzyme superfamily
5338:Enzyme promiscuity
4304:Class V, type 10:
4293:Class II, type 9:
3531:(1777): 20180246.
2795:10.1093/ndt/gfn325
2607:Burnier M (2008).
1635:10.1007/bf00374294
1055:cardiac glycosides
867:
523:interstitial fluid
466:in cytosol is 100
441:Reversal potential
413:
305:-ATPase enzyme is
234:
226:
5561:
5560:
5270:
5269:
5266:
5265:
4767:
4766:
4732:
4731:
4477:Adenylylsulfatase
4378:
4377:
4267:Class I, type 8:
4074:Na/K transporting
4058:
4057:
3416:(22): 8400–8408.
2922:10.1159/000363301
2620:978-0-8493-3978-3
2515:10.1002/ana.21381
1886:10.1242/jeb.02052
1480:978-0-470-57095-1
1455:978-0-7216-0240-0
1162:cardiac glycoside
932:dephosphorylation
577:species and many
541:uses the created
417:Resting potential
409:resting potential
395:Resting potential
345:resting potential
218:
217:
214:
213:
133:metabolic pathway
16:(Redirected from
5608:
5578:
5577:
5569:
5441:Hanes–Woolf plot
5384:Enzyme activator
5379:Enzyme inhibitor
5353:Enzyme catalysis
5297:
5290:
5283:
5274:
4782:
4512:
4505:
4405:
4398:
4391:
4382:
4371:ATPase disorders
4242:Other/ungrouped:
3796:
3759:
3752:
3745:
3736:
3697:
3696:
3686:
3676:
3652:
3646:
3645:
3613:
3607:
3606:
3565:
3559:
3558:
3548:
3516:
3510:
3509:
3499:
3451:
3442:
3441:
3401:
3395:
3394:
3384:
3360:
3354:
3353:
3343:
3302:
3296:
3295:
3285:
3267:
3243:
3234:
3233:
3193:
3187:
3186:
3176:
3135:
3126:
3123:
3114:
3113:
3103:
3063:
3054:
3053:
3051:
3049:
3038:
3032:
3031:
3020:
3014:
3013:
2977:
2971:
2970:
2968:
2967:
2958:. Archived from
2948:
2942:
2941:
2905:
2899:
2898:
2862:
2856:
2855:
2845:
2813:
2807:
2806:
2778:
2772:
2771:
2735:
2729:
2728:
2700:
2694:
2693:
2675:
2650:(44): eabb8542.
2644:Science Advances
2631:
2625:
2624:
2604:
2598:
2597:
2587:
2577:
2541:
2535:
2534:
2498:
2492:
2491:
2489:
2487:
2472:
2466:
2465:
2455:
2445:
2430:BMC Neuroscience
2421:
2415:
2414:
2404:
2372:
2366:
2365:
2355:
2323:
2317:
2316:
2306:
2296:
2272:
2266:
2265:
2255:
2245:
2221:
2215:
2214:
2204:
2194:
2185:(12): e1002319.
2170:
2164:
2163:
2153:
2143:
2111:
2105:
2104:
2094:
2077:(Pt 2): 377–85.
2062:
2056:
2055:
2045:
2035:
2026:(31): 21066–76.
2011:
2005:
2004:
1994:
1962:
1956:
1955:
1945:
1913:
1907:
1906:
1888:
1879:(Pt 4): 677–88.
1860:
1849:
1848:
1838:
1828:
1796:
1790:
1789:
1771:
1769:10.1113/JP278543
1747:
1741:
1740:
1715:(3–4): 353–364.
1704:
1698:
1697:
1661:
1655:
1654:
1618:
1612:
1611:
1583:
1577:
1576:
1540:
1534:
1533:
1523:
1491:
1485:
1484:
1466:
1460:
1459:
1441:
1432:
1431:
1421:
1411:
1387:
1381:
1380:
1370:
1360:
1334:
1282:
1255:
1201:
1178:
1167:
1159:
1139:
1124:
1120:
1116:
1112:
1108:
1104:
1100:
1089:
1085:
1080:
1052:
1040:triiodothyronine
1037:
1025:
1021:
1017:
999:
981:
970:
962:
944:
937:
929:
922:
918:
914:
888:
880:
876:
851:
847:
839:
835:
819:
815:
799:
795:
766:Purkinje neurons
760:
756:
724:
720:
716:
712:
708:
704:
700:
696:
684:
680:
676:
672:
662:(IP3) receptor (
636:
616:
612:
608:
604:
600:
596:
568:
564:
548:
544:
536:
532:
528:
516:
512:
508:
481:
477:
473:
465:
461:
457:
452:
448:
435:Nernst potential
431:§ Mechanism
406:
390:
378:
366:
362:
342:
304:
265:
258:
254:, also known as
171:
45:
35:
30:
21:
5616:
5615:
5611:
5610:
5609:
5607:
5606:
5605:
5586:
5585:
5584:
5572:
5564:
5562:
5557:
5469:Oxidoreductases
5455:
5431:Enzyme kinetics
5419:
5415:List of enzymes
5388:
5357:
5328:Catalytic triad
5306:
5301:
5271:
5262:
5207:
5166:
4917:Ras superfamily
4906:
4890:
4870:
4816:
4806:
4798:
4763:
4728:
4684:
4665:
4611:
4568:Plasma membrane
4531:
4486:
4463:
4437:Pyrophosphatase
4423:
4409:
4379:
4374:
4362:
4259:Mg transporting
4172:Ca transporting
4054:
4053:found in Archea
4042:
3898:
3785:
3763:
3706:
3701:
3700:
3654:
3653:
3649:
3615:
3614:
3610:
3581:(S1): S29–S43.
3568:
3566:
3562:
3518:
3517:
3513:
3453:
3452:
3445:
3403:
3402:
3398:
3362:
3361:
3357:
3318:(12): msac258.
3304:
3303:
3299:
3258:(8): e1010323.
3245:
3244:
3237:
3195:
3194:
3190:
3145:Current Biology
3137:
3136:
3129:
3124:
3117:
3072:Biology Letters
3065:
3064:
3057:
3047:
3045:
3040:
3039:
3035:
3022:
3021:
3017:
2979:
2978:
2974:
2965:
2963:
2950:
2949:
2945:
2907:
2906:
2902:
2864:
2863:
2859:
2828:(12): 1219–29.
2815:
2814:
2810:
2780:
2779:
2775:
2737:
2736:
2732:
2702:
2701:
2697:
2639:
2633:
2632:
2628:
2621:
2606:
2605:
2601:
2543:
2542:
2538:
2500:
2499:
2495:
2485:
2483:
2474:
2473:
2469:
2423:
2422:
2418:
2393:10.1038/nn.2753
2374:
2373:
2369:
2335:
2331:
2325:
2324:
2320:
2274:
2273:
2269:
2223:
2222:
2218:
2172:
2171:
2167:
2113:
2112:
2108:
2064:
2063:
2059:
2013:
2012:
2008:
1964:
1963:
1959:
1915:
1914:
1910:
1862:
1861:
1852:
1811:(10): 6257–62.
1798:
1797:
1793:
1749:
1748:
1744:
1706:
1705:
1701:
1663:
1662:
1658:
1623:Pflügers Archiv
1620:
1619:
1615:
1585:
1584:
1580:
1542:
1541:
1537:
1493:
1492:
1488:
1481:
1468:
1467:
1463:
1456:
1443:
1442:
1435:
1389:
1388:
1384:
1336:
1335:
1328:
1323:
1310:Thyroid hormone
1301:
1280:
1271:
1262:
1253:
1208:
1199:
1176:
1174:
1165:
1157:
1154:
1137:
1135:
1122:
1118:
1114:
1110:
1109:pump to remove
1106:
1102:
1098:
1087:
1083:
1078:
1050:
1035:
1032:
1023:
1019:
1015:
997:
987:
979:
976:
968:
960:
957:
952:
942:
935:
927:
920:
916:
912:
898:phosphorylation
886:
878:
874:
858:
849:
845:
837:
833:
817:
813:
797:
793:
780:element in the
758:
754:
751:
722:
718:
714:
710:
706:
702:
698:
694:
682:
678:
674:
670:
656:phospholipase C
634:
623:
614:
610:
606:
602:
598:
594:
566:
562:
561:Failure of the
559:
546:
542:
534:
530:
526:
525:) side via the
514:
510:
506:
494:, which import
488:
479:
475:
471:
463:
459:
455:
450:
446:
443:
419:
404:
397:
388:
376:
364:
360:
340:
337:
302:
296:cell physiology
286:) found in the
263:
256:
52:
33:
28:
23:
22:
15:
12:
11:
5:
5614:
5612:
5604:
5603:
5598:
5588:
5587:
5583:
5582:
5559:
5558:
5556:
5555:
5542:
5529:
5516:
5503:
5490:
5477:
5463:
5461:
5457:
5456:
5454:
5453:
5448:
5443:
5438:
5433:
5427:
5425:
5421:
5420:
5418:
5417:
5412:
5407:
5402:
5396:
5394:
5393:Classification
5390:
5389:
5387:
5386:
5381:
5376:
5371:
5365:
5363:
5359:
5358:
5356:
5355:
5350:
5345:
5340:
5335:
5330:
5325:
5320:
5314:
5312:
5308:
5307:
5302:
5300:
5299:
5292:
5285:
5277:
5268:
5267:
5264:
5263:
5261:
5260:
5255:
5254:
5253:
5248:
5239:
5234:
5229:
5218:
5216:
5209:
5208:
5206:
5205:
5200:
5199:
5198:
5193:
5188:
5177:
5175:
5168:
5167:
5165:
5164:
5159:
5154:
5149:
5144:
5143:
5142:
5137:
5132:
5127:
5117:
5116:
5115:
5110:
5100:
5099:
5098:
5093:
5088:
5076:
5075:
5074:
5073:
5068:
5058:
5057:
5056:
5051:
5046:
5036:
5035:
5034:
5029:
5024:
5014:
5009:
5008:
5007:
5002:
4992:
4991:
4990:
4985:
4980:
4975:
4965:
4964:
4963:
4958:
4948:
4947:
4946:
4941:
4936:
4921:
4919:
4908:
4907:
4905:
4904:
4903:
4902:
4897:
4888:
4884:
4883:
4882:
4877:
4868:
4864:
4863:
4862:
4861:
4860:
4850:
4849:
4848:
4843:
4833:
4828:
4823:
4814:
4810:
4809:
4808:
4804:
4796:
4791:
4789:
4779:
4769:
4768:
4765:
4764:
4762:
4761:
4756:
4751:
4746:
4740:
4738:
4734:
4733:
4730:
4729:
4727:
4726:
4721:
4716:
4711:
4706:
4701:
4695:
4693:
4686:
4685:
4683:
4682:
4676:
4674:
4667:
4666:
4664:
4663:
4658:
4653:
4648:
4643:
4638:
4633:
4628:
4622:
4620:
4613:
4612:
4610:
4609:
4608:
4607:
4602:
4592:
4591:
4590:
4585:
4580:
4575:
4565:
4564:
4563:
4558:
4553:
4542:
4540:
4533:
4532:
4530:
4529:
4524:
4518:
4516:
4515:Cu++ (3.6.3.4)
4509:
4502:
4488:
4487:
4485:
4484:
4479:
4473:
4471:
4465:
4464:
4462:
4461:
4456:
4451:
4450:
4449:
4444:
4433:
4431:
4425:
4424:
4410:
4408:
4407:
4400:
4393:
4385:
4376:
4375:
4367:
4364:
4363:
4361:
4360:
4355:
4350:
4345:
4340:
4334:
4329:
4324:
4318:
4313:
4308:
4302:
4297:
4291:
4286:
4281:
4276:
4271:
4265:
4255:
4254:
4244:
4243:
4239:
4238:
4227:
4226:
4221:
4214:
4213:
4208:
4203:
4198:
4193:
4188:
4183:
4178:
4151:
4150:
4143:
4142:
4137:
4131:H/K exchanging
4128:
4121:
4120:
4115:
4110:
4105:
4100:
4095:
4090:
4085:
4080:
4069:
4067:
4060:
4059:
4056:
4055:
4052:
4050:
4044:
4043:
4041:
4040:
4034:
4033:
4028:
4023:
4018:
4013:
4008:
4003:
3998:
3993:
3988:
3983:
3978:
3973:
3968:
3963:
3958:
3953:
3948:
3943:
3938:
3933:
3928:
3923:
3918:
3908:
3906:
3900:
3899:
3897:
3896:
3891:
3886:
3881:
3876:
3871:
3866:
3861:
3856:
3851:
3846:
3841:
3836:
3831:
3826:
3821:
3816:
3811:
3804:
3802:
3793:
3787:
3786:
3764:
3762:
3761:
3754:
3747:
3739:
3733:
3732:
3722:
3717:
3705:
3704:External links
3702:
3699:
3698:
3647:
3608:
3587:10.1086/691711
3560:
3511:
3443:
3396:
3375:(1): 289–294.
3355:
3297:
3235:
3188:
3127:
3115:
3078:(4): 513–516.
3055:
3033:
3028:NobelPrize.org
3015:
2988:(2): 394–401.
2972:
2943:
2916:(1–2): 11–21.
2900:
2857:
2808:
2773:
2746:(5): C165-73.
2730:
2711:(6): 375–382.
2695:
2637:
2626:
2619:
2599:
2536:
2493:
2467:
2416:
2367:
2333:
2329:
2318:
2267:
2216:
2165:
2126:(12): e51169.
2106:
2057:
2006:
1957:
1928:(9): 4034–45.
1908:
1850:
1791:
1762:(4): 789–803.
1742:
1699:
1672:(3): C967-77.
1656:
1613:
1578:
1535:
1506:(7): 1222–32.
1486:
1479:
1461:
1454:
1433:
1382:
1325:
1324:
1322:
1319:
1318:
1317:
1312:
1307:
1300:
1297:
1293:bufadienolides
1270:
1267:
1261:
1258:
1245:
1244:
1228:
1207:
1204:
1173:
1170:
1153:
1150:
1133:
1049:For instance,
1031:
1028:
985:
974:
956:
953:
951:
948:
947:
946:
939:
924:
909:
890:
857:
854:
750:
747:
622:
619:
558:
555:
487:
484:
442:
439:
437:of potassium.
396:
393:
336:
333:
329:P-type ATPases
252:triphosphatase
216:
215:
212:
211:
206:
200:
199:
194:
188:
187:
182:
176:
175:
167:
166:
155:
148:
147:
142:
136:
135:
130:
124:
123:
118:
112:
111:
106:
100:
99:
94:
88:
87:
82:
76:
75:
71:
70:
65:
59:
58:
54:
53:
46:
38:
37:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
5613:
5602:
5599:
5597:
5594:
5593:
5591:
5581:
5576:
5571:
5567:
5553:
5549:
5548:
5543:
5540:
5536:
5535:
5530:
5527:
5523:
5522:
5517:
5514:
5510:
5509:
5504:
5501:
5497:
5496:
5491:
5488:
5484:
5483:
5478:
5475:
5471:
5470:
5465:
5464:
5462:
5458:
5452:
5449:
5447:
5444:
5442:
5439:
5437:
5434:
5432:
5429:
5428:
5426:
5422:
5416:
5413:
5411:
5410:Enzyme family
5408:
5406:
5403:
5401:
5398:
5397:
5395:
5391:
5385:
5382:
5380:
5377:
5375:
5374:Cooperativity
5372:
5370:
5367:
5366:
5364:
5360:
5354:
5351:
5349:
5346:
5344:
5341:
5339:
5336:
5334:
5333:Oxyanion hole
5331:
5329:
5326:
5324:
5321:
5319:
5316:
5315:
5313:
5309:
5305:
5298:
5293:
5291:
5286:
5284:
5279:
5278:
5275:
5259:
5256:
5252:
5249:
5247:
5243:
5240:
5238:
5235:
5233:
5230:
5228:
5225:
5224:
5223:
5220:
5219:
5217:
5215:
5210:
5204:
5201:
5197:
5194:
5192:
5189:
5187:
5184:
5183:
5182:
5179:
5178:
5176:
5174:
5169:
5163:
5160:
5158:
5155:
5153:
5150:
5148:
5145:
5141:
5138:
5136:
5133:
5131:
5128:
5126:
5123:
5122:
5121:
5118:
5114:
5111:
5109:
5106:
5105:
5104:
5101:
5097:
5094:
5092:
5089:
5087:
5084:
5083:
5082:
5078:
5077:
5072:
5069:
5067:
5064:
5063:
5062:
5059:
5055:
5052:
5050:
5047:
5045:
5042:
5041:
5040:
5037:
5033:
5030:
5028:
5025:
5023:
5020:
5019:
5018:
5015:
5013:
5010:
5006:
5003:
5001:
4998:
4997:
4996:
4993:
4989:
4986:
4984:
4981:
4979:
4976:
4974:
4971:
4970:
4969:
4966:
4962:
4959:
4957:
4954:
4953:
4952:
4949:
4945:
4942:
4940:
4937:
4935:
4932:
4931:
4930:
4926:
4923:
4922:
4920:
4918:
4914:
4909:
4901:
4898:
4896:
4893:
4892:
4891:
4885:
4881:
4878:
4876:
4873:
4872:
4871:
4865:
4859:
4856:
4855:
4854:
4851:
4847:
4844:
4842:
4839:
4838:
4837:
4834:
4832:
4829:
4827:
4824:
4822:
4819:
4818:
4817:
4811:
4807:
4801:
4800:
4799:
4793:
4792:
4790:
4788:
4783:
4780:
4778:
4774:
4770:
4760:
4757:
4755:
4752:
4750:
4747:
4745:
4742:
4741:
4739:
4735:
4725:
4722:
4720:
4717:
4715:
4712:
4710:
4707:
4705:
4702:
4700:
4697:
4696:
4694:
4692:
4691:P-type ATPase
4687:
4681:
4678:
4677:
4675:
4672:
4668:
4662:
4659:
4657:
4654:
4652:
4649:
4647:
4644:
4642:
4639:
4637:
4634:
4632:
4629:
4627:
4624:
4623:
4621:
4618:
4614:
4606:
4603:
4601:
4598:
4597:
4596:
4593:
4589:
4586:
4584:
4581:
4579:
4576:
4574:
4571:
4570:
4569:
4566:
4562:
4559:
4557:
4554:
4552:
4549:
4548:
4547:
4544:
4543:
4541:
4538:
4534:
4528:
4525:
4523:
4520:
4519:
4517:
4513:
4510:
4506:
4503:
4501:
4497:
4493:
4489:
4483:
4480:
4478:
4475:
4474:
4472:
4470:
4466:
4460:
4457:
4455:
4452:
4448:
4445:
4443:
4440:
4439:
4438:
4435:
4434:
4432:
4430:
4426:
4421:
4417:
4413:
4406:
4401:
4399:
4394:
4392:
4387:
4386:
4383:
4373:
4372:
4365:
4359:
4356:
4354:
4351:
4349:
4346:
4344:
4341:
4339:
4335:
4333:
4330:
4328:
4325:
4323:
4319:
4317:
4314:
4312:
4309:
4307:
4303:
4301:
4298:
4296:
4292:
4290:
4287:
4285:
4282:
4280:
4277:
4275:
4272:
4270:
4266:
4264:
4260:
4257:
4256:
4253:
4249:
4246:
4245:
4241:
4240:
4237:
4233:
4229:
4228:
4225:
4222:
4220:
4216:
4215:
4212:
4209:
4207:
4204:
4202:
4199:
4197:
4194:
4192:
4189:
4187:
4184:
4182:
4179:
4177:
4173:
4169:
4165:
4161:
4157:
4153:
4152:
4149:
4145:
4144:
4141:
4138:
4136:
4132:
4129:
4127:
4123:
4122:
4119:
4116:
4114:
4111:
4109:
4106:
4104:
4101:
4099:
4096:
4094:
4091:
4089:
4086:
4084:
4081:
4079:
4075:
4071:
4070:
4068:
4065:
4064:P-type ATPase
4061:
4051:
4049:
4045:
4039:
4036:
4035:
4032:
4029:
4027:
4024:
4022:
4019:
4017:
4014:
4012:
4009:
4007:
4004:
4002:
3999:
3997:
3994:
3992:
3989:
3987:
3984:
3982:
3979:
3977:
3974:
3972:
3969:
3967:
3964:
3962:
3959:
3957:
3954:
3952:
3949:
3947:
3944:
3942:
3939:
3937:
3934:
3932:
3929:
3927:
3924:
3922:
3919:
3917:
3913:
3910:
3909:
3907:
3905:
3901:
3895:
3892:
3890:
3887:
3885:
3882:
3880:
3877:
3875:
3872:
3870:
3867:
3865:
3862:
3860:
3857:
3855:
3852:
3850:
3847:
3845:
3842:
3840:
3837:
3835:
3832:
3830:
3827:
3825:
3822:
3820:
3817:
3815:
3812:
3810:
3806:
3805:
3803:
3801:
3797:
3794:
3792:
3788:
3783:
3779:
3775:
3771:
3767:
3760:
3755:
3753:
3748:
3746:
3741:
3740:
3737:
3730:
3726:
3723:
3721:
3718:
3715:
3711:
3708:
3707:
3703:
3694:
3690:
3685:
3680:
3675:
3670:
3666:
3662:
3658:
3651:
3648:
3643:
3639:
3635:
3631:
3628:(2): 218–21.
3627:
3623:
3619:
3612:
3609:
3604:
3600:
3596:
3592:
3588:
3584:
3580:
3576:
3572:
3564:
3561:
3556:
3552:
3547:
3542:
3538:
3534:
3530:
3526:
3522:
3515:
3512:
3507:
3503:
3498:
3493:
3489:
3485:
3481:
3477:
3473:
3469:
3465:
3461:
3457:
3450:
3448:
3444:
3439:
3435:
3431:
3427:
3423:
3419:
3415:
3411:
3407:
3400:
3397:
3392:
3388:
3383:
3378:
3374:
3370:
3366:
3359:
3356:
3351:
3347:
3342:
3337:
3333:
3329:
3325:
3321:
3317:
3313:
3309:
3301:
3298:
3293:
3289:
3284:
3279:
3275:
3271:
3266:
3261:
3257:
3253:
3252:PLOS Genetics
3249:
3242:
3240:
3236:
3231:
3227:
3223:
3219:
3215:
3211:
3207:
3203:
3199:
3192:
3189:
3184:
3180:
3175:
3170:
3166:
3162:
3158:
3154:
3150:
3146:
3142:
3134:
3132:
3128:
3122:
3120:
3116:
3111:
3107:
3102:
3097:
3093:
3089:
3085:
3081:
3077:
3073:
3069:
3062:
3060:
3056:
3043:
3037:
3034:
3029:
3025:
3019:
3016:
3011:
3007:
3003:
2999:
2995:
2991:
2987:
2983:
2976:
2973:
2962:on 2020-09-28
2961:
2957:
2953:
2947:
2944:
2939:
2935:
2931:
2927:
2923:
2919:
2915:
2911:
2904:
2901:
2896:
2892:
2888:
2884:
2880:
2876:
2872:
2868:
2861:
2858:
2853:
2849:
2844:
2839:
2835:
2831:
2827:
2823:
2819:
2812:
2809:
2804:
2800:
2796:
2792:
2789:(9): 2723–9.
2788:
2784:
2777:
2774:
2769:
2765:
2761:
2757:
2753:
2749:
2745:
2741:
2734:
2731:
2726:
2722:
2718:
2714:
2710:
2706:
2705:Life Sciences
2699:
2696:
2691:
2687:
2683:
2679:
2674:
2669:
2665:
2661:
2657:
2653:
2649:
2645:
2641:
2630:
2627:
2622:
2616:
2612:
2611:
2603:
2600:
2595:
2591:
2586:
2581:
2576:
2571:
2567:
2563:
2559:
2555:
2551:
2549:
2540:
2537:
2532:
2528:
2524:
2520:
2516:
2512:
2509:(4): 428–35.
2508:
2504:
2497:
2494:
2482:
2478:
2471:
2468:
2463:
2459:
2454:
2449:
2444:
2439:
2435:
2431:
2427:
2420:
2417:
2412:
2408:
2403:
2398:
2394:
2390:
2387:(3): 357–65.
2386:
2382:
2378:
2371:
2368:
2363:
2359:
2354:
2349:
2345:
2341:
2337:
2322:
2319:
2314:
2310:
2305:
2300:
2295:
2290:
2286:
2282:
2278:
2271:
2268:
2263:
2259:
2254:
2249:
2244:
2239:
2235:
2231:
2227:
2220:
2217:
2212:
2208:
2203:
2198:
2193:
2188:
2184:
2180:
2176:
2169:
2166:
2161:
2157:
2152:
2147:
2142:
2137:
2133:
2129:
2125:
2121:
2117:
2110:
2107:
2102:
2098:
2093:
2088:
2084:
2080:
2076:
2072:
2068:
2061:
2058:
2053:
2049:
2044:
2039:
2034:
2029:
2025:
2021:
2017:
2010:
2007:
2002:
1998:
1993:
1988:
1984:
1980:
1977:(1): 317–26.
1976:
1972:
1968:
1961:
1958:
1953:
1949:
1944:
1939:
1935:
1931:
1927:
1923:
1919:
1912:
1909:
1904:
1900:
1896:
1892:
1887:
1882:
1878:
1874:
1870:
1868:
1859:
1857:
1855:
1851:
1846:
1842:
1837:
1832:
1827:
1822:
1818:
1814:
1810:
1806:
1802:
1795:
1792:
1787:
1783:
1779:
1775:
1770:
1765:
1761:
1757:
1753:
1746:
1743:
1738:
1734:
1730:
1726:
1722:
1718:
1714:
1710:
1703:
1700:
1695:
1691:
1687:
1683:
1679:
1675:
1671:
1667:
1660:
1657:
1652:
1648:
1644:
1640:
1636:
1632:
1628:
1624:
1617:
1614:
1609:
1605:
1601:
1597:
1593:
1589:
1582:
1579:
1574:
1570:
1566:
1562:
1558:
1554:
1550:
1546:
1539:
1536:
1531:
1527:
1522:
1517:
1513:
1509:
1505:
1501:
1497:
1490:
1487:
1482:
1476:
1472:
1465:
1462:
1457:
1451:
1447:
1440:
1438:
1434:
1429:
1425:
1420:
1415:
1410:
1405:
1401:
1397:
1393:
1386:
1383:
1378:
1374:
1369:
1364:
1359:
1354:
1350:
1346:
1345:
1340:
1333:
1331:
1327:
1320:
1316:
1313:
1311:
1308:
1306:
1303:
1302:
1298:
1296:
1294:
1290:
1286:
1278:
1277:
1268:
1266:
1259:
1257:
1251:
1250:
1243:
1239:
1238:
1233:
1229:
1226:
1222:
1218:
1214:
1210:
1209:
1205:
1203:
1197:
1192:
1190:
1186:
1182:
1171:
1169:
1163:
1151:
1149:
1147:
1143:
1132:
1128:
1125:level in the
1095:
1093:
1081:
1074:
1072:
1068:
1064:
1060:
1057:(for example
1056:
1047:
1045:
1041:
1029:
1027:
1012:
1010:
1007:
1003:
994:
992:
988:
977:
966:
954:
949:
940:
933:
925:
910:
907:
903:
899:
896:, leading to
895:
891:
884:
872:
871:
870:
862:
855:
853:
843:
831:
827:
823:
811:
807:
803:
791:
787:
783:
779:
775:
771:
767:
764:
748:
746:
744:
740:
736:
732:
728:
692:
688:
667:
665:
661:
657:
653:
649:
645:
640:
632:
628:
620:
618:
592:
588:
584:
580:
576:
572:
556:
554:
552:
540:
524:
520:
503:
501:
497:
493:
485:
483:
469:
445:Even if both
440:
438:
436:
432:
428:
424:
423:intracellular
418:
410:
401:
394:
392:
386:
382:
374:
370:
358:
354:
350:
346:
334:
332:
330:
325:
323:
319:
314:
312:
308:
299:
297:
293:
289:
285:
282:
281:transmembrane
279:
275:
271:
267:
260:
253:
250:
247:
243:
239:
230:
222:
210:
207:
205:
201:
198:
195:
193:
189:
186:
183:
181:
177:
172:
168:
165:
162:
159:
156:
153:
149:
146:
143:
141:
137:
134:
131:
129:
125:
122:
119:
117:
113:
110:
109:NiceZyme view
107:
105:
101:
98:
95:
93:
89:
86:
83:
81:
77:
72:
69:
66:
64:
60:
55:
50:
49:lipid bilayer
44:
39:
31:
19:
18:Na+/K+ ATPase
5547:Translocases
5544:
5531:
5518:
5505:
5492:
5482:Transferases
5479:
5466:
5323:Binding site
5060:
4994:
4950:
4928:
4913:Small GTPase
4527:Wilson/ATP7B
4522:Menkes/ATP7A
4368:
3778:ATP synthase
3729:Khan Academy
3664:
3660:
3650:
3625:
3621:
3611:
3578:
3574:
3563:
3528:
3524:
3514:
3463:
3459:
3413:
3410:Biochemistry
3409:
3399:
3372:
3368:
3358:
3315:
3311:
3300:
3255:
3251:
3205:
3201:
3191:
3148:
3144:
3075:
3071:
3046:. Retrieved
3036:
3027:
3018:
2985:
2981:
2975:
2964:. Retrieved
2960:the original
2956:Pharmacorama
2955:
2946:
2913:
2909:
2903:
2873:(1): 35–42.
2870:
2866:
2860:
2825:
2821:
2811:
2786:
2782:
2776:
2743:
2739:
2733:
2708:
2704:
2698:
2647:
2643:
2629:
2609:
2602:
2557:
2553:
2547:
2539:
2506:
2502:
2496:
2484:. Retrieved
2480:
2470:
2433:
2429:
2419:
2384:
2380:
2370:
2346:(2): 153–4.
2343:
2339:
2321:
2287:(472): 472.
2284:
2280:
2270:
2233:
2229:
2219:
2182:
2179:PLOS Biology
2178:
2168:
2123:
2119:
2109:
2074:
2070:
2060:
2023:
2019:
2009:
1974:
1970:
1960:
1925:
1921:
1911:
1876:
1872:
1867:Otala lactea
1866:
1808:
1804:
1794:
1759:
1755:
1745:
1712:
1708:
1702:
1669:
1665:
1659:
1629:(4): 380–5.
1626:
1622:
1616:
1591:
1587:
1581:
1551:(3): 263–6.
1548:
1544:
1538:
1503:
1499:
1489:
1471:Biochemistry
1470:
1464:
1445:
1399:
1395:
1385:
1348:
1342:
1289:cardenolides
1274:
1272:
1263:
1247:
1246:
1235:
1193:
1175:
1155:
1146:EGF receptor
1130:
1096:
1075:
1048:
1033:
1013:
995:
990:
958:
868:
806:parkinsonism
788:. Indeed, a
752:
668:
643:
639:Otala lactea
638:
624:
560:
504:
489:
444:
420:
353:MAPK pathway
338:
326:
315:
300:
278:electrogenic
269:
262:
255:
237:
235:
224:Flow of ions
97:BRENDA entry
36:-ATPase pump
5318:Active site
5237:Mitofusin-1
5212:3.6.5.5-6:
5181:Prokaryotic
4230:3.A.3.8.8:
4124:3.A.3.1.2:
4072:3.A.3.1.1:
2481:Science 2.0
778:computation
774:homeostatic
500:amino acids
322:nerve cells
85:IntEnz view
57:Identifiers
5590:Categories
5521:Isomerases
5495:Hydrolases
5362:Regulation
5203:Eukaryotic
4836:Transducin
4673:(3.6.3.10)
4412:Hydrolases
3904:H (V-type)
3800:H (F-type)
3782:TC 3A2-3A3
2966:2019-11-08
2436:(27): 27.
1351:: 588664.
1321:References
1269:In insects
1202:-ATPase."
955:Endogenous
950:Regulation
894:hydrolyzed
877:ions than
782:cerebellum
763:cerebellar
658:(PLC) and
583:osmolarity
571:osmolarity
415:See also:
385:glycolysis
369:glycolysis
154:structures
121:KEGG entry
5400:EC number
5171:3.6.5.3:
4911:3.6.5.2:
4853:Gustducin
4785:3.6.5.1:
4619:(3.6.3.9)
4539:(3.6.3.8)
4442:Inorganic
4369:see also
4336:type 13:
4154:3.A.3.2:
3770:ion pumps
3488:0036-8075
3430:0006-2960
3369:Evolution
3332:0737-4038
3274:1553-7390
3222:0041-0101
3208:: 13–15.
3165:0960-9822
3092:1744-9561
2690:226036261
1786:209317559
1737:195329741
1172:Discovery
1067:inotropic
1046:hormone.
1030:Exogenous
902:aspartate
856:Mechanism
812:block of
739:PLCgamma1
644:O. lactea
539:symporter
537:-glucose
486:Transport
249:adenosine
246:potassium
74:Databases
5596:EC 3.6.3
5424:Kinetics
5348:Cofactor
5311:Activity
4447:Thiamine
4232:flippase
4048:A-ATPase
4031:ATP6V0E1
4021:ATP6V0D2
4016:ATP6V0D1
4001:ATP6V0A4
3996:ATP6V0A2
3991:ATP6V0A1
3981:ATP6V1G3
3976:ATP6V1G2
3971:ATP6V1G1
3961:ATP6V1E2
3956:ATP6V1E1
3946:ATP6V1C2
3941:ATP6V1C1
3936:ATP6V1B2
3931:ATP6V1B1
3693:22826239
3642:12949136
3595:28731826
3555:31154978
3506:23019645
3391:23289579
3350:36472530
3292:35972957
3230:26905927
3183:33887183
3110:19465576
3010:32516710
3002:13412736
2930:25341357
2895:20180376
2887:21642827
2852:20211726
2803:18556748
2682:33115740
2594:34161264
2531:14658965
2523:18438950
2462:25928094
2411:21297628
2362:15260948
2336:-ATPase"
2313:25566080
2262:28970791
2211:26674618
2160:23284664
2120:PLOS ONE
2101:11139403
2052:19506077
2001:16267270
1952:15975899
1903:39271006
1895:16449562
1845:12730376
1778:31823376
1729:31236763
1686:17553934
1651:25076348
1573:22543559
1530:22434069
1428:28634454
1377:33716756
1315:V-ATPase
1299:See also
1142:caveolae
991:opposite
842:axolemma
826:dystonia
802:dystonia
790:mutation
784:and the
579:proteins
381:glycogen
335:Function
288:membrane
272:) is an
209:proteins
197:articles
185:articles
158:RCSB PDB
68:7.2.2.13
5580:Biology
5534:Ligases
5304:Enzymes
5258:Tubulin
5227:Dynamin
5079:other:
4759:Katanin
4749:Kinesin
4724:ATP13A3
4719:ATP13A2
4454:Apyrase
4358:ATP13A5
4353:ATP13A4
4348:ATP13A3
4343:ATP13A2
4338:ATP13A1
4066:(3.A.3)
4026:ATP6V0E
4011:ATP6V0C
4006:ATP6V0B
3986:ATP6V1H
3966:ATP6V1F
3951:ATP6V1D
3926:ATP6V1A
3921:ATP6AP2
3916:ATP6AP1
3774:ATPases
3725:A video
3684:3420205
3603:3908073
3546:6560278
3497:3770729
3468:Bibcode
3460:Science
3438:2853965
3341:9778839
3283:9462791
3202:Toxicon
3174:8281379
3101:2781935
3048:26 June
2938:2066801
2843:2909369
2768:9814212
2725:9031683
2673:7608788
2652:Bibcode
2585:8237684
2562:Bibcode
2453:4417229
2402:3430603
2304:4274886
2253:5609115
2202:4684409
2151:3527461
2128:Bibcode
2092:1221581
2043:2742871
1992:1345669
1943:1196317
1813:Bibcode
1694:2291836
1643:8382364
1608:3030131
1565:6833401
1521:3390818
1419:5459889
1402:: 371.
1368:7947867
1211:Alpha:
1189:Denmark
1063:ouabain
1059:digoxin
1044:thyroid
892:ATP is
830:Alcohol
810:ouabain
792:in the
743:cofilin
727:ankyrin
627:ouabain
587:osmosis
496:glucose
290:of all
259:-ATPase
145:profile
128:MetaCyc
5566:Portal
5508:Lyases
5135:ARL13B
4995:RhoBTB
4889:α12/13
4777:GTPase
4754:Myosin
4744:Dynein
4714:ATP12A
4709:ATP11B
4704:ATP10A
4699:ATP8B1
4689:Other
4661:ATP1B4
4656:ATP1B3
4651:ATP1B2
4646:ATP1B1
4641:ATP1A4
4636:ATP1A3
4631:ATP1A2
4626:ATP1A1
4617:Na+/K+
4605:ATP2C2
4600:ATP2C1
4588:ATP2B4
4583:ATP2B3
4578:ATP2B2
4573:ATP2B1
4561:ATP2A3
4556:ATP2A2
4551:ATP2A1
4500:ATPase
4332:ATP11C
4327:ATP11B
4322:ATP11A
4316:ATP10D
4311:ATP10B
4306:ATP10A
4289:ATP8B4
4284:ATP8B3
4279:ATP8B2
4274:ATP8B1
4269:ATP8A1
4236:ATP8A2
4211:ATP2C1
4206:ATP2B4
4201:ATP2B3
4196:ATP2B2
4191:ATP2B1
4186:ATP2A3
4181:ATP2A2
4176:ATP2A1
4148:ATP12A
4118:ATP1G1
4113:ATP1B4
4108:ATP1B3
4103:ATP1B2
4098:ATP1B1
4093:ATP1A4
4088:ATP1A3
4083:ATP1A2
4078:ATP1A1
4038:TCIRG1
3884:ATP5L2
3874:ATP5J2
3854:ATP5G3
3849:ATP5G2
3844:ATP5G1
3839:ATP5F1
3824:ATP5C2
3819:ATP5C1
3809:ATP5A1
3716:(MeSH)
3691:
3681:
3640:
3601:
3593:
3553:
3543:
3504:
3494:
3486:
3436:
3428:
3389:
3348:
3338:
3330:
3290:
3280:
3272:
3228:
3220:
3181:
3171:
3163:
3108:
3098:
3090:
3044:. HGNC
3008:
3000:
2936:
2928:
2893:
2885:
2850:
2840:
2801:
2766:
2760:324293
2758:
2723:
2688:
2680:
2670:
2617:
2592:
2582:
2548:ATP1A3
2529:
2521:
2486:30 May
2460:
2450:
2409:
2399:
2360:
2340:Neuron
2311:
2301:
2260:
2250:
2236:: 85.
2209:
2199:
2158:
2148:
2099:
2089:
2050:
2040:
1999:
1989:
1950:
1940:
1901:
1893:
1843:
1836:156359
1833:
1784:
1776:
1735:
1727:
1692:
1684:
1649:
1641:
1606:
1571:
1563:
1528:
1518:
1477:
1452:
1426:
1416:
1375:
1365:
1249:ATP1B4
1242:ATP1B3
1237:ATP1B2
1232:ATP1B1
1230:Beta:
1225:ATP1A4
1221:ATP1A3
1217:ATP1A2
1213:ATP1A1
1127:muscle
822:ataxia
593:. The
349:volume
307:active
292:animal
284:ATPase
274:enzyme
242:sodium
192:PubMed
174:Search
164:PDBsum
104:ExPASy
92:BRENDA
80:IntEnz
63:EC no.
5460:Types
5191:EF-Tu
5130:SAR1B
5113:RAB27
5108:RAB23
5061:RhoDF
4951:RhoUV
4934:CDC42
4929:Cdc42
4915:>
4900:GNA13
4895:GNA12
4880:GNA11
4869:αq/11
4858:GNAT3
4846:GNAT2
4841:GNAT1
4831:GNAI3
4826:GNAI2
4821:GNAI1
4773:3.6.5
4737:3.6.4
4680:ATP4A
4671:H+/K+
4546:SERCA
4508:3.6.3
4492:3.6.3
4469:3.6.2
4429:3.6.1
4300:ATP9B
4295:ATP9A
4224:ATP7B
4219:ATP7A
4160:SERCA
4140:ATP4B
4135:ATP4A
3894:ATP5S
3889:ATP5O
3879:ATP5L
3869:ATP5J
3864:ATP5I
3859:ATP5H
3834:ATP5E
3829:ATP5D
3814:ATP5B
3599:S2CID
3006:S2CID
2934:S2CID
2891:S2CID
2764:S2CID
2686:S2CID
2527:S2CID
1899:S2CID
1782:S2CID
1733:S2CID
1690:S2CID
1647:S2CID
1569:S2CID
1206:Genes
1099:Na/Ca
1071:heart
1002:IP6K1
923:ions.
889:ions.
865:cell.
786:brain
429:(see
268:, or
140:PRIAM
5552:list
5545:EC7
5539:list
5532:EC6
5526:list
5519:EC5
5513:list
5506:EC4
5500:list
5493:EC3
5487:list
5480:EC2
5474:list
5467:EC1
5251:OPA1
5244:and
5196:EF-G
5186:IF-2
5152:Rheb
5140:ARL6
5125:ARF6
5096:NRAS
5091:KRAS
5086:HRAS
5071:RhoD
5066:RhoF
5012:RhoH
4988:RhoG
4973:Rac1
4961:RhoV
4956:RhoU
4939:TC10
4875:GNAQ
4595:SPCA
4422:3.6)
4263:ATP3
4248:Na/K
4170:) /
4168:SPCA
4164:PMCA
3689:PMID
3638:PMID
3591:PMID
3551:PMID
3502:PMID
3484:ISSN
3434:PMID
3426:ISSN
3387:PMID
3346:PMID
3328:ISSN
3288:PMID
3270:ISSN
3226:PMID
3218:ISSN
3179:PMID
3161:ISSN
3106:PMID
3088:ISSN
3050:2024
2998:PMID
2926:PMID
2883:PMID
2848:PMID
2826:1802
2799:PMID
2756:PMID
2721:PMID
2678:PMID
2615:ISBN
2590:PMID
2519:PMID
2488:2018
2458:PMID
2407:PMID
2358:PMID
2309:PMID
2258:PMID
2207:PMID
2156:PMID
2097:PMID
2048:PMID
1997:PMID
1948:PMID
1891:PMID
1841:PMID
1774:PMID
1725:PMID
1682:PMID
1639:PMID
1604:PMID
1561:PMID
1526:PMID
1475:ISBN
1450:ISBN
1424:PMID
1373:PMID
1291:and
1281:Na/K
1254:Na/K
1200:Na,K
1177:Na/K
1166:Na/K
1158:Na/K
1061:and
1051:Na/K
1042:, a
1036:Na/K
1034:The
1024:Na/K
1020:Na/K
1016:Na/K
1014:The
1009:p85α
1006:PI3K
998:Na/K
996:The
980:Na/K
969:Na/K
965:cAMP
961:Na/K
959:The
824:and
753:The
741:and
735:PI3K
731:IP3R
689:, a
664:IP3R
635:Na/K
591:lyse
458:and
449:and
405:Na/K
403:The
389:Na/K
377:Na/K
365:Na/K
361:Na/K
341:Na/K
339:The
303:Na/K
301:The
276:(an
266:pump
264:Na/K
257:Na/K
236:The
204:NCBI
161:PDBe
116:KEGG
34:Na/K
5246:MX2
5242:MX1
5162:RGK
5157:Rap
5147:Ran
5120:Arf
5103:Rab
5081:Ras
5039:Rnd
5017:Rho
4968:Rac
4944:TCL
4805:olf
4537:Ca+
4126:H/K
3727:by
3679:PMC
3669:doi
3665:109
3630:doi
3583:doi
3579:190
3541:PMC
3533:doi
3529:374
3492:PMC
3476:doi
3464:337
3418:doi
3377:doi
3336:PMC
3320:doi
3278:PMC
3260:doi
3210:doi
3206:114
3169:PMC
3153:doi
3096:PMC
3080:doi
2990:doi
2918:doi
2914:128
2875:doi
2838:PMC
2830:doi
2791:doi
2748:doi
2744:232
2713:doi
2668:PMC
2660:doi
2580:PMC
2570:doi
2558:118
2511:doi
2448:PMC
2438:doi
2397:PMC
2389:doi
2348:doi
2299:PMC
2289:doi
2248:PMC
2238:doi
2197:PMC
2187:doi
2146:PMC
2136:doi
2087:PMC
2079:doi
2075:353
2038:PMC
2028:doi
2024:284
1987:PMC
1979:doi
1938:PMC
1930:doi
1881:doi
1877:209
1831:PMC
1821:doi
1809:100
1764:doi
1760:598
1717:doi
1674:doi
1670:293
1631:doi
1627:422
1596:doi
1592:252
1553:doi
1549:114
1516:PMC
1508:doi
1414:PMC
1404:doi
1363:PMC
1353:doi
1273:In
1065:),
906:ADP
883:ATP
687:Src
575:ion
519:gut
311:ATP
180:PMC
152:PDB
5592::
4927::
4815:αi
4797:αs
4775::
4498::
4420:EC
4414::
4261::
4250:–
4234::
4174::
4166:,
4162:,
4156:Ca
4133::
4076::
3914::
3776:/
3772:,
3768::
3687:.
3677:.
3663:.
3659:.
3636:.
3626:21
3624:.
3620:.
3597:.
3589:.
3577:.
3573:.
3549:.
3539:.
3527:.
3523:.
3500:.
3490:.
3482:.
3474:.
3462:.
3458:.
3446:^
3432:.
3424:.
3414:27
3412:.
3408:.
3385:.
3373:67
3371:.
3367:.
3344:.
3334:.
3326:.
3316:39
3314:.
3310:.
3286:.
3276:.
3268:.
3256:18
3254:.
3250:.
3238:^
3224:.
3216:.
3204:.
3200:.
3177:.
3167:.
3159:.
3149:31
3147:.
3143:.
3130:^
3118:^
3104:.
3094:.
3086:.
3074:.
3070:.
3058:^
3026:.
3004:.
2996:.
2986:23
2984:.
2954:.
2932:.
2924:.
2912:.
2889:.
2881:.
2871:21
2869:.
2846:.
2836:.
2824:.
2820:.
2797:.
2787:23
2785:.
2762:.
2754:.
2742:.
2719:.
2709:60
2707:.
2684:.
2676:.
2666:.
2658:.
2646:.
2642:.
2588:.
2578:.
2568:.
2556:.
2552:.
2525:.
2517:.
2507:63
2505:.
2479:.
2456:.
2446:.
2434:16
2432:.
2428:.
2405:.
2395:.
2385:14
2383:.
2379:.
2356:.
2344:43
2342:.
2338:.
2332:/K
2307:.
2297:.
2283:.
2279:.
2256:.
2246:.
2234:11
2232:.
2228:.
2205:.
2195:.
2183:13
2181:.
2177:.
2154:.
2144:.
2134:.
2122:.
2118:.
2095:.
2085:.
2073:.
2069:.
2046:.
2036:.
2022:.
2018:.
1995:.
1985:.
1975:17
1973:.
1969:.
1946:.
1936:.
1926:16
1924:.
1920:.
1897:.
1889:.
1875:.
1871:.
1853:^
1839:.
1829:.
1819:.
1807:.
1803:.
1780:.
1772:.
1758:.
1754:.
1731:.
1723:.
1713:40
1711:.
1688:.
1680:.
1668:.
1645:.
1637:.
1625:.
1602:.
1590:.
1567:.
1559:.
1547:.
1524:.
1514:.
1504:32
1502:.
1498:.
1436:^
1422:.
1412:.
1398:.
1394:.
1371:.
1361:.
1349:11
1347:.
1341:.
1329:^
1240:,
1234:,
1223:,
1219:,
1215:,
1187:,
1148:.
1138:Na
1123:Ca
1115:Na
1111:Ca
1103:Na
1088:Ca
1084:Ca
1079:Ca
943:Na
917:Na
913:Na
887:Na
875:Na
846:Na
834:Na
828:.
814:Na
794:Na
768:,
755:Na
745:.
737:,
733:,
729:,
719:Na
711:Na
703:Na
695:Na
679:Na
671:Na
611:Na
603:Na
595:Na
563:Na
553:.
547:Na
543:Na
535:Na
527:Na
515:Na
507:Na
498:,
480:Na
472:Na
468:mM
456:Na
451:Na
355:,
331:.
298:.
261:,
5568::
5554:)
5550:(
5541:)
5537:(
5528:)
5524:(
5515:)
5511:(
5502:)
5498:(
5489:)
5485:(
5476:)
5472:(
5296:e
5289:t
5282:v
5054:3
5049:2
5044:1
5032:C
5027:B
5022:A
5005:2
5000:1
4983:3
4978:2
4887:G
4867:G
4813:G
4803:G
4795:G
4496:4
4494:-
4418:(
4404:e
4397:t
4390:v
4252:H
4158:(
3784:)
3780:(
3758:e
3751:t
3744:v
3731:.
3695:.
3671::
3644:.
3632::
3605:.
3585::
3557:.
3535::
3508:.
3478::
3470::
3440:.
3420::
3393:.
3379::
3352:.
3322::
3294:.
3262::
3232:.
3212::
3185:.
3155::
3112:.
3082::
3076:5
3052:.
3012:.
2992::
2969:.
2940:.
2920::
2897:.
2877::
2854:.
2832::
2805:.
2793::
2770:.
2750::
2727:.
2715::
2692:.
2662::
2654::
2648:6
2638:7
2623:.
2596:.
2572::
2564::
2533:.
2513::
2490:.
2464:.
2440::
2413:.
2391::
2364:.
2350::
2334:+
2330:+
2315:.
2291::
2285:5
2264:.
2240::
2213:.
2189::
2162:.
2138::
2130::
2124:7
2103:.
2081::
2054:.
2030::
2003:.
1981::
1954:.
1932::
1905:.
1883::
1869:"
1847:.
1823::
1815::
1788:.
1766::
1739:.
1719::
1696:.
1676::
1653:.
1633::
1610:.
1598::
1575:.
1555::
1532:.
1510::
1483:.
1458:.
1430:.
1406::
1400:8
1379:.
1355::
1134:d
1131:K
1119:K
1117:-
1107:K
1105:-
986:i
984:G
975:s
973:G
936:K
928:K
921:K
879:K
850:K
848:-
838:K
836:-
818:K
816:-
804:-
798:K
796:-
759:K
757:-
723:K
721:-
715:K
713:-
707:K
705:-
699:K
697:-
683:K
681:-
675:K
673:-
615:K
613:-
607:K
605:-
599:K
597:-
567:K
565:-
531:K
529:-
511:K
509:-
476:K
464:K
460:K
447:K
244:–
240:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.