548:
retroelements, and are no longer responsible for intron gain. Tandem genomic duplication is the only proposed mechanism with supporting in vivo experimental evidence: a short intragenic tandem duplication can insert a novel intron into a protein-coding gene, leaving the corresponding peptide sequence unchanged. This mechanism also has extensive indirect evidence lending support to the idea that tandem genomic duplication is a prevalent mechanism for intron gain. The testing of other proposed mechanisms in vivo, particularly intron gain during DSBR, intron transfer, and intronization, is possible, although these mechanisms must be demonstrated in vivo to solidify them as actual mechanisms of intron gain. Further genomic analyses, especially when executed at the population level, may then quantify the relative contribution of each mechanism, possibly identifying species-specific biases that may shed light on varied rates of intron gain amongst different species.
528:
transcriptase-mediated intron loss (RTMIL) and genomic deletions, have been identified, and are known to occur. The definitive mechanisms of intron gain, however, remain elusive and controversial. At least seven mechanisms of intron gain have been reported thus far: intron transposition, transposon insertion, tandem genomic duplication, intron transfer, intron gain during double-strand break repair (DSBR), insertion of a group II intron, and intronization. In theory it should be easiest to deduce the origin of recently gained introns due to the lack of host-induced mutations, yet even introns gained recently did not arise from any of the aforementioned mechanisms. These findings thus raise the question of whether or not the proposed mechanisms of intron gain fail to describe the mechanistic origin of many novel introns because they are not accurate mechanisms of intron gain, or if there are other, yet to be discovered, processes generating novel introns.
373:, that is, the intron-containing RNA molecule can rearrange its own covalent structure so as to precisely remove the intron and link the exons together in the correct order. In some cases, particular intron-binding proteins are involved in splicing, acting in such a way that they assist the intron in folding into the three-dimensional structure that is necessary for self-splicing activity. Group I and group II introns are distinguished by different sets of internal conserved sequences and folded structures, and by the fact that splicing of RNA molecules containing group II introns generates branched introns (like those of spliceosomal RNAs), while group I introns use a non-encoded guanosine nucleotide (typically GTP) to initiate splicing, adding it on to the 5'-end of the excised intron.
540:
sequence between the original and duplicated AGGT will be spliced, resulting in the creation of an intron without alteration of the coding sequence of the gene. Double-stranded break repair via non-homologous end joining was recently identified as a source of intron gain when researchers identified short direct repeats flanking 43% of gained introns in
Daphnia. These numbers must be compared to the number of conserved introns flanked by repeats in other organisms, though, for statistical relevance. For group II intron insertion, the retrohoming of a group II intron into a nuclear gene was proposed to cause recent spliceosomal intron gain.
404:
in a heterozygous state this will result in production of two abundant splice variants; one functional and one non-functional. In the homozygous state the mutant alleles may cause a genetic disease such as the hemophilia found in descendants of Queen
Victoria where a mutation in one of the introns in a blood clotting factor gene creates a cryptic 3' splice site resulting in aberrant splicing. A significant fraction of human deaths by disease may be caused by mutations that interfere with normal splicing; mostly by creating cryptic splice sites.
386:
competing cryptic splice site sequences within the introns and those conditions are rarely met in large eukaryotic genes that may cover more than 40 kilobase pairs. Recent studies have shown that the actual error rate can be considerably higher than 10 and may be as high as 2% or 3% errors (error rate of 2 or 3 x 10) per gene. Additional studies suggest that the error rate is no less than 0.1% per intron. This relatively high level of splicing errors explains why most splice variants are rapidly degraded by nonsense-mediated decay.
408:
processing noise due to splicing errors. One of the central issues in the field of alternative splicing is working out the differences between these two possibilities. Many scientists have argued that the null hypothesis should be splicing noise, putting the burden of proof on those who claim biologically relevant alternative splicing. According to those scientists, the claim of function must be accompanied by convincing evidence that multiple functional products are produced from the same gene.
440:(the introns-first hypothesis). There is still considerable debate about the extent to which of these hypotheses is most correct but the popular consensus at the moment is that following the formation of the first eukaryotic cell, group II introns from the bacterial endosymbiont invaded the host genome. In the beginning these self-splicing introns excised themselves from the mRNA precursor but over time some of them lost that ability and their excision had to be aided in
536:
sequence when a transposon inserts into the sequence AGGT or encodes the splice sites within the transposon sequence. Where intron-generating transposons do not create target site duplications, elements include both splice sites GT (5') and AG (3') thereby splicing precisely without affecting the protein-coding sequence. It is not yet understood why these elements are spliced, whether by chance, or by some preferential action by the transposon.
215:
400:
rate of 10 – 10 is high enough that one in every 25,000 transcribed exons will have an incorporation error in one of the splice sites leading to a skipped intron or a skipped exon. Almost all multi-exon genes will produce incorrectly spliced transcripts but the frequency of this background noise will depend on the size of the genes, the number of introns, and the quality of the splice site sequences.
503:. In highly expressed yeast genes, introns inhibit R-loop formation and the occurrence of DNA damage. Genome-wide analysis in both yeast and humans revealed that intron-containing genes have decreased R-loop levels and decreased DNA damage compared to intronless genes of similar expression. Insertion of an intron within an R-loop prone gene can also suppress R-loop formation and
382:
nucleotides apart. All biochemical reactions are associated with known error rates and the more complicated the reaction the higher the error rate. Therefore, it is not surprising that the splicing reaction catalyzed by the spliceosome has a significant error rate even though there are spliceosome accessory factors that suppress the accidental cleavage of cryptic splice sites.
469:), there must have been extensive gain or loss of introns during evolutionary time. This process is thought to be subject to selection, with a tendency towards intron gain in larger species due to their smaller population sizes, and the converse in smaller (particularly unicellular) species. Biological factors also influence which genes in a genome lose or accumulate introns.
531:
In intron transposition, the most commonly purported intron gain mechanism, a spliced intron is thought to reverse splice into either its own mRNA or another mRNA at a previously intron-less position. This intron-containing mRNA is then reverse transcribed and the resulting intron-containing cDNA may
475:
of exons within a gene after intron excision acts to introduce greater variability of protein sequences translated from a single gene, allowing multiple related proteins to be generated from a single gene and a single precursor mRNA transcript. The control of alternative RNA splicing is performed by
431:
After the initial discovery of introns in protein-coding genes of the eukaryotic nucleus, there was significant debate as to whether introns in modern-day organisms were inherited from a common ancient ancestor (termed the introns-early hypothesis), or whether they appeared in genes rather recently
407:
Incorrectly spliced transcripts can easily be detected and their sequences entered into the online databases. They are usually described as "alternatively spliced" transcripts, which can be confusing because the term does not distinguish between real, biologically relevant, alternative splicing and
403:
In some cases, splice variants will be produced by mutations in the gene (DNA). These can be SNP polymorphisms that create a cryptic splice site or mutate a functional site. They can also be somatic cell mutations that affect splicing in a particular tissue or a cell line. When the mutant allele is
543:
Intron transfer has been hypothesized to result in intron gain when a paralog or pseudogene gains an intron and then transfers this intron via recombination to an intron-absent location in its sister paralog. Intronization is the process by which mutations create novel introns from formerly exonic
334:
Transfer RNA introns that depend upon proteins for removal occur at a specific location within the anticodon loop of unspliced tRNA precursors, and are removed by a tRNA splicing endonuclease. The exons are then linked together by a second protein, the tRNA splicing ligase. Note that self-splicing
259:
Splicing of all intron-containing RNA molecules is superficially similar, as described above. However, different types of introns were identified through the examination of intron structure by DNA sequence analysis, together with genetic and biochemical analysis of RNA splicing reactions. At least
399:
While the catalytic reaction may be accurate enough for effective processing most of the time, the overall error rate may be partly limited by the fidelity of transcription because transcription errors will introduce mutations that create cryptic splice sites. In addition, the transcription error
394:
Although mutations which create or disrupt binding sites may be slightly deleterious, the large number of possible such mutations makes it inevitable that some will reach fixation in a population. This is particularly relevant in species, such as humans, with relatively small long-term effective
535:
Transposon insertions have been shown to generate thousands of new introns across diverse eukaryotic species. Transposon insertions sometimes result in the duplication of this sequence on each side of the transposon. Such an insertion could intronize the transposon without disrupting the coding
317:
Nuclear pre-mRNA introns (spliceosomal introns) are characterized by specific intron sequences located at the boundaries between introns and exons. These sequences are recognized by spliceosomal RNA molecules when the splicing reactions are initiated. In addition, they contain a branch point, a
539:
In tandem genomic duplication, due to the similarity between consensus donor and acceptor splice sites, which both closely resemble AGGT, the tandem genomic duplication of an exonic segment harboring an AGGT sequence generates two potential splice sites. When recognized by the spliceosome, the
385:
Under ideal circumstances, the splicing reaction is likely to be 99.999% accurate (error rate of 10) and the correct exons will be joined and the correct intron will be deleted. However, these ideal conditions require very close matches to the best splice site sequences and the absence of any
381:
The spliceosome is a very complex structure containing up to one hundred proteins and five different RNAs. The substrate of the reaction is a long RNA molecule and the transesterification reactions catalyzed by the spliceosome require the bringing together of sites that may be thousands of
547:
The only hypothesized mechanism of recent intron gain lacking any direct evidence is that of group II intron insertion, which when demonstrated in vivo, abolishes gene expression. Group II introns are therefore likely the presumed ancestors of spliceosomal introns, acting as site-specific
527:
genes. Subsequent analyses have identified thousands of examples of intron loss and gain events, and it has been proposed that the emergence of eukaryotes, or the initial stages of eukaryotic evolution, involved an intron invasion. Two definitive mechanisms of intron loss, reverse
679:"The notion of the cistron ... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978)
152:... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978)
395:
population sizes. It is plausible, then, that the human genome carries a substantial load of suboptimal sequences which cause the generation of aberrant transcript isoforms. In this study, we present direct evidence that this is indeed the case.
458:
have now shown that the lengths and density (introns/gene) of introns varies considerably between related species. For example, while the human genome contains an average of 8.4 introns/gene (139,418 in the genome), the unicellular fungus
389:
The presence of sloppy binding sites within genes causes splicing errors and it may seem strange that these sites haven't been eliminated by natural selection. The argument for their persistence is similar to the argument for junk DNA.
102:, and were subsequently identified in genes encoding transfer RNA and ribosomal RNA genes. Introns are now known to occur within a wide variety of genes throughout organisms, bacteria, and viruses within all of the biological kingdoms.
479:
Introns contain several short sequences that are important for efficient splicing, such as acceptor and donor sites at either end of the intron as well as a branch point site, which are required for proper splicing by the
199:(e.g. humans, mice, and pufferfish (fugu)), where protein-coding genes almost always contain multiple introns, while introns are rare within the nuclear genes of some eukaryotic microorganisms, for example
302:
are proposed to be a fifth family, but little is known about the biochemical apparatus that mediates their splicing. They appear to be related to group II introns, and possibly to spliceosomal introns.
507:. Bonnet et al. (2017) speculated that the function of introns in maintaining genetic stability may explain their evolutionary maintenance at certain locations, particularly in highly expressed genes.
218:
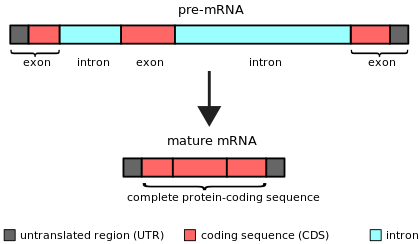
Simple illustration of an unspliced mRNA precursor, with two introns and three exons (top). After the introns have been removed via splicing, the mature mRNA sequence is ready for translation (bottom).
451:
Early studies of genomic DNA sequences from a wide range of organisms show that the intron-exon structure of homologous genes in different organisms can vary widely. More recent studies of entire
416:
While introns do not encode protein products, they are integral to gene expression regulation. Some introns themselves encode functional RNAs through further processing after splicing to generate
365:
in a very wide range of living organisms. Following transcription into RNA, group I and group II introns also make extensive internal interactions that allow them to fold into a specific, complex
195:
The frequency of introns within different genomes is observed to vary widely across the spectrum of biological organisms. For example, introns are extremely common within the nuclear genome of
4014:
326:
intron. Apart from these three short conserved elements, nuclear pre-mRNA intron sequences are highly variable. Nuclear pre-mRNA introns are often much longer than their surrounding exons.
90:). There are four main types of introns: tRNA introns, group I introns, group II introns, and spliceosomal introns (see below). Introns are rare in Bacteria and Archaea (prokaryotes).
922:, Pedersen-Lane J, West D, Ehrenman K, Maley G, Chu F, Maley F (June 1985). "Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level".
424:
is widely used to generate multiple proteins from a single gene. Furthermore, some introns play essential roles in a wide range of gene expression regulatory functions such as
318:
particular nucleotide sequence near the 3' end of the intron that becomes covalently linked to the 5' end of the intron during the splicing process, generating a branched
4058:
4007:
515:
The physical presence of introns promotes cellular resistance to starvation via intron enhanced repression of ribosomal protein genes of nutrient-sensing pathways.
175:, the term "intervening sequence" can refer to any of several families of internal nucleic acid sequences that are not present in the final gene product, including
86:
Introns are found in the genes of most eukaryotes and many eukaryotic viruses and they can be located in both protein-coding genes and genes that function as RNA (
1299:"Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei"
544:
sequence. Thus, unlike other proposed mechanisms of intron gain, this mechanism does not require the insertion or generation of DNA to create a novel intron.
4000:
3462:
Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S (January 2019). "Introns are mediators of cell response to starvation".
1444:
Copertino DW, Hallick RB (December 1993). "Group II and group III introns of twintrons: potential relationships with nuclear pre-mRNA introns".
3963:
817:
Chow LT, Gelinas RE, Broker TR, Roberts RJ (September 1977). "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA".
118:
572:
741:
Kinniburgh AJ, Mertz JE, Ross J (July 1978). "The precursor of mouse beta-globin messenger RNA contains two intervening RNA sequences".
448:
of the spliceosome. The efficiency of splicing was improved by association with stabilizing proteins to form the primitive spliceosome.
4068:
4023:
793:
500:
444:
by other group II introns. Eventually a number of specific trans-acting introns evolved and these became the precursors to the
366:
233:(Mb) intron, which takes roughly three days to transcribe. On the other extreme, a 2015 study suggests that the shortest known
3949:
1549:
Greer CL, Peebles CL, Gegenheimer P, Abelson J (February 1983). "Mechanism of action of a yeast RNA ligase in tRNA splicing".
1013:
1348:"Identification of minimal eukaryotic introns through GeneBase, a user-friendly tool for parsing the NCBI Gene databank"
2806:"Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes"
4063:
2179:"Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay"
485:
980:
476:
a complex network of signaling molecules that respond to a wide range of intracellular and extracellular signals.
211:
of vertebrates are entirely devoid of introns, while those of eukaryotic microorganisms may contain many introns.
122:
2228:
Bitton DA, Atkinson SR, Rallis C, Smith GC, Ellis DA, Chen YY, Malecki M, Codlin S, Lemay JF, Cotobal C (2015).
484:. Some introns are known to enhance the expression of the gene that they are contained in by a process known as
4324:
2441:"Expression changes confirm genomic variants predicted to result in allele-specific, alternative mRNA splicing"
1213:
Taanman JW (February 1999). "The mitochondrial genome: structure, transcription, translation and replication".
461:
344:
282:
76:
965:
465:
contains only 0.0075 introns/gene (15 introns in the genome). Since eukaryotes arose from a common ancestor (
4344:
4269:
4090:
425:
3833:"Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites"
1592:
Reinhold-Hurek B, Shub DA (May 1992). "Self-splicing introns in tRNA genes of widely divergent bacteria".
1479:
Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA (1986). "Splicing of messenger RNA precursors".
3154:
Roy SW, Gilbert W (March 2006). "The evolution of spliceosomal introns: patterns, puzzles and progress".
1799:
Sales-Lee J, Perry DS, Bowser BA, Diedrich JK, Rao B, Beusch I, Yates III JR, Roy SW, Madhani HD (2021).
125:
for the researchers and collaborators in their labs that did the experiments resulting in the discovery,
4339:
2855:
Penny D, Hoeppner MP, Poole AM, Jeffares DC (November 2009). "An overview of the introns-first theory".
504:
50:
79:. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called
3788:
Cech TR (January 1986). "The generality of self-splicing RNA: relationship to nuclear mRNA splicing".
4334:
4153:
3903:
3844:
3581:
3471:
3253:
2864:
2560:
2503:
1974:
1601:
1116:
1057:
873:
697:
658:
589:
472:
421:
3982:
2804:
Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, Derti A, et al. (April 2011). Snyder M (ed.).
4146:
1103:
Tilghman SM, Tiemeier DC, Seidman JG, Peterlin BM, Sullivan M, Maizel JV, Leder P (February 1978).
1018:
180:
110:
1713:
Wan R, Bai R, Zhan X, Shi Y (2020). "How is precursor messenger RNA spliced by the spliceosome?".
3813:
3605:
3495:
3398:
3222:
3179:
3136:
3076:
Rodríguez-Trelles F, Tarrío R, Ayala FJ (2006). "Origins and evolution of spliceosomal introns".
3009:
2966:
2888:
2786:
2529:
2421:
1895:
1838:
1781:
1738:
1625:
1574:
1238:
985:
947:
842:
766:
723:
114:
1395:
Slabodnick MM, Ruby JG, Reiff SB, Swart EC, Gosai S, Prabakaran S, et al. (February 2017).
3421:
Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, et al. (August 2017).
532:
then cause intron gain via complete or partial recombination with its original genomic locus.
4141:
3959:
3929:
3872:
3805:
3770:
3717:
3663:
3597:
3550:
3487:
3444:
3390:
3351:
3316:
3281:
3214:
3171:
3128:
3093:
3058:
3001:
2958:
2923:
2880:
2837:
2778:
2737:
2688:
2637:
2588:
2521:
2472:
2413:
2359:
2310:
2259:
2210:
2159:
2107:
2051:
2002:
1940:
1887:
1830:
1773:
1730:
1695:
1660:
1617:
1566:
1531:
1496:
1461:
1426:
1377:
1328:
1279:
1230:
1195:
1144:
1105:"Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene"
1085:
939:
901:
834:
799:
789:
758:
715:
599:
523:
Introns may be lost or gained over evolutionary time, as shown by many comparative studies of
247:
208:
4221:
4216:
3977:
3954:
Bruce
Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter
432:
in the evolutionary process (termed the introns-late hypothesis). Another theory is that the
4226:
4201:
3967:
3919:
3911:
3862:
3852:
3797:
3760:
3752:
3707:
3697:
3653:
3643:
3589:
3540:
3530:
3479:
3434:
3382:
3343:
3334:
Jeffares DC, Penkett CJ, Bähler J (August 2008). "Rapidly regulated genes are intron poor".
3308:
3271:
3261:
3206:
3163:
3120:
3085:
3048:
3040:
2993:
2950:
2915:
2872:
2827:
2817:
2768:
2727:
2719:
2678:
2668:
2627:
2619:
2578:
2568:
2511:
2462:
2452:
2403:
2393:
2349:
2341:
2300:
2290:
2249:
2241:
2230:"Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast"
2200:
2190:
2149:
2141:
2097:
2087:
2041:
2033:
1992:
1982:
1930:
1922:
1877:
1869:
1820:
1812:
1765:
1722:
1687:
1652:
1609:
1558:
1523:
1488:
1453:
1416:
1408:
1367:
1359:
1318:
1310:
1269:
1222:
1185:
1175:
1134:
1124:
1075:
1065:
931:
891:
881:
826:
750:
705:
299:
200:
196:
106:
3089:
2279:"The fitness cost of mis-splicing is the main determinant of alternative splicing patterns"
2277:
Saudemont B, Popa A, Parmley JL, Rocher V, Blugeon C, Necsulea A, Meyer E, Duret L (2017).
1046:"Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain"
4299:
4100:
4083:
4078:
4053:
3736:
1769:
1726:
919:
348:
292:
39:
35:
3741:"Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry"
2657:"Systematic evaluation of isoform function in literature reports of alternative splicing"
3907:
3848:
3585:
3475:
3257:
2868:
2564:
2507:
1978:
1691:
1656:
1605:
1527:
1492:
1120:
1061:
877:
701:
75:
refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA
4264:
4163:
4048:
4043:
3924:
3891:
3765:
3740:
3712:
3685:
3658:
3631:
3545:
3518:
3299:
Jeffares DC, Mourier T, Penny D (January 2006). "The biology of intron gain and loss".
3053:
3028:
2832:
2805:
2732:
2707:
2683:
2656:
2632:
2607:
2583:
2548:
2467:
2440:
2408:
2381:
2354:
2329:
2305:
2278:
2254:
2229:
2205:
2178:
2154:
2129:
2102:
2075:
2046:
2021:
1997:
1962:
1935:
1914:
1882:
1857:
1825:
1800:
1421:
1396:
1372:
1347:
1323:
1298:
1190:
1163:
1030:
681:
628:
577:
466:
141:
87:
3423:"Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability"
3276:
3241:
1274:
1257:
1226:
1139:
1104:
1080:
1045:
935:
896:
861:
4318:
3867:
3832:
3801:
2984:
Sharp PA (1991). ""Five easy pieces."(role of RNA catalysis in cellular processes)".
2954:
2919:
2533:
2425:
1842:
1785:
1742:
1562:
1457:
1346:
Piovesan A, Caracausi M, Ricci M, Strippoli P, Vitale L, Pelleri MC (December 2015).
830:
754:
643:
638:
417:
362:
354:
3992:
3817:
3499:
3197:
de Souza SJ (July 2003). "The emergence of a synthetic theory of intron evolution".
3183:
2970:
2892:
1899:
1578:
1242:
951:
770:
4329:
4110:
4031:
3609:
3514:
3402:
3226:
3140:
2790:
2655:
Bhuiyan SA, Ly S, Phan M, Huntington B, Hogan E, Liu CC, Liu J, Pavlidis P (2018).
2398:
1858:"Spliceosomes walk the line: splicing errors and their impact on cellular function"
1629:
1397:"The Macronuclear Genome of Stentor coeruleus Reveals Tiny Introns in a Giant Cell"
846:
727:
358:
312:
275:
264:
214:
126:
3831:
Dickson L, Huang HR, Liu L, Matsuura M, Lambowitz AM, Perlman PS (November 2001).
3684:
Gozashti L, Roy S, Thornlow B, Kramer A, Ares M, Corbett-Detig R (November 2022).
3369:
Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (August 2002).
3013:
2706:
Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A (March 2011).
3439:
3422:
2822:
2092:
1314:
4289:
4136:
4095:
3044:
2382:"Pan-cancer repository of validated natural and cryptic mRNA splicing mutations"
648:
524:
481:
433:
268:
242:
184:
130:
3837:
Proceedings of the
National Academy of Sciences of the United States of America
3246:
Proceedings of the
National Academy of Sciences of the United States of America
1109:
Proceedings of the
National Academy of Sciences of the United States of America
1050:
Proceedings of the
National Academy of Sciences of the United States of America
866:
Proceedings of the
National Academy of Sciences of the United States of America
3483:
3347:
3312:
3210:
2876:
2673:
2295:
1816:
1756:
Wilkinson ME, Charenton C, Nagai K (2020). "RNA splicing by the spliceosome".
1412:
1180:
1164:"Comparative genomic analysis of fungal genomes reveals intron-rich ancestors"
224:
138:
99:
17:
3976:
5th edition, 2002, W H Freeman. Available online through the NCBI Bookshelf:
2457:
3702:
3124:
2997:
2573:
2516:
2491:
2022:"Widespread alternative and aberrant splicing revealed by lariat sequencing"
1987:
1363:
803:
452:
437:
369:. These complex architectures allow some group I and group II introns to be
3933:
3876:
3857:
3774:
3721:
3667:
3648:
3554:
3535:
3491:
3448:
3394:
3355:
3320:
3285:
3266:
3218:
3175:
3132:
3097:
3062:
2962:
2884:
2841:
2782:
2773:
2756:
2741:
2692:
2641:
2592:
2525:
2476:
2417:
2363:
2314:
2263:
2214:
2195:
2163:
2145:
2111:
2055:
2006:
1944:
1891:
1834:
1777:
1734:
1678:
Michel F, Ferat JL (1995). "Structure and activities of group II introns".
1430:
1381:
1332:
1283:
1234:
1199:
1070:
886:
3809:
3601:
3005:
2927:
2723:
2490:
Rogaev EI, Grigorenko AP, Faskhutdinova G, Kittler EL, Moliaka YK (2009).
2245:
1699:
1664:
1621:
1570:
1535:
1500:
1465:
1129:
943:
2623:
2037:
1926:
1873:
1148:
1089:
905:
838:
762:
719:
653:
616:
286:
234:
230:
31:
3756:
4256:
4236:
4231:
3915:
2345:
594:
176:
165:
149:
30:
For the interferon-based drug used in viral and cancer treatments, see
57:
that is not expressed or operative in the final RNA product. The word
4211:
4206:
4186:
4073:
3593:
2134:
Mutation
Research/Fundamental and Molecular Mechanisms of Mutagenesis
1613:
710:
685:
633:
604:
496:
455:
3569:
3370:
3167:
2130:"Estimation of the minimum mRNA splicing error rate in vertebrates"
1044:
Tonegawa S, Maxam AM, Tizard R, Bernard O, Gilbert W (March 1978).
353:
Group I and group II introns are found in genes encoding proteins (
4279:
4274:
4196:
4191:
4181:
4176:
4171:
4131:
3386:
2755:
Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ (December 2012).
2020:
Stepankiw N, Raghavan M, Fogarty EA, Grimson A, Pleiss JA (2015).
445:
213:
3966:. Fourth edition is available online through the NCBI Bookshelf:
4294:
4284:
4126:
3987:
3632:"Identifying the mechanisms of intron gain: progress and trends"
567:
562:
80:
54:
3996:
3892:"A segmental genomic duplication generates a functional intron"
3686:"Transposable elements drive intron gain in diverse eukaryotes"
2906:
Cavalier-Smith T (1991). "Intron phylogeny: a new hypothesis".
2492:"Genotype analysis identifies the cause of the "royal disease""
2177:
Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, Hurst LD (2009).
1963:"Splice-site pairing is an intrinsically high fidelity process"
1297:
Reugels AM, Kurek R, Lammermann U, Bünemann H (February 2000).
862:"Spliced segments at the 5' terminus of adenovirus 2 late mRNA"
2549:"Rate, molecular spectrum, and consequences of human mutation"
492:
3111:
Mourier T, Jeffares DC (May 2003). "Eukaryotic intron loss".
2076:"Noisy splicing drives mRNA isoform diversity in human cells"
237:
intron length is 30 base pairs (bp) belonging to the human
3950:
A search engine for exon/intron sequences defined by NCBI
3890:
Hellsten U, Aspden JL, Rio DC, Rokhsar DS (August 2011).
3029:"Origin of spliceosomal introns and alternative splicing"
436:
and the intron-exon structure of genes is a relic of the
251:, in which most (> 95%) introns are 15 or 16 bp long.
98:
Introns were first discovered in protein-coding genes of
1801:"Coupling of spliceosome complexity to intron diversity"
788:(3rd ed.). New York: Wiley. pp. 159–179, 386.
164:, i.e., an additional piece of DNA that arises within a
3371:"Selection for short introns in highly expressed genes"
2608:"The state of play in higher eukaryote gene annotation"
260:
four distinct classes of introns have been identified:
1514:
Guthrie C, Patterson B (1988). "Spliceosomal snRNAs".
1643:
Cech TR (1990). "Self-splicing of group I introns".
335:
introns are also sometimes found within tRNA genes.
4255:
4162:
4109:
4030:
3972:Jeremy M Berg, John L Tymoczko, and Lubert Stryer,
3679:
3677:
3570:"A role for reverse transcripts in gene conversion"
2757:"Introns in UTRs: why we should stop ignoring them"
2074:Pickrell JK, Pai AA, Gilad Y, Pritchard JK (2010).
1215:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
109:by introns was discovered independently in 1977 by
3242:"Intron evolution as a population-genetic process"
278:genes that are removed by proteins (tRNA introns)
1014:"Nobel Prize in medicine brews resentment, envy"
966:"The Nobel Prize in Physiology or Medicine 1993"
3983:Intron finding tool for plant genomic sequences
3625:
3623:
3621:
3619:
3416:
3414:
3412:
2941:Doolittle WF (1991). "The origins of introns".
2553:Proceedings of the National Academy of Sciences
1967:Proceedings of the National Academy of Sciences
241:gene. The shortest known introns belong to the
3519:"Origin and evolution of spliceosomal introns"
4008:
675:
673:
8:
2708:"Critical association of ncRNA with introns"
2123:
2121:
860:Berget SM, Moore C, Sharp PA (August 1977).
2069:
2067:
2065:
979:Abir-Am, Pnina Geraldine (September 2020).
4115:
4036:
4015:
4001:
3993:
3033:Cold Spring Harbor Perspectives in Biology
2375:
2373:
27:Specific base pair sequences within a gene
3923:
3866:
3856:
3764:
3711:
3701:
3657:
3647:
3544:
3534:
3438:
3275:
3265:
3052:
2831:
2821:
2772:
2731:
2682:
2672:
2631:
2582:
2572:
2515:
2466:
2456:
2407:
2397:
2353:
2304:
2294:
2253:
2204:
2194:
2153:
2101:
2091:
2045:
1996:
1986:
1934:
1881:
1824:
1420:
1371:
1322:
1273:
1256:Tollervey D, Caceres JF (November 2000).
1189:
1179:
1138:
1128:
1079:
1069:
895:
885:
709:
71:, i.e., a region inside a gene. The term
2439:Mucaki EJ, Shirley BC, Rogan PK (2020).
1915:"Stochastic noise in splicing machinery"
1162:Stajich JE, Dietrich FS, Roy SW (2007).
981:"The Women Who Discovered RNA Splicing"
669:
265:Introns in nuclear protein-coding genes
3568:Derr LK, Strathern JN (January 1993).
3090:10.1146/annurev.genet.40.110405.090625
1956:
1954:
171:Although introns are sometimes called
3630:Yenerall P, Zhou L (September 2012).
2380:Shirley B, Mucaki E, Rogan P (2019).
1770:10.1146/annurev-biochem-091719-064225
1727:10.1146/annurev-biochem-013118-111024
119:Nobel Prize in Physiology or Medicine
7:
3517:, Csuros M, Koonin EV (April 2012).
573:Eukaryotic chromosome fine structure
1692:10.1146/annurev.bi.64.070195.002251
1657:10.1146/annurev.bi.59.070190.002551
1528:10.1146/annurev.ge.22.120188.002131
1493:10.1146/annurev.bi.55.070186.005351
222:A particularly extreme case is the
1012:Flint, Anthony (8 November 1993).
412:Biological functions and evolution
183:(UTR), and nucleotides removed by
25:
4024:Post-transcriptional modification
295:that are removed by RNA catalysis
1961:Fox-Walsh KL, Hertel KJ (2009).
491:Actively transcribed regions of
313:RNA splicing § Spliceosomal
274:Introns in nuclear and archaeal
2857:Journal of Molecular Evolution
2399:10.12688/f1000research.17204.3
2328:Scotti MM, Swanson MS (2016).
1446:Trends in Biochemical Sciences
367:three-dimensional architecture
1:
3956:Molecular Biology of the Cell
3027:Irimia M, and Roy SW (2014).
2330:"RNA mis-splicing in disease"
1758:Annual Review of Biochemistry
1715:Annual Review of Biochemistry
1680:Annual Review of Biochemistry
1645:Annual Review of Biochemistry
1481:Annual Review of Biochemistry
1275:10.1016/S0092-8674(00)00174-4
1227:10.1016/s0005-2728(98)00161-3
936:10.1016/s0092-8674(85)80010-6
3802:10.1016/0092-8674(86)90751-8
3735:Chalamcharla VR, Curcio MJ,
3440:10.1016/j.molcel.2017.07.002
2955:10.1016/0960-9822(91)90214-h
2920:10.1016/0168-9525(91)90377-3
2823:10.1371/journal.pgen.1001366
2093:10.1371/journal.pgen.1001236
1563:10.1016/0092-8674(83)90473-7
1458:10.1016/0968-0004(93)90008-b
831:10.1016/0092-8674(77)90180-5
755:10.1016/0092-8674(78)90251-9
339:Group I and group II introns
117:, for which they shared the
3045:10.1101/cshperspect.a016071
2606:Mudge JM, Harrow J (2016).
1913:Melamud E, Moult J (2009).
1258:"RNA processing marches on"
486:intron-mediated enhancement
377:On the accuracy of splicing
137:was introduced by American
121:in 1993, though credit was
4361:
1921:. gkp471 (14): 4873–4886.
1856:Hsu SN, Hertel KJ (2009).
1315:10.1093/genetics/154.2.759
519:As mobile genetic elements
342:
310:
187:, in addition to introns.
29:
4118:
4039:
3988:Exon-intron graphic maker
3484:10.1038/s41586-018-0859-7
3348:10.1016/j.tig.2008.05.006
3313:10.1016/j.tig.2005.10.006
3078:Annual Review of Genetics
2877:10.1007/s00239-009-9279-5
2674:10.1186/s12864-018-5013-2
2296:10.1186/s13059-017-1344-6
1817:10.1016/j.cub.2021.09.004
1516:Annual Review of Genetics
1413:10.1016/j.cub.2016.12.057
1181:10.1186/gb-2007-8-10-r223
105:The fact that genes were
61:is derived from the term
3156:Nature Reviews. Genetics
2458:10.3389/fgene.2020.00109
462:Encephalitozoon cuniculi
345:Group I catalytic intron
205:Saccharomyces cerevisiae
4091:Poly(A)-binding protein
3745:Genes & Development
3703:10.1073/pnas.2209766119
3211:10.1023/A:1024193323397
3125:10.1126/science.1080559
2998:10.1126/science.1948046
2612:Nature Reviews Genetics
2574:10.1073/pnas.0912629107
2517:10.1126/science.1180660
2334:Nature Reviews Genetics
1988:10.1073/pnas.0813128106
1811:(22): 4898–4910 e4894.
499:that are vulnerable to
426:nonsense-mediated decay
229:gene containing a ≥3.6
94:Discovery and etymology
3858:10.1073/pnas.231494498
3649:10.1186/1745-6150-7-29
3536:10.1186/1745-6150-7-11
3267:10.1073/pnas.092595699
3240:Lynch M (April 2002).
2774:10.1002/bies.201200073
2712:Nucleic Acids Research
2196:10.1186/1741-7007-7-23
2146:10.1098/rstb.2015.0474
2026:Nucleic Acids Research
1919:Nucleic Acids Research
1071:10.1073/pnas.75.3.1485
887:10.1073/pnas.74.8.3171
686:"Why genes in pieces?"
397:
271:(spliceosomal introns)
219:
201:baker's/brewer's yeast
154:
3896:Nature Communications
2445:Frontiers in Genetics
2246:10.1101/gr.185371.114
1364:10.1093/dnares/dsv028
1130:10.1073/pnas.75.2.725
511:Starvation adaptation
392:
217:
209:mitochondrial genomes
173:intervening sequences
146:
4154:Alternative splicing
2624:10.1038/nrg.2016.119
2128:Skandalis A (2016).
1874:10.4161/rna.6.5.9860
659:Exon-intron database
590:Alternative splicing
473:Alternative splicing
422:Alternative splicing
307:Spliceosomal introns
285:that are removed by
267:that are removed by
207:). In contrast, the
181:untranslated regions
107:split or interrupted
3908:2011NatCo...2..454H
3849:2001PNAS...9813207D
3843:(23): 13207–13212.
3757:10.1101/gad.1905010
3586:1993Natur.361..170D
3476:2019Natur.565..612P
3258:2002PNAS...99.6118L
2869:2009JMolE..69..527P
2724:10.1093/nar/gkq1080
2565:2010PNAS..107..961L
2508:2009Sci...326..817R
1979:2009PNAS..106.1766F
1606:1992Natur.357..173R
1121:1978PNAS...75..725T
1062:1978PNAS...75.1485T
1019:The Idaho Statesman
878:1977PNAS...74.3171B
702:1978Natur.271..501G
148:"The notion of the
111:Phillip Allen Sharp
51:nucleotide sequence
34:. For the album by
4265:5′ cap methylation
3916:10.1038/ncomms1461
3336:Trends in Genetics
3301:Trends in Genetics
2908:Trends in Genetics
2346:10.1038/nrg.2015.3
2038:10.1093/nar/gkv763
1927:10.1093/nar/gkp471
986:American Scientist
245:ciliates, such as
220:
115:Richard J. Roberts
4310:
4309:
4251:
4250:
4247:
4246:
4164:pre-mRNA factors
3964:978-0-8153-4105-5
3580:(6408): 170–173.
3470:(7741): 612–617.
3433:(4): 608–621.e6.
2992:(5032): 663–664.
2767:(12): 1025–1034.
2032:(17): 8488–8501.
1600:(6374): 173–176.
684:(February 1978).
600:Minor spliceosome
428:and mRNA export.
300:Group III introns
248:Stentor coeruleus
197:jawed vertebrates
16:(Redirected from
4352:
4116:
4049:5′ cap formation
4037:
4017:
4010:
4003:
3994:
3938:
3937:
3927:
3887:
3881:
3880:
3870:
3860:
3828:
3822:
3821:
3785:
3779:
3778:
3768:
3732:
3726:
3725:
3715:
3705:
3681:
3672:
3671:
3661:
3651:
3627:
3614:
3613:
3594:10.1038/361170a0
3565:
3559:
3558:
3548:
3538:
3510:
3504:
3503:
3459:
3453:
3452:
3442:
3418:
3407:
3406:
3366:
3360:
3359:
3331:
3325:
3324:
3296:
3290:
3289:
3279:
3269:
3252:(9): 6118–6123.
3237:
3231:
3230:
3205:(2–3): 117–121.
3194:
3188:
3187:
3151:
3145:
3144:
3108:
3102:
3101:
3073:
3067:
3066:
3056:
3024:
3018:
3017:
2981:
2975:
2974:
2938:
2932:
2931:
2903:
2897:
2896:
2852:
2846:
2845:
2835:
2825:
2801:
2795:
2794:
2776:
2752:
2746:
2745:
2735:
2718:(6): 2357–2366.
2703:
2697:
2696:
2686:
2676:
2652:
2646:
2645:
2635:
2603:
2597:
2596:
2586:
2576:
2547:Lynch M (2010).
2544:
2538:
2537:
2519:
2487:
2481:
2480:
2470:
2460:
2436:
2430:
2429:
2411:
2401:
2377:
2368:
2367:
2357:
2325:
2319:
2318:
2308:
2298:
2274:
2268:
2267:
2257:
2225:
2219:
2218:
2208:
2198:
2174:
2168:
2167:
2157:
2125:
2116:
2115:
2105:
2095:
2086:(12): e1001236.
2071:
2060:
2059:
2049:
2017:
2011:
2010:
2000:
1990:
1973:(6): 1766–1771.
1958:
1949:
1948:
1938:
1910:
1904:
1903:
1885:
1853:
1847:
1846:
1828:
1796:
1790:
1789:
1753:
1747:
1746:
1710:
1704:
1703:
1675:
1669:
1668:
1640:
1634:
1633:
1614:10.1038/357173a0
1589:
1583:
1582:
1546:
1540:
1539:
1511:
1505:
1504:
1476:
1470:
1469:
1441:
1435:
1434:
1424:
1392:
1386:
1385:
1375:
1343:
1337:
1336:
1326:
1294:
1288:
1287:
1277:
1253:
1247:
1246:
1210:
1204:
1203:
1193:
1183:
1159:
1153:
1152:
1142:
1132:
1100:
1094:
1093:
1083:
1073:
1056:(3): 1485–1489.
1041:
1035:
1034:
1028:
1026:
1009:
1003:
1002:
1000:
998:
976:
970:
969:
962:
956:
955:
916:
910:
909:
899:
889:
872:(8): 3171–3175.
857:
851:
850:
814:
808:
807:
784:Lewin B (1987).
781:
775:
774:
738:
732:
731:
713:
711:10.1038/271501a0
677:
495:frequently form
325:
293:group II introns
21:
4360:
4359:
4355:
4354:
4353:
4351:
4350:
4349:
4325:Gene expression
4315:
4314:
4311:
4306:
4243:
4158:
4105:
4101:Polyuridylation
4054:Polyadenylation
4026:
4021:
3946:
3941:
3889:
3888:
3884:
3830:
3829:
3825:
3787:
3786:
3782:
3734:
3733:
3729:
3683:
3682:
3675:
3629:
3628:
3617:
3567:
3566:
3562:
3512:
3511:
3507:
3461:
3460:
3456:
3420:
3419:
3410:
3375:Nature Genetics
3368:
3367:
3363:
3333:
3332:
3328:
3298:
3297:
3293:
3239:
3238:
3234:
3196:
3195:
3191:
3168:10.1038/nrg1807
3153:
3152:
3148:
3110:
3109:
3105:
3075:
3074:
3070:
3026:
3025:
3021:
2983:
2982:
2978:
2943:Current Biology
2940:
2939:
2935:
2905:
2904:
2900:
2854:
2853:
2849:
2816:(4): e1001366.
2803:
2802:
2798:
2754:
2753:
2749:
2705:
2704:
2700:
2654:
2653:
2649:
2618:(12): 758–772.
2605:
2604:
2600:
2546:
2545:
2541:
2489:
2488:
2484:
2438:
2437:
2433:
2379:
2378:
2371:
2327:
2326:
2322:
2276:
2275:
2271:
2234:Genome Research
2227:
2226:
2222:
2176:
2175:
2171:
2140:(1713): 34–38.
2127:
2126:
2119:
2073:
2072:
2063:
2019:
2018:
2014:
1960:
1959:
1952:
1912:
1911:
1907:
1855:
1854:
1850:
1805:Current Biology
1798:
1797:
1793:
1755:
1754:
1750:
1712:
1711:
1707:
1677:
1676:
1672:
1642:
1641:
1637:
1591:
1590:
1586:
1548:
1547:
1543:
1513:
1512:
1508:
1478:
1477:
1473:
1452:(12): 467–471.
1443:
1442:
1438:
1401:Current Biology
1394:
1393:
1389:
1345:
1344:
1340:
1296:
1295:
1291:
1255:
1254:
1250:
1212:
1211:
1207:
1161:
1160:
1156:
1102:
1101:
1097:
1043:
1042:
1038:
1024:
1022:
1011:
1010:
1006:
996:
994:
978:
977:
973:
964:
963:
959:
918:
917:
913:
859:
858:
854:
816:
815:
811:
796:
783:
782:
778:
740:
739:
735:
680:
678:
671:
667:
554:
521:
513:
414:
379:
351:
349:Group II intron
341:
332:
319:
315:
309:
283:group I introns
257:
193:
160:also refers to
96:
88:noncoding genes
43:
40:Introns (album)
36:LCD Soundsystem
28:
23:
22:
15:
12:
11:
5:
4358:
4356:
4348:
4347:
4345:Non-coding DNA
4342:
4337:
4332:
4327:
4317:
4316:
4308:
4307:
4305:
4304:
4303:
4302:
4297:
4292:
4287:
4282:
4277:
4270:mRNA decapping
4267:
4261:
4259:
4253:
4252:
4249:
4248:
4245:
4244:
4242:
4241:
4240:
4239:
4234:
4229:
4224:
4219:
4214:
4209:
4204:
4199:
4194:
4189:
4184:
4179:
4168:
4166:
4160:
4159:
4157:
4156:
4151:
4150:
4149:
4144:
4134:
4129:
4119:
4113:
4107:
4106:
4104:
4103:
4098:
4093:
4088:
4087:
4086:
4081:
4076:
4071:
4066:
4061:
4051:
4046:
4044:Precursor mRNA
4040:
4034:
4028:
4027:
4022:
4020:
4019:
4012:
4005:
3997:
3991:
3990:
3985:
3980:
3970:
3952:
3945:
3944:External links
3942:
3940:
3939:
3882:
3823:
3796:(2): 207–210.
3780:
3751:(8): 827–836.
3739:(April 2010).
3727:
3673:
3636:Biology Direct
3615:
3560:
3523:Biology Direct
3505:
3454:
3427:Molecular Cell
3408:
3381:(4): 415–418.
3361:
3342:(8): 375–378.
3326:
3291:
3232:
3189:
3162:(3): 211–221.
3146:
3119:(5624): 1393.
3103:
3068:
3039:(6): a016071.
3019:
2976:
2949:(3): 145–146.
2933:
2914:(5): 145–148.
2898:
2863:(5): 527–540.
2847:
2796:
2747:
2698:
2647:
2598:
2559:(3): 961–968.
2539:
2482:
2431:
2369:
2320:
2283:Genome Biology
2269:
2240:(6): 884–896.
2220:
2169:
2117:
2061:
2012:
1950:
1905:
1868:(5): 526–530.
1848:
1791:
1748:
1705:
1670:
1635:
1584:
1557:(2): 537–546.
1541:
1506:
1471:
1436:
1407:(4): 569–575.
1387:
1358:(6): 495–503.
1338:
1309:(2): 759–769.
1289:
1268:(5): 703–709.
1248:
1221:(2): 103–123.
1205:
1168:Genome Biology
1154:
1115:(2): 725–729.
1095:
1036:
1031:Newspapers.com
1004:
971:
957:
930:(2): 375–382.
911:
852:
809:
794:
776:
749:(3): 681–693.
733:
668:
666:
663:
662:
661:
656:
651:
646:
641:
636:
631:
629:Exon shuffling
620:
619:
608:
607:
602:
597:
592:
581:
580:
578:Small t intron
575:
570:
565:
553:
550:
520:
517:
512:
509:
467:common descent
413:
410:
378:
375:
340:
337:
331:
328:
308:
305:
297:
296:
291:Self-splicing
289:
281:Self-splicing
279:
272:
256:
255:Classification
253:
192:
189:
142:Walter Gilbert
95:
92:
26:
24:
18:Non-gene locus
14:
13:
10:
9:
6:
4:
3:
2:
4357:
4346:
4343:
4341:
4338:
4336:
4333:
4331:
4328:
4326:
4323:
4322:
4320:
4313:
4301:
4298:
4296:
4293:
4291:
4288:
4286:
4283:
4281:
4278:
4276:
4273:
4272:
4271:
4268:
4266:
4263:
4262:
4260:
4258:
4254:
4238:
4235:
4233:
4230:
4228:
4225:
4223:
4220:
4218:
4215:
4213:
4210:
4208:
4205:
4203:
4200:
4198:
4195:
4193:
4190:
4188:
4185:
4183:
4180:
4178:
4175:
4174:
4173:
4170:
4169:
4167:
4165:
4161:
4155:
4152:
4148:
4145:
4143:
4140:
4139:
4138:
4135:
4133:
4130:
4128:
4124:
4121:
4120:
4117:
4114:
4112:
4108:
4102:
4099:
4097:
4094:
4092:
4089:
4085:
4082:
4080:
4077:
4075:
4072:
4070:
4067:
4065:
4062:
4060:
4057:
4056:
4055:
4052:
4050:
4047:
4045:
4042:
4041:
4038:
4035:
4033:
4029:
4025:
4018:
4013:
4011:
4006:
4004:
3999:
3998:
3995:
3989:
3986:
3984:
3981:
3979:
3975:
3971:
3969:
3965:
3961:
3957:
3953:
3951:
3948:
3947:
3943:
3935:
3931:
3926:
3921:
3917:
3913:
3909:
3905:
3901:
3897:
3893:
3886:
3883:
3878:
3874:
3869:
3864:
3859:
3854:
3850:
3846:
3842:
3838:
3834:
3827:
3824:
3819:
3815:
3811:
3807:
3803:
3799:
3795:
3791:
3784:
3781:
3776:
3772:
3767:
3762:
3758:
3754:
3750:
3746:
3742:
3738:
3731:
3728:
3723:
3719:
3714:
3709:
3704:
3699:
3695:
3691:
3687:
3680:
3678:
3674:
3669:
3665:
3660:
3655:
3650:
3645:
3641:
3637:
3633:
3626:
3624:
3622:
3620:
3616:
3611:
3607:
3603:
3599:
3595:
3591:
3587:
3583:
3579:
3575:
3571:
3564:
3561:
3556:
3552:
3547:
3542:
3537:
3532:
3528:
3524:
3520:
3516:
3509:
3506:
3501:
3497:
3493:
3489:
3485:
3481:
3477:
3473:
3469:
3465:
3458:
3455:
3450:
3446:
3441:
3436:
3432:
3428:
3424:
3417:
3415:
3413:
3409:
3404:
3400:
3396:
3392:
3388:
3387:10.1038/ng940
3384:
3380:
3376:
3372:
3365:
3362:
3357:
3353:
3349:
3345:
3341:
3337:
3330:
3327:
3322:
3318:
3314:
3310:
3306:
3302:
3295:
3292:
3287:
3283:
3278:
3273:
3268:
3263:
3259:
3255:
3251:
3247:
3243:
3236:
3233:
3228:
3224:
3220:
3216:
3212:
3208:
3204:
3200:
3193:
3190:
3185:
3181:
3177:
3173:
3169:
3165:
3161:
3157:
3150:
3147:
3142:
3138:
3134:
3130:
3126:
3122:
3118:
3114:
3107:
3104:
3099:
3095:
3091:
3087:
3083:
3079:
3072:
3069:
3064:
3060:
3055:
3050:
3046:
3042:
3038:
3034:
3030:
3023:
3020:
3015:
3011:
3007:
3003:
2999:
2995:
2991:
2987:
2980:
2977:
2972:
2968:
2964:
2960:
2956:
2952:
2948:
2944:
2937:
2934:
2929:
2925:
2921:
2917:
2913:
2909:
2902:
2899:
2894:
2890:
2886:
2882:
2878:
2874:
2870:
2866:
2862:
2858:
2851:
2848:
2843:
2839:
2834:
2829:
2824:
2819:
2815:
2811:
2810:PLOS Genetics
2807:
2800:
2797:
2792:
2788:
2784:
2780:
2775:
2770:
2766:
2762:
2758:
2751:
2748:
2743:
2739:
2734:
2729:
2725:
2721:
2717:
2713:
2709:
2702:
2699:
2694:
2690:
2685:
2680:
2675:
2670:
2666:
2662:
2658:
2651:
2648:
2643:
2639:
2634:
2629:
2625:
2621:
2617:
2613:
2609:
2602:
2599:
2594:
2590:
2585:
2580:
2575:
2570:
2566:
2562:
2558:
2554:
2550:
2543:
2540:
2535:
2531:
2527:
2523:
2518:
2513:
2509:
2505:
2502:(5954): 817.
2501:
2497:
2493:
2486:
2483:
2478:
2474:
2469:
2464:
2459:
2454:
2450:
2446:
2442:
2435:
2432:
2427:
2423:
2419:
2415:
2410:
2405:
2400:
2395:
2391:
2387:
2386:F1000Research
2383:
2376:
2374:
2370:
2365:
2361:
2356:
2351:
2347:
2343:
2339:
2335:
2331:
2324:
2321:
2316:
2312:
2307:
2302:
2297:
2292:
2288:
2284:
2280:
2273:
2270:
2265:
2261:
2256:
2251:
2247:
2243:
2239:
2235:
2231:
2224:
2221:
2216:
2212:
2207:
2202:
2197:
2192:
2188:
2184:
2180:
2173:
2170:
2165:
2161:
2156:
2151:
2147:
2143:
2139:
2135:
2131:
2124:
2122:
2118:
2113:
2109:
2104:
2099:
2094:
2089:
2085:
2081:
2077:
2070:
2068:
2066:
2062:
2057:
2053:
2048:
2043:
2039:
2035:
2031:
2027:
2023:
2016:
2013:
2008:
2004:
1999:
1994:
1989:
1984:
1980:
1976:
1972:
1968:
1964:
1957:
1955:
1951:
1946:
1942:
1937:
1932:
1928:
1924:
1920:
1916:
1909:
1906:
1901:
1897:
1893:
1889:
1884:
1879:
1875:
1871:
1867:
1863:
1859:
1852:
1849:
1844:
1840:
1836:
1832:
1827:
1822:
1818:
1814:
1810:
1806:
1802:
1795:
1792:
1787:
1783:
1779:
1775:
1771:
1767:
1763:
1759:
1752:
1749:
1744:
1740:
1736:
1732:
1728:
1724:
1720:
1716:
1709:
1706:
1701:
1697:
1693:
1689:
1685:
1681:
1674:
1671:
1666:
1662:
1658:
1654:
1650:
1646:
1639:
1636:
1631:
1627:
1623:
1619:
1615:
1611:
1607:
1603:
1599:
1595:
1588:
1585:
1580:
1576:
1572:
1568:
1564:
1560:
1556:
1552:
1545:
1542:
1537:
1533:
1529:
1525:
1521:
1517:
1510:
1507:
1502:
1498:
1494:
1490:
1487:: 1119–1150.
1486:
1482:
1475:
1472:
1467:
1463:
1459:
1455:
1451:
1447:
1440:
1437:
1432:
1428:
1423:
1418:
1414:
1410:
1406:
1402:
1398:
1391:
1388:
1383:
1379:
1374:
1369:
1365:
1361:
1357:
1353:
1349:
1342:
1339:
1334:
1330:
1325:
1320:
1316:
1312:
1308:
1304:
1300:
1293:
1290:
1285:
1281:
1276:
1271:
1267:
1263:
1259:
1252:
1249:
1244:
1240:
1236:
1232:
1228:
1224:
1220:
1216:
1209:
1206:
1201:
1197:
1192:
1187:
1182:
1177:
1173:
1169:
1165:
1158:
1155:
1150:
1146:
1141:
1136:
1131:
1126:
1122:
1118:
1114:
1110:
1106:
1099:
1096:
1091:
1087:
1082:
1077:
1072:
1067:
1063:
1059:
1055:
1051:
1047:
1040:
1037:
1032:
1021:
1020:
1015:
1008:
1005:
992:
988:
987:
982:
975:
972:
967:
961:
958:
953:
949:
945:
941:
937:
933:
929:
925:
921:
915:
912:
907:
903:
898:
893:
888:
883:
879:
875:
871:
867:
863:
856:
853:
848:
844:
840:
836:
832:
828:
824:
820:
813:
810:
805:
801:
797:
795:0-471-83278-2
791:
787:
780:
777:
772:
768:
764:
760:
756:
752:
748:
744:
737:
734:
729:
725:
721:
717:
712:
707:
703:
699:
696:(5645): 501.
695:
691:
687:
683:
676:
674:
670:
664:
660:
657:
655:
652:
650:
647:
645:
644:Noncoding RNA
642:
640:
639:Noncoding DNA
637:
635:
632:
630:
627:
626:
625:
624:
618:
615:
614:
613:
612:
606:
603:
601:
598:
596:
593:
591:
588:
587:
586:
585:
579:
576:
574:
571:
569:
566:
564:
561:
560:
559:
558:
551:
549:
545:
541:
537:
533:
529:
526:
518:
516:
510:
508:
506:
505:recombination
502:
498:
494:
489:
487:
483:
477:
474:
470:
468:
464:
463:
457:
454:
449:
447:
443:
439:
435:
429:
427:
423:
419:
418:noncoding RNA
411:
409:
405:
401:
396:
391:
387:
383:
376:
374:
372:
371:self-splicing
368:
364:
363:ribosomal RNA
360:
356:
355:messenger RNA
350:
346:
338:
336:
329:
327:
323:
314:
306:
304:
301:
294:
290:
288:
287:RNA catalysis
284:
280:
277:
273:
270:
266:
263:
262:
261:
254:
252:
250:
249:
244:
240:
236:
232:
228:
226:
216:
212:
210:
206:
202:
198:
190:
188:
186:
182:
178:
174:
169:
167:
163:
159:
153:
151:
145:
143:
140:
136:
132:
128:
124:
120:
116:
112:
108:
103:
101:
93:
91:
89:
84:
82:
78:
74:
70:
69:
65:
60:
56:
52:
48:
41:
37:
33:
19:
4340:RNA splicing
4312:
4125: /
4122:
4111:RNA splicing
3974:Biochemistry
3973:
3955:
3899:
3895:
3885:
3840:
3836:
3826:
3793:
3789:
3783:
3748:
3744:
3730:
3693:
3689:
3639:
3635:
3577:
3573:
3563:
3526:
3522:
3513:Rogozin IB,
3508:
3467:
3463:
3457:
3430:
3426:
3378:
3374:
3364:
3339:
3335:
3329:
3307:(1): 16–22.
3304:
3300:
3294:
3249:
3245:
3235:
3202:
3198:
3192:
3159:
3155:
3149:
3116:
3112:
3106:
3081:
3077:
3071:
3036:
3032:
3022:
2989:
2985:
2979:
2946:
2942:
2936:
2911:
2907:
2901:
2860:
2856:
2850:
2813:
2809:
2799:
2764:
2760:
2750:
2715:
2711:
2701:
2664:
2661:BMC Genomics
2660:
2650:
2615:
2611:
2601:
2556:
2552:
2542:
2499:
2495:
2485:
2448:
2444:
2434:
2389:
2385:
2340:(1): 19–32.
2337:
2333:
2323:
2286:
2282:
2272:
2237:
2233:
2223:
2186:
2182:
2172:
2137:
2133:
2083:
2079:
2029:
2025:
2015:
1970:
1966:
1918:
1908:
1865:
1861:
1851:
1808:
1804:
1794:
1761:
1757:
1751:
1718:
1714:
1708:
1683:
1679:
1673:
1648:
1644:
1638:
1597:
1593:
1587:
1554:
1550:
1544:
1519:
1515:
1509:
1484:
1480:
1474:
1449:
1445:
1439:
1404:
1400:
1390:
1355:
1352:DNA Research
1351:
1341:
1306:
1302:
1292:
1265:
1261:
1251:
1218:
1214:
1208:
1174:(10): R223.
1171:
1167:
1157:
1112:
1108:
1098:
1053:
1049:
1039:
1029:– via
1023:. Retrieved
1017:
1007:
995:. Retrieved
993:(5): 298–305
990:
984:
974:
960:
927:
923:
914:
869:
865:
855:
822:
818:
812:
785:
779:
746:
742:
736:
693:
689:
622:
621:
610:
609:
583:
582:
556:
555:
546:
542:
538:
534:
530:
522:
514:
490:
478:
471:
460:
450:
441:
430:
415:
406:
402:
398:
393:
388:
384:
380:
370:
359:transfer RNA
352:
333:
330:tRNA introns
321:
316:
298:
276:transfer RNA
269:spliceosomes
258:
246:
238:
223:
221:
204:
194:
191:Distribution
172:
170:
162:intracistron
161:
157:
155:
147:
134:
127:Susan Berget
104:
97:
85:
72:
67:
63:
62:
58:
46:
44:
4335:Spliceosome
4137:Spliceosome
4096:RNA editing
2183:BMC Biology
1862:RNA Biology
1764:: 359–388.
1721:: 333–358.
1686:: 435–461.
1651:: 543–568.
1522:: 387–419.
649:Selfish DNA
525:orthologous
482:spliceosome
434:spliceosome
420:molecules.
243:heterotrich
185:RNA editing
133:. The term
131:Louise Chow
77:transcripts
66:agenic regi
4319:Categories
3696:(48): 48.
2667:(1): 637.
2289:(1): 208.
2080:PLOS Genet
1025:12 January
997:12 January
825:(1): 1–8.
665:References
557:Structure:
501:DNA damage
453:eukaryotic
343:See also:
311:See also:
225:Drosophila
139:biochemist
100:adenovirus
4257:Cytosolic
3737:Belfort M
3084:: 47–76.
2761:BioEssays
2534:206522975
2426:202702147
1843:237603074
1786:208626110
1743:209167227
920:Belfort M
682:Gilbert W
584:Splicing:
438:RNA world
156:The term
53:within a
3958:, 2007,
3934:21878908
3877:11687644
3818:11652546
3775:20351053
3722:36417430
3668:22963364
3555:22507701
3515:Carmel L
3500:58014466
3492:30651641
3449:28757210
3395:12134150
3356:18586348
3321:16290250
3286:11983904
3219:12868602
3199:Genetica
3184:33672491
3176:16485020
3133:12775832
3098:17094737
3063:24890509
2971:35790897
2963:15336149
2893:22386774
2885:19777149
2842:21533221
2783:23108796
2742:21071396
2693:30153812
2642:27773922
2593:20080596
2526:19815722
2477:32211018
2418:31275557
2392:: 1908.
2364:26593421
2315:29084568
2264:25883323
2215:19442261
2164:27994117
2112:21151575
2056:26261211
2007:19179398
1945:19546110
1900:22592978
1892:19829058
1835:34555349
1778:31794245
1735:31815536
1579:44978152
1431:28190732
1382:26581719
1333:10655227
1303:Genetics
1284:11114327
1243:19229072
1235:10076021
1200:17949488
952:27127017
804:14069165
771:21897383
654:Twintron
617:MicroRNA
611:Function
552:See also
235:metazoan
231:megabase
123:excluded
32:Intron A
4237:PRPF40B
4232:PRPF40A
4222:PRPF38B
4217:PRPF38A
4032:Nuclear
3925:3265369
3904:Bibcode
3902:: 454.
3845:Bibcode
3810:2417724
3766:2854396
3713:9860276
3659:3443670
3610:4364102
3602:8380627
3582:Bibcode
3546:3488318
3472:Bibcode
3403:9057609
3254:Bibcode
3227:7539892
3141:7235937
3113:Science
3054:4031966
3006:1948046
2986:Science
2928:2068786
2865:Bibcode
2833:3077370
2791:5808466
2733:3064772
2684:6114036
2633:5876476
2584:2824313
2561:Bibcode
2504:Bibcode
2496:Science
2468:7066660
2451:: 109.
2409:6544075
2355:5993438
2306:5663052
2255:4448684
2206:2697156
2155:5182408
2103:3000347
2047:4787815
1998:2644112
1975:Bibcode
1936:2724286
1883:3912188
1826:8967684
1700:7574489
1665:2197983
1630:4370160
1622:1579169
1602:Bibcode
1571:6297798
1536:2977088
1501:2943217
1466:8108859
1422:5659724
1373:4675715
1324:1460963
1191:2246297
1117:Bibcode
1058:Bibcode
944:3986907
874:Bibcode
847:2099968
728:4216649
698:Bibcode
623:Others:
595:Exitron
497:R-loops
488:(IME).
456:genomes
177:inteins
166:cistron
150:cistron
49:is any
4227:PRPF39
4212:PRPF31
4207:PRPF19
4202:PRPF18
4187:PRPF4B
4123:Intron
3962:
3932:
3922:
3875:
3865:
3816:
3808:
3773:
3763:
3720:
3710:
3666:
3656:
3642:: 29.
3608:
3600:
3574:Nature
3553:
3543:
3529:: 11.
3498:
3490:
3464:Nature
3447:
3401:
3393:
3354:
3319:
3284:
3277:122912
3274:
3225:
3217:
3182:
3174:
3139:
3131:
3096:
3061:
3051:
3014:508870
3012:
3004:
2969:
2961:
2926:
2891:
2883:
2840:
2830:
2789:
2781:
2740:
2730:
2691:
2681:
2640:
2630:
2591:
2581:
2532:
2524:
2475:
2465:
2424:
2416:
2406:
2362:
2352:
2313:
2303:
2262:
2252:
2213:
2203:
2189:: 23.
2162:
2152:
2110:
2100:
2054:
2044:
2005:
1995:
1943:
1933:
1898:
1890:
1880:
1841:
1833:
1823:
1784:
1776:
1741:
1733:
1698:
1663:
1628:
1620:
1594:Nature
1577:
1569:
1534:
1499:
1464:
1429:
1419:
1380:
1370:
1331:
1321:
1282:
1241:
1233:
1198:
1188:
1149:273235
1147:
1140:411329
1137:
1090:418414
1088:
1081:411497
1078:
950:
942:
906:269380
904:
897:431482
894:
845:
839:902310
837:
802:
792:
769:
763:688388
761:
726:
720:622185
718:
690:Nature
634:Intein
605:Outron
446:snRNAs
322:lariat
158:intron
135:intron
73:intron
59:intron
47:intron
38:, see
4280:DCP1B
4275:DCP1A
4197:PRPF8
4192:PRPF6
4182:PRPF4
4177:PRPF3
4172:PLRG1
4142:minor
4132:snRNP
3868:60849
3814:S2CID
3606:S2CID
3496:S2CID
3399:S2CID
3223:S2CID
3180:S2CID
3137:S2CID
3010:S2CID
2967:S2CID
2889:S2CID
2787:S2CID
2530:S2CID
2422:S2CID
1896:S2CID
1839:S2CID
1782:S2CID
1739:S2CID
1626:S2CID
1575:S2CID
1239:S2CID
948:S2CID
843:S2CID
786:Genes
767:S2CID
724:S2CID
442:trans
239:MST1L
81:exons
4300:EDC4
4295:EDC3
4290:DCPS
4285:DCP2
4127:Exon
4084:CFII
4074:PAB2
4064:CstF
4059:CPSF
3978:link
3968:link
3960:ISBN
3930:PMID
3873:PMID
3806:PMID
3790:Cell
3771:PMID
3718:PMID
3690:PNAS
3664:PMID
3598:PMID
3551:PMID
3488:PMID
3445:PMID
3391:PMID
3352:PMID
3317:PMID
3282:PMID
3215:PMID
3172:PMID
3129:PMID
3094:PMID
3059:PMID
3002:PMID
2959:PMID
2924:PMID
2881:PMID
2838:PMID
2779:PMID
2738:PMID
2689:PMID
2638:PMID
2589:PMID
2522:PMID
2473:PMID
2414:PMID
2360:PMID
2311:PMID
2260:PMID
2211:PMID
2160:PMID
2108:PMID
2052:PMID
2003:PMID
1941:PMID
1888:PMID
1831:PMID
1774:PMID
1731:PMID
1696:PMID
1661:PMID
1618:PMID
1567:PMID
1551:Cell
1532:PMID
1497:PMID
1462:PMID
1427:PMID
1378:PMID
1329:PMID
1280:PMID
1262:Cell
1231:PMID
1219:1410
1196:PMID
1145:PMID
1086:PMID
1027:2024
999:2024
940:PMID
924:Cell
902:PMID
835:PMID
819:Cell
800:OCLC
790:ISBN
759:PMID
743:Cell
716:PMID
568:mRNA
563:Exon
361:and
347:and
227:dhc7
129:and
113:and
64:intr
55:gene
4330:DNA
4079:CFI
4069:PAP
3920:PMC
3912:doi
3863:PMC
3853:doi
3798:doi
3761:PMC
3753:doi
3708:PMC
3698:doi
3694:119
3654:PMC
3644:doi
3590:doi
3578:361
3541:PMC
3531:doi
3480:doi
3468:565
3435:doi
3383:doi
3344:doi
3309:doi
3272:PMC
3262:doi
3207:doi
3203:118
3164:doi
3121:doi
3117:300
3086:doi
3049:PMC
3041:doi
2994:doi
2990:254
2951:doi
2916:doi
2873:doi
2828:PMC
2818:doi
2769:doi
2728:PMC
2720:doi
2679:PMC
2669:doi
2628:PMC
2620:doi
2579:PMC
2569:doi
2557:107
2512:doi
2500:326
2463:PMC
2453:doi
2404:PMC
2394:doi
2350:PMC
2342:doi
2301:PMC
2291:doi
2250:PMC
2242:doi
2201:PMC
2191:doi
2150:PMC
2142:doi
2138:784
2098:PMC
2088:doi
2042:PMC
2034:doi
1993:PMC
1983:doi
1971:106
1931:PMC
1923:doi
1878:PMC
1870:doi
1821:PMC
1813:doi
1766:doi
1723:doi
1688:doi
1653:doi
1610:doi
1598:357
1559:doi
1524:doi
1489:doi
1454:doi
1417:PMC
1409:doi
1368:PMC
1360:doi
1319:PMC
1311:doi
1307:154
1270:doi
1266:103
1223:doi
1186:PMC
1176:doi
1135:PMC
1125:doi
1076:PMC
1066:doi
991:108
932:doi
892:PMC
882:doi
827:doi
751:doi
706:doi
694:271
493:DNA
357:),
45:An
4321::
4147:U1
3928:.
3918:.
3910:.
3898:.
3894:.
3871:.
3861:.
3851:.
3841:98
3839:.
3835:.
3812:.
3804:.
3794:44
3792:.
3769:.
3759:.
3749:24
3747:.
3743:.
3716:.
3706:.
3692:.
3688:.
3676:^
3662:.
3652:.
3638:.
3634:.
3618:^
3604:.
3596:.
3588:.
3576:.
3572:.
3549:.
3539:.
3525:.
3521:.
3494:.
3486:.
3478:.
3466:.
3443:.
3431:67
3429:.
3425:.
3411:^
3397:.
3389:.
3379:31
3377:.
3373:.
3350:.
3340:24
3338:.
3315:.
3305:22
3303:.
3280:.
3270:.
3260:.
3250:99
3248:.
3244:.
3221:.
3213:.
3201:.
3178:.
3170:.
3158:.
3135:.
3127:.
3115:.
3092:.
3082:40
3080:.
3057:.
3047:.
3035:.
3031:.
3008:.
3000:.
2988:.
2965:.
2957:.
2945:.
2922:.
2910:.
2887:.
2879:.
2871:.
2861:69
2859:.
2836:.
2826:.
2812:.
2808:.
2785:.
2777:.
2765:34
2763:.
2759:.
2736:.
2726:.
2716:39
2714:.
2710:.
2687:.
2677:.
2665:19
2663:.
2659:.
2636:.
2626:.
2616:17
2614:.
2610:.
2587:.
2577:.
2567:.
2555:.
2551:.
2528:.
2520:.
2510:.
2498:.
2494:.
2471:.
2461:.
2449:11
2447:.
2443:.
2420:.
2412:.
2402:.
2388:.
2384:.
2372:^
2358:.
2348:.
2338:17
2336:.
2332:.
2309:.
2299:.
2287:18
2285:.
2281:.
2258:.
2248:.
2238:25
2236:.
2232:.
2209:.
2199:.
2185:.
2181:.
2158:.
2148:.
2136:.
2132:.
2120:^
2106:.
2096:.
2082:.
2078:.
2064:^
2050:.
2040:.
2030:43
2028:.
2024:.
2001:.
1991:.
1981:.
1969:.
1965:.
1953:^
1939:.
1929:.
1917:.
1894:.
1886:.
1876:.
1864:.
1860:.
1837:.
1829:.
1819:.
1809:31
1807:.
1803:.
1780:.
1772:.
1762:89
1760:.
1737:.
1729:.
1719:89
1717:.
1694:.
1684:64
1682:.
1659:.
1649:59
1647:.
1624:.
1616:.
1608:.
1596:.
1573:.
1565:.
1555:32
1553:.
1530:.
1520:22
1518:.
1495:.
1485:55
1483:.
1460:.
1450:18
1448:.
1425:.
1415:.
1405:27
1403:.
1399:.
1376:.
1366:.
1356:22
1354:.
1350:.
1327:.
1317:.
1305:.
1301:.
1278:.
1264:.
1260:.
1237:.
1229:.
1217:.
1194:.
1184:.
1170:.
1166:.
1143:.
1133:.
1123:.
1113:75
1111:.
1107:.
1084:.
1074:.
1064:.
1054:75
1052:.
1048:.
1016:.
989:.
983:.
946:.
938:.
928:41
926:.
900:.
890:.
880:.
870:74
868:.
864:.
841:.
833:.
823:12
821:.
798:.
765:.
757:.
747:14
745:.
722:.
714:.
704:.
692:.
688:.
672:^
179:,
168:.
144::
83:.
68:on
4016:e
4009:t
4002:v
3936:.
3914::
3906::
3900:2
3879:.
3855::
3847::
3820:.
3800::
3777:.
3755::
3724:.
3700::
3670:.
3646::
3640:7
3612:.
3592::
3584::
3557:.
3533::
3527:7
3502:.
3482::
3474::
3451:.
3437::
3405:.
3385::
3358:.
3346::
3323:.
3311::
3288:.
3264::
3256::
3229:.
3209::
3186:.
3166::
3160:7
3143:.
3123::
3100:.
3088::
3065:.
3043::
3037:6
3016:.
2996::
2973:.
2953::
2947:1
2930:.
2918::
2912:7
2895:.
2875::
2867::
2844:.
2820::
2814:7
2793:.
2771::
2744:.
2722::
2695:.
2671::
2644:.
2622::
2595:.
2571::
2563::
2536:.
2514::
2506::
2479:.
2455::
2428:.
2396::
2390:7
2366:.
2344::
2317:.
2293::
2266:.
2244::
2217:.
2193::
2187:7
2166:.
2144::
2114:.
2090::
2084:6
2058:.
2036::
2009:.
1985::
1977::
1947:.
1925::
1902:.
1872::
1866:6
1845:.
1815::
1788:.
1768::
1745:.
1725::
1702:.
1690::
1667:.
1655::
1632:.
1612::
1604::
1581:.
1561::
1538:.
1526::
1503:.
1491::
1468:.
1456::
1433:.
1411::
1384:.
1362::
1335:.
1313::
1286:.
1272::
1245:.
1225::
1202:.
1178::
1172:8
1151:.
1127::
1119::
1092:.
1068::
1060::
1033:.
1001:.
968:.
954:.
934::
908:.
884::
876::
849:.
829::
806:.
773:.
753::
730:.
708::
700::
324:)
320:(
203:(
42:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.