399:
694:
6050:
927:(3 of each), arranged like segments of an orange around a rotating asymmetrical gamma subunit. According to the current model of ATP synthesis (known as the alternating catalytic model), the transmembrane potential created by (H+) proton cations supplied by the electron transport chain, drives the (H+) proton cations from the intermembrane space through the membrane via the F
871:
40:
437:
457:
1189:
994:
The binding change mechanism involves the active site of a β subunit's cycling between three states. In the "loose" state, ADP and phosphate enter the active site; in the adjacent diagram, this is shown in pink. The enzyme then undergoes a change in shape and forces these molecules together, with the
1124:
region shows significant structural similarity to hexameric DNA helicases; both form a ring with 3-fold rotational symmetry with a central pore. Both have roles dependent on the relative rotation of a macromolecule within the pore; the DNA helicases use the helical shape of DNA to drive their motion
1069:
of ATP synthase is thought to have been modular whereby two functionally independent subunits became associated and gained new functionality. This association appears to have occurred early in evolutionary history, because essentially the same structure and activity of ATP synthase enzymes are
1258:
A variety of natural and synthetic inhibitors of ATP synthase have been discovered. These have been used to probe the structure and mechanism of ATP synthase. Some may be of therapeutic use. There are several classes of ATP synthase inhibitors, including peptide inhibitors, polyphenolic
1358:) heart mitochondria is, in terms of biochemistry and structure, the best-characterized ATP synthase. Beef heart is used as a source for the enzyme because of the high concentration of mitochondria in cardiac muscle. Their genes have close homology to human ATP synthases.
543:
subunits γ, δ, and ε are a part of a rotational motor mechanism (rotor/axle). The γ subunit allows β to go through conformational changes (i.e., closed, half open, and open states) that allow for ATP to be bound and released once synthesized. The
911:
catalytic-domain of ATP synthase. The structure, at the time the largest asymmetric protein structure known, indicated that Boyer's rotary-catalysis model was, in essence, correct. For elucidating this, Boyer and Walker shared half of the 1997
1216:
motor, were able to bind, and the rotation of the motor drove the ATPase activity of the helicase in reverse. This complex then evolved greater efficiency and eventually developed into today's intricate ATP synthases. Alternatively, the DNA
1259:
phytochemicals, polyketides, organotin compounds, polyenic α-pyrone derivatives, cationic inhibitors, substrate analogs, amino acid modifiers, and other miscellaneous chemicals. Some of the most commonly used ATP synthase inhibitors are
894:
Professor, developed the binding change, or flip-flop, mechanism theory, which postulated that ATP synthesis is dependent on a conformational change in ATP synthase generated by rotation of the gamma subunit. The research group of
1192:
1196:
1195:
1191:
1190:
1197:
859:
733:
region of ATP synthase is a proton pore that is embedded in the mitochondrial membrane. It consists of three main subunits, a, b, and c. Six c subunits make up the rotor ring, and subunit b makes up a stalk connecting to
1194:
1902:"A macromolecular repeating unit of mitochondrial structure and function. Correlated electron microscopic and biochemical studies of isolated mitochondria and submitochondrial particles of beef heart muscle"
1366:
1342:) and ATP synthesis take place. The overall structure and the catalytic mechanism of the chloroplast ATP synthase are almost the same as those of the bacterial enzyme. However, in chloroplasts, the
2572:
Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezón E, Champagne E, et al. (January 2003). "Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis".
2005:
Carbajo RJ, Silvester JA, Runswick MJ, Walker JE, Neuhaus D (September 2004). "Solution structure of subunit F(6) from the peripheral stalk region of ATP synthase from bovine heart mitochondria".
1346:
is generated not by respiratory electron transport chain but by primary photosynthetic proteins. The synthase has a 40-aa insert in the gamma-subunit to inhibit wasteful activity when dark.
548:
particle is large and can be seen in the transmission electron microscope by negative staining. These are particles of 9 nm diameter that pepper the inner mitochondrial membrane.
1694:"Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase"
4877:
3256:
Matzke NJ, Lin A, Stone M, Baker MA (July 2021). "Flagellar export apparatus and ATP synthetase: Homology evidenced by synteny predating the Last
Universal Common Ancestor".
390:
unit of ATP synthase. These functional regions consist of different protein subunits — refer to tables. This enzyme is used in synthesis of ATP through aerobic respiration.
979:(cryo-EM) studies of the complex. The cryo-EM model of ATP synthase suggests that the peripheral stalk is a flexible structure that wraps around the complex as it joins F
4072:
209:
762:(or A6L). This part of the enzyme is located in the mitochondrial inner membrane and couples proton translocation to the rotation that causes ATP synthesis in the F
3346:
2380:
999:. Finally, the active site cycles back to the open state (orange), releasing ATP and binding more ADP and phosphate, ready for the next cycle of ATP production.
1290:
Bacterial F-ATPases can occasionally operate in reverse, turning them into an ATPase. Some bacteria have no F-ATPase, using an A/V-type ATPase bidirectionally.
1022:, this is used by fermenting bacteria that do not have an electron transport chain, but rather hydrolyze ATP to make a proton gradient, which they use to drive
228:
1521:
F-ATPase gene linkage and gene order are widely conserved across ancient prokaryote lineages, implying that this system already existed at a date before the
1193:
1205:
The modular evolution theory for the origin of ATP synthase suggests that two subunits with independent function, a DNA helicase with ATPase activity and a
1173:
potential gradient as an energy source. This link is tenuous, however, as the overall structure of flagellar motors is far more complex than that of the F
4553:
3360:
to Paul D. Boyer and John E. Walker for the enzymatic mechanism of synthesis of ATP; and to Jens C. Skou, for discovery of an ion-transporting enzyme,
3144:"Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis"
2220:
adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model"
5570:
4548:
3312:
721:
protein that goes through conformational changes when protonated and deprotonated, pushing neighboring subunits to rotate, causing the spinning of F
4639:
4221:
2821:"Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex"
4870:
1509:
Archaea do not generally have an F-ATPase. Instead, they synthesize ATP using the A-ATPase/synthase, a rotary machine structurally similar to the
1162:
motors that drive flagella. Both feature a ring of many small alpha-helical proteins that rotate relative to nearby stationary proteins, using a
5100:
3099:
Gibbons C, Montgomery MG, Leslie AG, Walker JE (November 2000). "The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution".
4307:
1580:
940:
891:
5069:
4116:
3837:
4065:
959:
causing the 3 catalytic nucleotide binding sites to go through a series of conformational changes that lead to ATP synthesis. The major F
6090:
3451:
3048:
Abrahams JP, Leslie AG, Lutter R, Walker JE (August 1994). "Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria".
3833:
4863:
4407:
4342:
3322:
947:
is tightly attached to the asymmetric central stalk (consisting primarily of the gamma subunit), causing it to rotate within the alpha
632:
5769:
4352:
4177:
4678:
4476:
4111:
3426:
1264:
649:
594:
572:
221:
5660:
4845:
4673:
4337:
4058:
2682:"The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio"
900:
666:
1861:"Organisation of the yeast ATP synthase F(0):a study based on cysteine mutants, thiol modification and cross-linking reagents"
5655:
4668:
4454:
4412:
3404:
2734:"ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas"
1522:
148:
5925:
172:
398:
6075:
5677:
3395:
3385:
1306:
subunits, and seven associated proteins have been identified. Most of these proteins have homologues in other eukaryotes.
738:
OSCP that prevents the αβ hexamer from rotating. Subunit a connects b to the c ring. Humans have six additional subunits,
299:
can cross from areas of high concentration to areas of low concentration, imparting energy for the synthesis of ATP. This
6040:
536:
space. Subunits α and β make a hexamer with 6 binding sites. Three of them are catalytically inactive and they bind ADP.
5725:
5042:
4258:
4200:
4090:
963:
subunits are prevented from rotating in sympathy with the central stalk rotor by a peripheral stalk that joins the alpha
469:
320:
1033:
under physiological conditions, ATP synthase, in general, runs in the opposite direction, creating ATP while using the
866:(pink) shown being combined into ATP (red), while the rotating γ (gamma) subunit in black causes conformational change.
5647:
4312:
2512:"Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator"
1513:
but mainly functioning as an ATP synthase. Like the bacteria F-ATPase, it is believed to also function as an ATPase.
1177:
particle and the ring with about 30 rotating proteins is far larger than the 10, 11, or 14 helical proteins in the F
6095:
4486:
4481:
4285:
4151:
3435:
616:
5910:
1181:
complex. More recent structural data do however show that the ring and the stalk are structurally similar to the F
987:. Under the right conditions, the enzyme reaction can also be carried out in reverse, with ATP hydrolysis driving
6026:
6013:
6000:
5987:
5974:
5961:
5948:
5706:
5247:
5145:
4970:
4966:
4943:
4916:
4903:
4890:
4211:
4040:
3800:
3795:
3357:
1565:
1343:
1042:
1034:
995:
active site in the resulting "tight" state (shown in red) binding the newly produced ATP molecule with very high
879:
5920:
5874:
5817:
5261:
4956:
4921:
4894:
4402:
4280:
1070:
present in all kingdoms of life. The F-ATP synthase displays high functional and mechanistic similarity to the
976:
913:
304:
300:
166:
59:
2950:"The purification and characterization of ATP synthase complexes from the mitochondria of four fungal species"
1133:
hexamer uses the conformational changes through the rotation of the γ subunit to drive an enzymatic reaction.
2913:
Stock D, Leslie AG, Walker JE (November 1999). "Molecular architecture of the rotary motor in ATP synthase".
6080:
5822:
5631:
5594:
4933:
4449:
4236:
1550:
1038:
153:
5361:
5001:
4444:
4101:
3893:
944:
936:
826:
253:
5843:
5762:
5491:
5399:
4818:
4558:
4434:
4270:
4248:
996:
257:
233:
45:
5915:
1477:
Eukaryotes belonging to some divergent lineages have very special organizations of the ATP synthase. A
141:
1250:
motor in reverse. This may have evolved to carry out the reverse reaction and act as an ATP synthase.
4659:
4565:
4422:
4371:
4317:
3419:
3057:
2581:
2523:
2328:
2170:
2107:
1585:
1054:
533:
382:(written as a subscript letter "o", not "zero") derives its name from being the binding fraction for
76:
3928:
5879:
5696:
2460:"Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading"
71:
3236:
169:
5812:
5711:
5277:
4855:
4589:
4357:
4231:
3291:
3173:
3124:
3081:
2711:
2662:
2605:
2489:
465:
328:
296:
93:
2948:
Liu S, Charlesworth TJ, Bason JV, Montgomery MG, Harbour ME, Fearnley IM, Walker JE (May 2015).
6085:
4951:
4579:
4382:
4163:
3318:
3283:
3224:
3165:
3116:
3073:
3030:
2979:
2930:
2895:
2842:
2801:
2763:
2703:
2654:
2597:
2551:
2481:
2440:
2354:
2290:
2241:
2196:
2135:
2076:
2022:
1995:
1977:
1931:
1882:
1859:
Velours J, Paumard P, Soubannier V, Spannagel C, Vaillier J, Arselin G, Graves PV (May 2000).
1811:
1768:
1750:
1715:
1674:
1633:
1015:
ATP synthase is reversible. Large-enough quantities of ATP cause it to create a transmembrane
410:, axle, and stator regions are color coded magenta, green, orange, and cyan respectively i.e.
269:
160:
4027:
4022:
4007:
3695:
2784:
Kühlbrandt W, Davies KM (January 2016). "Rotary ATPases: A New Twist to an
Ancient Machine".
1338:, where dark reactions of photosynthesis (also called the light-independent reactions or the
6070:
5858:
5853:
5827:
5755:
5341:
5287:
5269:
5135:
5125:
5079:
4392:
4185:
3991:
3985:
3980:
3958:
3948:
3938:
3905:
3787:
3782:
3772:
3553:
3493:
3273:
3265:
3214:
3204:
3155:
3108:
3065:
3020:
3010:
2969:
2961:
2922:
2885:
2877:
2832:
2793:
2753:
2745:
2693:
2644:
2589:
2541:
2531:
2471:
2430:
2419:"The V-type H+ ATPase: molecular structure and function, physiological roles and regulation"
2344:
2336:
2280:
2272:
2231:
2186:
2178:
2125:
2115:
2096:"Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows"
2066:
2056:
2014:
1967:
1921:
1913:
1872:
1838:
1801:
1791:
1742:
1705:
1664:
1623:
718:
5154:
3969:
3809:
3804:
2376:
1778:
Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, et al. (October 2015).
5905:
5889:
5802:
5688:
5577:
5391:
5091:
4911:
4297:
4190:
3917:
3743:
3439:
3412:
3399:
1669:
1652:
1498:
924:
773:
forms membrane-bending dimers. These dimers self-arrange into long rows at the end of the
312:
129:
1780:"Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM"
1733:
Mccarty RE (November 1992). "A PLANT BIOCHEMIST'S VIEW OF H+-ATPases AND ATP SYNTHASES".
1287:
ATP synthase is the simplest known form of ATP synthase, with 8 different subunit types.
3061:
2585:
2527:
2332:
2174:
2111:
1078:
generates a proton gradient at the expense of ATP, generating pH values of as low as 1.
1074:. However, whereas the F-ATP synthase generates ATP by utilising a proton gradient, the
1041:
as a source of energy. The overall process of creating energy in this fashion is termed
358:. This article deals mainly with this type. An F-ATPase consists of two main subunits, F
105:
6054:
5943:
5884:
5011:
3841:
3825:
3352:
3342:
3219:
3192:
3025:
2998:
2974:
2949:
2890:
2861:
2758:
2733:
2546:
2511:
2399:
2349:
2316:
2285:
2260:
2191:
2154:
2130:
2095:
2071:
2044:
1926:
1901:
1806:
1779:
1335:
896:
777:, possibly the first step of cristae formation. An atomic model for the dimeric yeast F
324:
204:
64:
3160:
3143:
2862:"Novel features of the rotary catalytic mechanism revealed in the structure of yeast F
2837:
2820:
2476:
2459:
2340:
2236:
2215:
1877:
1860:
1710:
1693:
693:
184:
6064:
5848:
5807:
5555:
5165:
4081:
3921:
3733:
3295:
2649:
2624:
1560:
1535:
1057:. By pumping proton cations into the matrix, the ATP-synthase converts ADP into ATP.
975:. The structure of the intact ATP synthase is currently known at low-resolution from
887:
340:
179:
3128:
2715:
2666:
5797:
5540:
5513:
5442:
5435:
5418:
5413:
5387:
4714:
4226:
4139:
3177:
3085:
2609:
2493:
1842:
1555:
1339:
1315:
1086:
1046:
875:
2926:
2698:
2681:
2045:"Role of Charged Residues in the Catalytic Sites of Escherichia coli ATP Synthase"
1298:
Yeast ATP synthase is one of the best-studied eukaryotic ATP synthases; and five F
3353:
Proton and Sodium translocating F-type, V-type and A-type ATPases in OPM database
2625:"Gene duplication as a means for altering H+/ATP ratios during the evolution of F
6021:
5956:
5792:
5636:
1575:
1545:
1490:
988:
525:
336:
188:
6049:
3392:
2797:
2516:
Proceedings of the
National Academy of Sciences of the United States of America
2100:
Proceedings of the
National Academy of Sciences of the United States of America
5621:
5310:
4466:
4128:
2276:
2018:
1972:
1955:
1478:
1260:
1090:
383:
308:
2881:
1049:, where ATP synthase is located in the inner mitochondrial membrane and the F
17:
5995:
5969:
5327:
4886:
4397:
3015:
2536:
2182:
2120:
1327:
1109:
1066:
904:
710:
344:
316:
261:
3287:
3269:
3228:
3169:
3120:
3034:
2983:
2934:
2899:
2860:
Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM (November 2006).
2846:
2805:
2767:
2707:
2601:
2555:
2485:
2444:
2294:
2200:
2139:
2080:
2026:
1981:
1935:
1886:
1815:
1678:
1637:
3077:
2749:
2658:
2358:
2245:
2061:
1754:
1746:
1719:
870:
39:
4543:
4417:
4050:
3901:
3700:
3690:
3685:
3670:
3665:
3660:
3650:
3645:
3640:
3630:
3625:
3615:
3610:
3605:
3600:
3581:
3573:
3469:
3193:"Structure of a mitochondrial ATP synthase with bound native cardiolipin"
1590:
1570:
1510:
1075:
1071:
1030:
1023:
1019:
714:
351:
3209:
2593:
1917:
1796:
1628:
1611:
456:
436:
295:
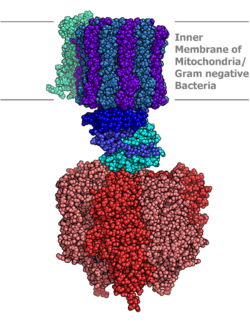
ATP synthase lies across a cellular membrane and forms an aperture that
5732:
5701:
5479:
5474:
5233:
5223:
5198:
5193:
4928:
4703:
4698:
4618:
4017:
4012:
3680:
3675:
3655:
3635:
3620:
3595:
3590:
3585:
2965:
1465:
1459:
806:
774:
759:
136:
117:
3314:
The Vital
Question: Energy, Evolution, and the Origins of Complex Life
3278:
2999:"Structure, mechanism, and regulation of the chloroplast ATP synthase"
2435:
2418:
1999:
1772:
386:, a type of naturally derived antibiotic that is able to inhibit the F
366:, which has a rotational motor mechanism allowing for ATP production.
6008:
5778:
5609:
5251:
5228:
5218:
5188:
5183:
5178:
5173:
5130:
5120:
5115:
5110:
5105:
5074:
5062:
5057:
5052:
5047:
5035:
5030:
5025:
4974:
4808:
4803:
4773:
4768:
4648:
4628:
4623:
4613:
4608:
4603:
4598:
4001:
3996:
3975:
3953:
3943:
3880:
3875:
3870:
3865:
3860:
3855:
3850:
3845:
3817:
3777:
3767:
3762:
3757:
3752:
3747:
3717:
3707:
3543:
3523:
3518:
3513:
3508:
3488:
3478:
3443:
3069:
1440:
1416:
1410:
1404:
1398:
1380:
1016:
844:
838:
832:
818:
747:
713:
protein with eight subunits and a transmembrane ring. The ring has a
622:
606:
584:
578:
355:
332:
249:
216:
112:
100:
88:
781:
region was determined by cryo-EM at an overall resolution of 3.6 Å.
729:, resulting in switching of states of alpha and beta subunits. The F
1239:
pump activity with the ATPase activity of the helicase driving the
5982:
5665:
5604:
5587:
5582:
5430:
5408:
5374:
5369:
5354:
5332:
5320:
5315:
5305:
5300:
5295:
5020:
4996:
4828:
4823:
4813:
4798:
4793:
4788:
4783:
4778:
4763:
4758:
4753:
4748:
4743:
4738:
4733:
4728:
4723:
4519:
4506:
3964:
3888:
3829:
3563:
3558:
3548:
3538:
3533:
3528:
3503:
3498:
3483:
3112:
2510:
Imada K, Minamino T, Uchida Y, Kinoshita M, Namba K (March 2016).
1830:
1452:
1446:
1434:
1428:
1422:
1392:
1386:
1373:
1187:
858:
857:
755:
751:
743:
739:
672:
655:
638:
600:
5670:
5626:
5614:
5599:
5565:
5560:
5545:
5528:
5523:
5518:
5506:
5501:
5496:
5486:
5462:
5457:
5452:
5447:
5349:
3932:
2094:
Blum TB, Hahn A, Meier T, Davies KM, Kühlbrandt W (March 2019).
1125:
along the DNA molecule and to detect supercoiling, whereas the α
1105:
124:
5751:
4859:
4054:
3408:
3336:
5720:
5716:
2997:
Hahn A, Vonck J, Mills DJ, Meier T, Kühlbrandt W (May 2018).
701:
subunit F6 from the peripheral stalk region of ATP synthase.
539:
Three other subunits catalyze the ATP synthesis. The other F
508:
creates a pathway for protons movement across the membrane.
252:
that catalyzes the formation of the energy storage molecule
2259:
Nakamoto RK, Baylis
Scanlon JA, Al-Shawi MK (August 2008).
2216:"Catalytic site cooperativity of beef heart mitochondrial F
307:
and allows cells to store energy in ATP for later use. In
5747:
1900:
Fernandez Moran H, Oda T, Blair PV, Green DE (July 1964).
644:
Mitochondrial "delta" is bacterial/chloroplastic epsilon.
378:
fraction derives its name from the term "Fraction 1" and F
1093:), and the entire enzyme region shows some similarity to
3325:(Link points to Figure 10 showing model of ATP synthase)
2819:
Devenish RJ, Prescott M, Roucou X, Nagley P (May 2000).
678:
Called "delta" in bacterial and chloroplastic versions.
1954:
Stewart AG, Laming EM, Sobti M, Stock D (April 2014).
1485:
head like other mitochondrial ATP synthases, but the F
1085:
region also shows significant similarity to hexameric
6038:
1361:
Human genes that encode components of ATP synthases:
1201:
Conformation changes of ATP synthase during synthesis
272:. The overall reaction catalyzed by ATP synthase is:
2394:
2392:
2390:
1481:
ATP synthase forms a dimer with a boomerang-shaped F
5934:
5898:
5867:
5836:
5785:
5686:
5645:
5385:
5259:
5246:
5211:
5163:
5144:
5090:
5010:
4989:
4982:
4965:
4942:
4902:
4712:
4687:
4657:
4637:
4587:
4578:
4532:
4495:
4463:
4431:
4379:
4370:
4326:
4294:
4267:
4245:
4208:
4199:
4176:
4148:
4125:
4098:
4089:
3732:
3716:
3572:
3468:
3459:
2825:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
1865:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
500:
on the other hand has mainly hydrophobic regions. F
227:
215:
203:
198:
178:
159:
147:
135:
123:
111:
99:
87:
82:
70:
58:
53:
32:
2315:Doering C, Ermentrout B, Oster G (December 1995).
1151:particle shows great functional similarity to the
496:has a water-soluble part that can hydrolyze ATP. F
488:and is made of c-ring and subunits a, two b, F6. F
3191:Mühleip A, McComas SE, Amunts A (November 2019).
2370:
2368:
2153:Guo H, Bueler SA, Rubinstein JL (November 2017).
452:. Subunits of the enzyme are labeled accordingly.
2779:
2777:
2214:Gresser MJ, Myers JA, Boyer PD (October 1982).
878:proton gradient to power ATP synthesis through
1949:
1947:
1945:
1854:
1852:
1326:-ATP synthase). The enzyme is integrated into
1026:and the transport of nutrients into the cell.
943:as the protons pass through the membrane. The
44:Molecular model of ATP synthase determined by
5763:
4871:
4066:
3420:
3337:"ATP synthase — a splendid molecular machine"
3142:Menz RI, Walker JE, Leslie AG (August 2001).
2727:
2725:
2567:
2565:
1612:"Rotation and structure of FoF1-ATP synthase"
1497:also binds differently, in a way shared with
1489:subcomplex has many unique subunits. It uses
8:
2310:
2308:
2306:
2304:
1956:"Rotary ATPases--dynamic molecular machines"
3386:Harvard Multimedia Production Site — Videos
2417:Beyenbach KW, Wieczorek H (February 2006).
1542:sector of the mitochondrial ATPase complex.
1314:In plants, ATP synthase is also present in
5770:
5756:
5748:
5256:
4986:
4979:
4878:
4864:
4856:
4584:
4554:Mitochondrial permeability transition pore
4536:
4376:
4205:
4095:
4073:
4059:
4051:
3465:
3427:
3413:
3405:
3347:University of Illinois at Urbana–Champaign
2738:Microbiology and Molecular Biology Reviews
2381:University of Illinois at Urbana–Champaign
2261:"The rotary mechanism of the ATP synthase"
931:region of ATP synthase. A portion of the F
528:and responsible for hydrolyzing ATP. The F
195:
3816:3.A.3.1.4: H/K transporting, nongastric:
3277:
3218:
3208:
3159:
3024:
3014:
2973:
2889:
2836:
2757:
2697:
2648:
2545:
2535:
2475:
2434:
2348:
2284:
2235:
2190:
2129:
2119:
2070:
2060:
1971:
1925:
1876:
1805:
1795:
1709:
1668:
1627:
725:which then also affects conformation of F
4549:Mitochondrial membrane transport protein
2038:
2036:
869:
783:
692:
550:
472:, ATP synthase consists of two regions F
455:
435:
402:Bovine mitochondrial ATP synthase. The F
397:
6045:
2505:
2503:
2265:Archives of Biochemistry and Biophysics
2043:Ahmad Z, Okafor F, Laughlin TF (2011).
1602:
1354:The ATP synthase isolated from bovine (
4308:Cholesterol side-chain cleavage enzyme
2458:Skordalakes E, Berger JM (July 2003).
1045:. The same process takes place in the
29:
3393:"ATP Synthase- Molecule of the Month"
2732:Hong S, Pedersen PL (December 2008).
2159:region of mitochondrial ATP synthase"
1960:Current Opinion in Structural Biology
1670:10.1146/annurev-biochem-060614-034124
1610:Okuno D, Iino R, Noji H (June 2011).
1581:Rotating locomotion in living systems
1007:Like other enzymes, the activity of F
492:is made of α, β, γ, and δ subunits. F
7:
874:Depiction of ATP synthase using the
862:Mechanism of ATP synthase. ADP and P
4222:Coenzyme Q – cytochrome c reductase
2680:Cross RL, Müller V (October 2004).
2423:The Journal of Experimental Biology
2224:The Journal of Biological Chemistry
1735:The Journal of Experimental Biology
1698:The Journal of Biological Chemistry
901:MRC Laboratory of Molecular Biology
4408:Oxoglutarate dehydrogenase complex
4343:Glycerol-3-phosphate dehydrogenase
1538:required for the assembly of the F
923:showed alternating alpha and beta
327:also have ATP synthase across the
25:
4353:Carnitine palmitoyltransferase II
3461:F-, V-, and A-type ATPase (3.A.2)
3358:The Nobel Prize in Chemistry 1997
2744:(4): 590–641, Table of Contents.
2623:Cross RL, Taiz L (January 1990).
6048:
4477:Carbamoyl phosphate synthetase I
4117:Long-chain-fatty-acid—CoA ligase
4112:Carnitine palmitoyltransferase I
971:to the non-rotating portion of F
886:In the 1960s through the 1970s,
460:Rotation engine of ATP synthase.
38:
4338:Glutamate aspartate transporter
3477:H transporting, mitochondrial:
2155:"Atomic model for the dimeric F
1692:Kagawa Y, Racker E (May 1966).
1651:Junge W, Nelson N (June 2015).
4455:Pyruvate dehydrogenase complex
4413:Succinyl coenzyme A synthetase
2786:Trends in Biochemical Sciences
1523:last universal common ancestor
919:The crystal structure of the F
769:In eukaryotes, mitochondrial F
448:-ATPase alias ATP synthase of
354:, running "in reverse" for an
1:
3161:10.1016/s0092-8674(01)00452-4
2927:10.1126/science.286.5445.1700
2838:10.1016/S0005-2728(00)00092-X
2699:10.1016/j.febslet.2004.08.065
2477:10.1016/S0092-8674(03)00512-9
2341:10.1016/S0006-3495(95)80096-2
2237:10.1016/S0021-9258(18)33672-X
1878:10.1016/S0005-2728(00)00093-1
1711:10.1016/S0021-9258(18)96640-8
1657:Annual Review of Biochemistry
350:Eukaryotic ATP synthases are
311:ATP synthase lies across the
4259:Dihydroorotate dehydrogenase
2650:10.1016/0014-5793(90)80014-a
2007:Journal of Molecular Biology
1843:10.2210/rcsb_pdb/mom_2005_12
1829:Goodsell D (December 2005).
470:inner mitochondrial membrane
321:inner mitochondrial membrane
5648:Protein-synthesizing GTPase
4313:Steroid 11-beta-hydroxylase
1906:The Journal of Cell Biology
1228:motor complex may have had
524:portion of ATP synthase is
48:. Stator is not shown here.
6112:
6091:Integral membrane proteins
4487:N-Acetylglutamate synthase
4482:Ornithine transcarbamylase
4286:Glycerol phosphate shuttle
4152:monoamine neurotransmitter
3887:3.A.3.5: Cu transporting:
3436:Membrane transport protein
2798:10.1016/j.tibs.2015.10.006
5926:Michaelis–Menten kinetics
5707:Guanylate-binding protein
4891:acid anhydride hydrolases
4841:
4539:
4515:
4212:oxidative phosphorylation
4036:
3582:H transporting, lysosomal
3388:– ATP synthesis animation
3317:, Ww Norton, 2015-07-20,
3237:"Different from the rest"
3101:Nature Structural Biology
2377:"Lecture 10:ATP synthase"
2277:10.1016/j.abb.2008.05.004
2019:10.1016/j.jmb.2004.07.013
1973:10.1016/j.sbi.2013.11.013
1566:Oxidative phosphorylation
1043:oxidative phosphorylation
880:oxidative phosphorylation
194:
37:
5818:Diffusion-limited enzyme
5262:Heterotrimeric G protein
4957:Phosphoadenylylsulfatase
4403:Isocitrate dehydrogenase
4281:Malate-aspartate shuttle
3345:by Antony Crofts of the
2882:10.1038/sj.emboj.7601410
1053:-part projects into the
1039:electron transport chain
977:electron cryo-microscopy
914:Nobel Prize in Chemistry
661:Unique to mitochondria.
532:unit protrudes into the
305:electron transport chain
301:electrochemical gradient
4934:Thiamine-triphosphatase
4450:Glutamate dehydrogenase
4237:Succinate dehydrogenase
3016:10.1126/science.aat4318
2954:The Biochemical Journal
2537:10.1073/pnas.1524025113
2379:. Life Sciences at the
2183:10.1126/science.aao4815
2121:10.1073/pnas.1816556116
1616:Journal of Biochemistry
1551:Electron transfer chain
323:. Organisms capable of
4846:mitochondrial diseases
4445:Aspartate transaminase
4102:fatty acid degradation
3270:10.1002/bies.202100004
2633:ATPases and synthases"
2049:Journal of Amino Acids
1271:In different organisms
1202:
883:
867:
702:
461:
453:
433:
394:Structure and function
254:adenosine triphosphate
5911:Eadie–Hofstee diagram
5844:Allosteric regulation
5689:Polymerization motors
5400:Rho family of GTPases
4559:Mitochondrial carrier
4435:anaplerotic reactions
4271:mitochondrial shuttle
4249:pyrimidine metabolism
2750:10.1128/MMBR.00016-08
1835:Molecule of the Month
1747:10.1242/jeb.172.1.431
1200:
991:across the membrane.
873:
861:
696:
459:
440:Simplified model of F
439:
401:
268:). ATP synthase is a
258:adenosine diphosphate
46:X-ray crystallography
6076:Cellular respiration
5921:Lineweaver–Burk plot
4566:Translocator protein
4423:Malate dehydrogenase
4318:Aldosterone synthase
3343:ATP synthase lecture
3243:. December 24, 2019.
1586:Transmembrane ATPase
1055:mitochondrial matrix
907:, crystallized the F
534:mitochondrial matrix
484:causes rotation of F
303:is generated by the
260:(ADP) and inorganic
5697:dynamin superfamily
4178:Intermembrane space
3990:Class VI, type 11:
3210:10.7554/eLife.51179
3062:1994Natur.370..621A
2921:(5445): 1700–1705.
2594:10.1038/nature01250
2586:2003Natur.421...75M
2528:2016PNAS..113.3633I
2333:1995BpJ....69.2256D
2321:Biophysical Journal
2317:"Rotary DNA motors"
2230:(20): 12030–12038.
2175:2017Sci...358..936G
2112:2019PNAS..116.4250B
2062:10.4061/2011/785741
1918:10.1083/jcb.22.1.63
1797:10.7554/eLife.10180
1493:. The inhibitory IF
1344:proton motive force
1035:proton motive force
790:
557:
464:Located within the
319:it lies across the
5880:Enzyme superfamily
5813:Enzyme promiscuity
4533:Other/to be sorted
4498:alcohol metabolism
4358:Uncoupling protein
4232:NADH dehydrogenase
3974:Class V, type 10:
3963:Class II, type 9:
3398:2015-09-05 at the
3335:Boris A. Feniouk:
3009:(6389): eaat4318.
2966:10.1042/BJ20150197
1334:-part sticks into
1203:
1003:Physiological role
884:
868:
784:
703:
551:
466:thylakoid membrane
462:
454:
434:
343:is located in the
335:is located in the
329:thylakoid membrane
6096:Protein complexes
6036:
6035:
5745:
5744:
5741:
5740:
5242:
5241:
5207:
5206:
4952:Adenylylsulfatase
4853:
4852:
4837:
4836:
4580:Mitochondrial DNA
4574:
4573:
4528:
4527:
4383:citric acid cycle
4366:
4365:
4172:
4171:
4164:Monoamine oxidase
4048:
4047:
3937:Class I, type 8:
3744:Na/K transporting
3728:
3727:
3341:Well illustrated
3107:(11): 1055–1061.
3056:(6491): 621–628.
2876:(22): 5433–5442.
2522:(13): 3633–3638.
2436:10.1242/jeb.02014
2429:(Pt 4): 577–589.
2404:InterPro Database
2169:(6365): 936–940.
2106:(10): 4250–4255.
1741:(Pt 1): 431–441.
1704:(10): 2461–2466.
1629:10.1093/jb/mvr049
1198:
851:
850:
682:
681:
309:prokaryotic cells
270:molecular machine
243:
242:
239:
238:
142:metabolic pathway
16:(Redirected from
6103:
6053:
6052:
6044:
5916:Hanes–Woolf plot
5859:Enzyme activator
5854:Enzyme inhibitor
5828:Enzyme catalysis
5772:
5765:
5758:
5749:
5257:
4987:
4980:
4880:
4873:
4866:
4857:
4717:
4692:
4662:
4642:
4592:
4585:
4537:
4500:
4470:
4438:
4393:Citrate synthase
4386:
4377:
4331:
4301:
4274:
4252:
4215:
4206:
4186:Adenylate kinase
4157:
4133:
4105:
4096:
4075:
4068:
4061:
4052:
4041:ATPase disorders
3912:Other/ungrouped:
3466:
3429:
3422:
3415:
3406:
3391:David Goodsell:
3381:
3380:
3379:
3370:
3369:
3368:
3300:
3299:
3281:
3253:
3247:
3244:
3232:
3222:
3212:
3188:
3182:
3181:
3163:
3139:
3133:
3132:
3096:
3090:
3089:
3070:10.1038/370621a0
3045:
3039:
3038:
3028:
3018:
2994:
2988:
2987:
2977:
2945:
2939:
2938:
2910:
2904:
2903:
2893:
2870:The EMBO Journal
2857:
2851:
2850:
2840:
2831:(2–3): 428–442.
2816:
2810:
2809:
2781:
2772:
2771:
2761:
2729:
2720:
2719:
2701:
2677:
2671:
2670:
2652:
2620:
2614:
2613:
2569:
2560:
2559:
2549:
2539:
2507:
2498:
2497:
2479:
2455:
2449:
2448:
2438:
2414:
2408:
2407:
2396:
2385:
2384:
2372:
2363:
2362:
2352:
2327:(6): 2256–2267.
2312:
2299:
2298:
2288:
2256:
2250:
2249:
2239:
2211:
2205:
2204:
2194:
2150:
2144:
2143:
2133:
2123:
2091:
2085:
2084:
2074:
2064:
2040:
2031:
2030:
2002:
1992:
1986:
1985:
1975:
1951:
1940:
1939:
1929:
1897:
1891:
1890:
1880:
1871:(2–3): 443–456.
1856:
1847:
1846:
1826:
1820:
1819:
1809:
1799:
1775:
1765:
1759:
1758:
1730:
1724:
1723:
1713:
1689:
1683:
1682:
1672:
1648:
1642:
1641:
1631:
1607:
1517:LUCA and earlier
1473:Other eukaryotes
1330:membrane; the CF
1285:
1284:
1249:
1248:
1247:
1238:
1237:
1236:
1227:
1226:
1225:
1215:
1214:
1213:
1199:
1172:
1171:
1170:
1161:
1160:
1159:
1146:
1145:
1144:
1120:hexamer of the F
1112:complexes. The α
1103:
1102:
1101:
1089:(especially the
791:
719:helix-loop-helix
558:
431:
427:
423:
416:
317:eukaryotic cells
196:
42:
30:
21:
6111:
6110:
6106:
6105:
6104:
6102:
6101:
6100:
6061:
6060:
6059:
6047:
6039:
6037:
6032:
5944:Oxidoreductases
5930:
5906:Enzyme kinetics
5894:
5890:List of enzymes
5863:
5832:
5803:Catalytic triad
5781:
5776:
5746:
5737:
5682:
5641:
5392:Ras superfamily
5381:
5365:
5345:
5291:
5281:
5273:
5238:
5203:
5159:
5140:
5086:
5043:Plasma membrane
5006:
4961:
4938:
4912:Pyrophosphatase
4898:
4884:
4854:
4849:
4833:
4713:
4708:
4688:
4683:
4658:
4653:
4638:
4633:
4588:
4570:
4524:
4511:
4496:
4491:
4464:
4459:
4432:
4427:
4380:
4362:
4327:
4322:
4298:steroidogenesis
4295:
4290:
4268:
4263:
4246:
4241:
4209:
4195:
4191:Creatine kinase
4168:
4154:
4149:
4144:
4126:
4121:
4099:
4085:
4079:
4049:
4044:
4032:
3929:Mg transporting
3842:Ca transporting
3724:
3723:found in Archea
3712:
3568:
3455:
3433:
3400:Wayback Machine
3378:
3376:
3375:
3374:
3372:
3367:
3365:
3364:
3363:
3361:
3332:
3308:
3306:Further reading
3303:
3264:(7): e2100004.
3255:
3254:
3250:
3235:
3190:
3189:
3185:
3141:
3140:
3136:
3098:
3097:
3093:
3047:
3046:
3042:
2996:
2995:
2991:
2947:
2946:
2942:
2912:
2911:
2907:
2865:
2859:
2858:
2854:
2818:
2817:
2813:
2783:
2782:
2775:
2731:
2730:
2723:
2679:
2678:
2674:
2632:
2628:
2622:
2621:
2617:
2580:(6918): 75–79.
2571:
2570:
2563:
2509:
2508:
2501:
2457:
2456:
2452:
2416:
2415:
2411:
2398:
2397:
2388:
2374:
2373:
2366:
2314:
2313:
2302:
2258:
2257:
2253:
2219:
2213:
2212:
2208:
2158:
2152:
2151:
2147:
2093:
2092:
2088:
2042:
2041:
2034:
2004:
1994:
1993:
1989:
1953:
1952:
1943:
1899:
1898:
1894:
1858:
1857:
1850:
1828:
1827:
1823:
1777:
1767:
1766:
1762:
1732:
1731:
1727:
1691:
1690:
1686:
1650:
1649:
1645:
1609:
1608:
1604:
1600:
1595:
1541:
1531:
1519:
1507:
1499:trypanosomatida
1496:
1488:
1484:
1475:
1352:
1333:
1325:
1321:
1312:
1305:
1301:
1296:
1282:
1281:
1278:
1273:
1256:
1246:
1244:
1243:
1242:
1240:
1235:
1233:
1232:
1231:
1229:
1224:
1222:
1221:
1220:
1218:
1212:
1210:
1209:
1208:
1206:
1188:
1184:
1180:
1176:
1169:
1167:
1166:
1165:
1163:
1158:
1156:
1155:
1154:
1152:
1150:
1143:
1141:
1140:
1139:
1137:
1132:
1128:
1123:
1119:
1115:
1110:flagellar motor
1100:
1098:
1097:
1096:
1094:
1084:
1063:
1052:
1037:created by the
1014:
1010:
1005:
986:
982:
974:
970:
966:
962:
958:
954:
950:
934:
930:
922:
910:
865:
856:
789:-Main subunits
788:
780:
772:
765:
737:
732:
728:
724:
708:
700:
691:
688:
555:
547:
542:
531:
523:
518:
515:
507:
503:
499:
495:
491:
487:
483:
479:
475:
447:
443:
429:
425:
422:
418:
415:
411:
409:
405:
396:
389:
381:
377:
372:
365:
361:
313:plasma membrane
291:
287:
283:
279:
267:
49:
28:
23:
22:
15:
12:
11:
5:
6109:
6107:
6099:
6098:
6093:
6088:
6083:
6081:Photosynthesis
6078:
6073:
6063:
6062:
6058:
6057:
6034:
6033:
6031:
6030:
6017:
6004:
5991:
5978:
5965:
5952:
5938:
5936:
5932:
5931:
5929:
5928:
5923:
5918:
5913:
5908:
5902:
5900:
5896:
5895:
5893:
5892:
5887:
5882:
5877:
5871:
5869:
5868:Classification
5865:
5864:
5862:
5861:
5856:
5851:
5846:
5840:
5838:
5834:
5833:
5831:
5830:
5825:
5820:
5815:
5810:
5805:
5800:
5795:
5789:
5787:
5783:
5782:
5777:
5775:
5774:
5767:
5760:
5752:
5743:
5742:
5739:
5738:
5736:
5735:
5730:
5729:
5728:
5723:
5714:
5709:
5704:
5693:
5691:
5684:
5683:
5681:
5680:
5675:
5674:
5673:
5668:
5663:
5652:
5650:
5643:
5642:
5640:
5639:
5634:
5629:
5624:
5619:
5618:
5617:
5612:
5607:
5602:
5592:
5591:
5590:
5585:
5575:
5574:
5573:
5568:
5563:
5551:
5550:
5549:
5548:
5543:
5533:
5532:
5531:
5526:
5521:
5511:
5510:
5509:
5504:
5499:
5489:
5484:
5483:
5482:
5477:
5467:
5466:
5465:
5460:
5455:
5450:
5440:
5439:
5438:
5433:
5423:
5422:
5421:
5416:
5411:
5396:
5394:
5383:
5382:
5380:
5379:
5378:
5377:
5372:
5363:
5359:
5358:
5357:
5352:
5343:
5339:
5338:
5337:
5336:
5335:
5325:
5324:
5323:
5318:
5308:
5303:
5298:
5289:
5285:
5284:
5283:
5279:
5271:
5266:
5264:
5254:
5244:
5243:
5240:
5239:
5237:
5236:
5231:
5226:
5221:
5215:
5213:
5209:
5208:
5205:
5204:
5202:
5201:
5196:
5191:
5186:
5181:
5176:
5170:
5168:
5161:
5160:
5158:
5157:
5151:
5149:
5142:
5141:
5139:
5138:
5133:
5128:
5123:
5118:
5113:
5108:
5103:
5097:
5095:
5088:
5087:
5085:
5084:
5083:
5082:
5077:
5067:
5066:
5065:
5060:
5055:
5050:
5040:
5039:
5038:
5033:
5028:
5017:
5015:
5008:
5007:
5005:
5004:
4999:
4993:
4991:
4990:Cu++ (3.6.3.4)
4984:
4977:
4963:
4962:
4960:
4959:
4954:
4948:
4946:
4940:
4939:
4937:
4936:
4931:
4926:
4925:
4924:
4919:
4908:
4906:
4900:
4899:
4885:
4883:
4882:
4875:
4868:
4860:
4851:
4850:
4842:
4839:
4838:
4835:
4834:
4832:
4831:
4826:
4821:
4816:
4811:
4806:
4801:
4796:
4791:
4786:
4781:
4776:
4771:
4766:
4761:
4756:
4751:
4746:
4741:
4736:
4731:
4726:
4720:
4718:
4710:
4709:
4707:
4706:
4701:
4695:
4693:
4685:
4684:
4682:
4681:
4676:
4671:
4665:
4663:
4655:
4654:
4652:
4651:
4645:
4643:
4635:
4634:
4632:
4631:
4626:
4621:
4616:
4611:
4606:
4601:
4595:
4593:
4582:
4576:
4575:
4572:
4571:
4569:
4568:
4563:
4562:
4561:
4556:
4546:
4540:
4534:
4530:
4529:
4526:
4525:
4523:
4522:
4516:
4513:
4512:
4510:
4509:
4503:
4501:
4493:
4492:
4490:
4489:
4484:
4479:
4473:
4471:
4461:
4460:
4458:
4457:
4452:
4447:
4441:
4439:
4429:
4428:
4426:
4425:
4420:
4415:
4410:
4405:
4400:
4395:
4389:
4387:
4374:
4368:
4367:
4364:
4363:
4361:
4360:
4355:
4350:
4345:
4340:
4334:
4332:
4324:
4323:
4321:
4320:
4315:
4310:
4304:
4302:
4292:
4291:
4289:
4288:
4283:
4277:
4275:
4265:
4264:
4262:
4261:
4255:
4253:
4243:
4242:
4240:
4239:
4234:
4229:
4224:
4218:
4216:
4203:
4201:Inner membrane
4197:
4196:
4194:
4193:
4188:
4182:
4180:
4174:
4173:
4170:
4169:
4167:
4166:
4160:
4158:
4146:
4145:
4143:
4142:
4136:
4134:
4123:
4122:
4120:
4119:
4114:
4108:
4106:
4093:
4091:Outer membrane
4087:
4086:
4080:
4078:
4077:
4070:
4063:
4055:
4046:
4045:
4037:
4034:
4033:
4031:
4030:
4025:
4020:
4015:
4010:
4004:
3999:
3994:
3988:
3983:
3978:
3972:
3967:
3961:
3956:
3951:
3946:
3941:
3935:
3925:
3924:
3914:
3913:
3909:
3908:
3897:
3896:
3891:
3884:
3883:
3878:
3873:
3868:
3863:
3858:
3853:
3848:
3821:
3820:
3813:
3812:
3807:
3801:H/K exchanging
3798:
3791:
3790:
3785:
3780:
3775:
3770:
3765:
3760:
3755:
3750:
3739:
3737:
3730:
3729:
3726:
3725:
3722:
3720:
3714:
3713:
3711:
3710:
3704:
3703:
3698:
3693:
3688:
3683:
3678:
3673:
3668:
3663:
3658:
3653:
3648:
3643:
3638:
3633:
3628:
3623:
3618:
3613:
3608:
3603:
3598:
3593:
3588:
3578:
3576:
3570:
3569:
3567:
3566:
3561:
3556:
3551:
3546:
3541:
3536:
3531:
3526:
3521:
3516:
3511:
3506:
3501:
3496:
3491:
3486:
3481:
3474:
3472:
3463:
3457:
3456:
3434:
3432:
3431:
3424:
3417:
3409:
3403:
3402:
3389:
3383:
3377:
3366:
3355:
3350:
3339:
3331:
3330:External links
3328:
3327:
3326:
3323:978-0393088816
3307:
3304:
3302:
3301:
3248:
3246:
3245:
3183:
3154:(3): 331–341.
3134:
3091:
3040:
2989:
2960:(1): 167–175.
2940:
2905:
2863:
2852:
2811:
2792:(1): 106–116.
2773:
2721:
2672:
2643:(2): 227–229.
2630:
2626:
2615:
2561:
2499:
2470:(1): 135–146.
2450:
2409:
2400:"ATP Synthase"
2386:
2364:
2300:
2251:
2217:
2206:
2156:
2145:
2086:
2032:
2013:(2): 593–603.
1987:
1941:
1892:
1848:
1831:"ATP Synthase"
1821:
1760:
1725:
1684:
1653:"ATP synthase"
1643:
1622:(6): 655–664.
1601:
1599:
1596:
1594:
1593:
1588:
1583:
1578:
1573:
1568:
1563:
1558:
1553:
1548:
1543:
1539:
1532:
1530:
1527:
1518:
1515:
1506:
1503:
1494:
1486:
1482:
1474:
1471:
1470:
1469:
1456:
1377:
1370:
1351:
1348:
1331:
1323:
1319:
1311:
1308:
1303:
1299:
1295:
1292:
1277:
1274:
1272:
1269:
1255:
1252:
1245:
1234:
1223:
1211:
1182:
1178:
1174:
1168:
1157:
1148:
1147:motor of the F
1142:
1130:
1126:
1121:
1117:
1113:
1099:
1082:
1062:
1059:
1050:
1012:
1008:
1004:
1001:
989:proton pumping
984:
980:
972:
968:
964:
960:
956:
952:
948:
932:
928:
920:
908:
899:, then at the
897:John E. Walker
863:
855:
852:
849:
848:
829:
823:
822:
815:
811:
810:
803:
799:
798:
795:
786:
778:
770:
763:
735:
730:
726:
722:
706:
698:
690:
686:
683:
680:
679:
676:
669:
663:
662:
659:
652:
646:
645:
642:
635:
629:
628:
626:
619:
613:
612:
610:
597:
591:
590:
588:
575:
569:
568:
565:
562:
553:
545:
540:
529:
521:
517:
513:
510:
505:
501:
497:
493:
489:
485:
481:
477:
473:
445:
441:
420:
413:
407:
403:
395:
392:
387:
379:
375:
371:
368:
363:
359:
325:photosynthesis
293:
292:
289:
285:
281:
277:
265:
241:
240:
237:
236:
231:
225:
224:
219:
213:
212:
207:
201:
200:
192:
191:
182:
176:
175:
164:
157:
156:
151:
145:
144:
139:
133:
132:
127:
121:
120:
115:
109:
108:
103:
97:
96:
91:
85:
84:
80:
79:
74:
68:
67:
62:
56:
55:
51:
50:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
6108:
6097:
6094:
6092:
6089:
6087:
6084:
6082:
6079:
6077:
6074:
6072:
6069:
6068:
6066:
6056:
6051:
6046:
6042:
6028:
6024:
6023:
6018:
6015:
6011:
6010:
6005:
6002:
5998:
5997:
5992:
5989:
5985:
5984:
5979:
5976:
5972:
5971:
5966:
5963:
5959:
5958:
5953:
5950:
5946:
5945:
5940:
5939:
5937:
5933:
5927:
5924:
5922:
5919:
5917:
5914:
5912:
5909:
5907:
5904:
5903:
5901:
5897:
5891:
5888:
5886:
5885:Enzyme family
5883:
5881:
5878:
5876:
5873:
5872:
5870:
5866:
5860:
5857:
5855:
5852:
5850:
5849:Cooperativity
5847:
5845:
5842:
5841:
5839:
5835:
5829:
5826:
5824:
5821:
5819:
5816:
5814:
5811:
5809:
5808:Oxyanion hole
5806:
5804:
5801:
5799:
5796:
5794:
5791:
5790:
5788:
5784:
5780:
5773:
5768:
5766:
5761:
5759:
5754:
5753:
5750:
5734:
5731:
5727:
5724:
5722:
5718:
5715:
5713:
5710:
5708:
5705:
5703:
5700:
5699:
5698:
5695:
5694:
5692:
5690:
5685:
5679:
5676:
5672:
5669:
5667:
5664:
5662:
5659:
5658:
5657:
5654:
5653:
5651:
5649:
5644:
5638:
5635:
5633:
5630:
5628:
5625:
5623:
5620:
5616:
5613:
5611:
5608:
5606:
5603:
5601:
5598:
5597:
5596:
5593:
5589:
5586:
5584:
5581:
5580:
5579:
5576:
5572:
5569:
5567:
5564:
5562:
5559:
5558:
5557:
5553:
5552:
5547:
5544:
5542:
5539:
5538:
5537:
5534:
5530:
5527:
5525:
5522:
5520:
5517:
5516:
5515:
5512:
5508:
5505:
5503:
5500:
5498:
5495:
5494:
5493:
5490:
5488:
5485:
5481:
5478:
5476:
5473:
5472:
5471:
5468:
5464:
5461:
5459:
5456:
5454:
5451:
5449:
5446:
5445:
5444:
5441:
5437:
5434:
5432:
5429:
5428:
5427:
5424:
5420:
5417:
5415:
5412:
5410:
5407:
5406:
5405:
5401:
5398:
5397:
5395:
5393:
5389:
5384:
5376:
5373:
5371:
5368:
5367:
5366:
5360:
5356:
5353:
5351:
5348:
5347:
5346:
5340:
5334:
5331:
5330:
5329:
5326:
5322:
5319:
5317:
5314:
5313:
5312:
5309:
5307:
5304:
5302:
5299:
5297:
5294:
5293:
5292:
5286:
5282:
5276:
5275:
5274:
5268:
5267:
5265:
5263:
5258:
5255:
5253:
5249:
5245:
5235:
5232:
5230:
5227:
5225:
5222:
5220:
5217:
5216:
5214:
5210:
5200:
5197:
5195:
5192:
5190:
5187:
5185:
5182:
5180:
5177:
5175:
5172:
5171:
5169:
5167:
5166:P-type ATPase
5162:
5156:
5153:
5152:
5150:
5147:
5143:
5137:
5134:
5132:
5129:
5127:
5124:
5122:
5119:
5117:
5114:
5112:
5109:
5107:
5104:
5102:
5099:
5098:
5096:
5093:
5089:
5081:
5078:
5076:
5073:
5072:
5071:
5068:
5064:
5061:
5059:
5056:
5054:
5051:
5049:
5046:
5045:
5044:
5041:
5037:
5034:
5032:
5029:
5027:
5024:
5023:
5022:
5019:
5018:
5016:
5013:
5009:
5003:
5000:
4998:
4995:
4994:
4992:
4988:
4985:
4981:
4978:
4976:
4972:
4968:
4964:
4958:
4955:
4953:
4950:
4949:
4947:
4945:
4941:
4935:
4932:
4930:
4927:
4923:
4920:
4918:
4915:
4914:
4913:
4910:
4909:
4907:
4905:
4901:
4896:
4892:
4888:
4881:
4876:
4874:
4869:
4867:
4862:
4861:
4858:
4848:
4847:
4840:
4830:
4827:
4825:
4822:
4820:
4817:
4815:
4812:
4810:
4807:
4805:
4802:
4800:
4797:
4795:
4792:
4790:
4787:
4785:
4782:
4780:
4777:
4775:
4772:
4770:
4767:
4765:
4762:
4760:
4757:
4755:
4752:
4750:
4747:
4745:
4742:
4740:
4737:
4735:
4732:
4730:
4727:
4725:
4722:
4721:
4719:
4716:
4711:
4705:
4702:
4700:
4697:
4696:
4694:
4691:
4686:
4680:
4677:
4675:
4672:
4670:
4667:
4666:
4664:
4661:
4656:
4650:
4647:
4646:
4644:
4641:
4636:
4630:
4627:
4625:
4622:
4620:
4617:
4615:
4612:
4610:
4607:
4605:
4602:
4600:
4597:
4596:
4594:
4591:
4586:
4583:
4581:
4577:
4567:
4564:
4560:
4557:
4555:
4552:
4551:
4550:
4547:
4545:
4542:
4541:
4538:
4535:
4531:
4521:
4518:
4517:
4514:
4508:
4505:
4504:
4502:
4499:
4494:
4488:
4485:
4483:
4480:
4478:
4475:
4474:
4472:
4469:
4468:
4462:
4456:
4453:
4451:
4448:
4446:
4443:
4442:
4440:
4437:
4436:
4430:
4424:
4421:
4419:
4416:
4414:
4411:
4409:
4406:
4404:
4401:
4399:
4396:
4394:
4391:
4390:
4388:
4385:
4384:
4378:
4375:
4373:
4369:
4359:
4356:
4354:
4351:
4349:
4346:
4344:
4341:
4339:
4336:
4335:
4333:
4330:
4325:
4319:
4316:
4314:
4311:
4309:
4306:
4305:
4303:
4300:
4299:
4293:
4287:
4284:
4282:
4279:
4278:
4276:
4273:
4272:
4266:
4260:
4257:
4256:
4254:
4251:
4250:
4244:
4238:
4235:
4233:
4230:
4228:
4225:
4223:
4220:
4219:
4217:
4214:
4213:
4207:
4204:
4202:
4198:
4192:
4189:
4187:
4184:
4183:
4181:
4179:
4175:
4165:
4162:
4161:
4159:
4156:
4153:
4147:
4141:
4138:
4137:
4135:
4132:
4130:
4124:
4118:
4115:
4113:
4110:
4109:
4107:
4104:
4103:
4097:
4094:
4092:
4088:
4083:
4082:Mitochondrial
4076:
4071:
4069:
4064:
4062:
4057:
4056:
4053:
4043:
4042:
4035:
4029:
4026:
4024:
4021:
4019:
4016:
4014:
4011:
4009:
4005:
4003:
4000:
3998:
3995:
3993:
3989:
3987:
3984:
3982:
3979:
3977:
3973:
3971:
3968:
3966:
3962:
3960:
3957:
3955:
3952:
3950:
3947:
3945:
3942:
3940:
3936:
3934:
3930:
3927:
3926:
3923:
3919:
3916:
3915:
3911:
3910:
3907:
3903:
3899:
3898:
3895:
3892:
3890:
3886:
3885:
3882:
3879:
3877:
3874:
3872:
3869:
3867:
3864:
3862:
3859:
3857:
3854:
3852:
3849:
3847:
3843:
3839:
3835:
3831:
3827:
3823:
3822:
3819:
3815:
3814:
3811:
3808:
3806:
3802:
3799:
3797:
3793:
3792:
3789:
3786:
3784:
3781:
3779:
3776:
3774:
3771:
3769:
3766:
3764:
3761:
3759:
3756:
3754:
3751:
3749:
3745:
3741:
3740:
3738:
3735:
3734:P-type ATPase
3731:
3721:
3719:
3715:
3709:
3706:
3705:
3702:
3699:
3697:
3694:
3692:
3689:
3687:
3684:
3682:
3679:
3677:
3674:
3672:
3669:
3667:
3664:
3662:
3659:
3657:
3654:
3652:
3649:
3647:
3644:
3642:
3639:
3637:
3634:
3632:
3629:
3627:
3624:
3622:
3619:
3617:
3614:
3612:
3609:
3607:
3604:
3602:
3599:
3597:
3594:
3592:
3589:
3587:
3583:
3580:
3579:
3577:
3575:
3571:
3565:
3562:
3560:
3557:
3555:
3552:
3550:
3547:
3545:
3542:
3540:
3537:
3535:
3532:
3530:
3527:
3525:
3522:
3520:
3517:
3515:
3512:
3510:
3507:
3505:
3502:
3500:
3497:
3495:
3492:
3490:
3487:
3485:
3482:
3480:
3476:
3475:
3473:
3471:
3467:
3464:
3462:
3458:
3453:
3449:
3445:
3441:
3437:
3430:
3425:
3423:
3418:
3416:
3411:
3410:
3407:
3401:
3397:
3394:
3390:
3387:
3384:
3359:
3356:
3354:
3351:
3348:
3344:
3340:
3338:
3334:
3333:
3329:
3324:
3320:
3316:
3315:
3310:
3309:
3305:
3297:
3293:
3289:
3285:
3280:
3275:
3271:
3267:
3263:
3259:
3252:
3249:
3242:
3238:
3234:
3233:
3230:
3226:
3221:
3216:
3211:
3206:
3202:
3198:
3194:
3187:
3184:
3179:
3175:
3171:
3167:
3162:
3157:
3153:
3149:
3145:
3138:
3135:
3130:
3126:
3122:
3118:
3114:
3113:10.1038/80981
3110:
3106:
3102:
3095:
3092:
3087:
3083:
3079:
3075:
3071:
3067:
3063:
3059:
3055:
3051:
3044:
3041:
3036:
3032:
3027:
3022:
3017:
3012:
3008:
3004:
3000:
2993:
2990:
2985:
2981:
2976:
2971:
2967:
2963:
2959:
2955:
2951:
2944:
2941:
2936:
2932:
2928:
2924:
2920:
2916:
2909:
2906:
2901:
2897:
2892:
2887:
2883:
2879:
2875:
2871:
2867:
2856:
2853:
2848:
2844:
2839:
2834:
2830:
2826:
2822:
2815:
2812:
2807:
2803:
2799:
2795:
2791:
2787:
2780:
2778:
2774:
2769:
2765:
2760:
2755:
2751:
2747:
2743:
2739:
2735:
2728:
2726:
2722:
2717:
2713:
2709:
2705:
2700:
2695:
2691:
2687:
2683:
2676:
2673:
2668:
2664:
2660:
2656:
2651:
2646:
2642:
2638:
2634:
2619:
2616:
2611:
2607:
2603:
2599:
2595:
2591:
2587:
2583:
2579:
2575:
2568:
2566:
2562:
2557:
2553:
2548:
2543:
2538:
2533:
2529:
2525:
2521:
2517:
2513:
2506:
2504:
2500:
2495:
2491:
2487:
2483:
2478:
2473:
2469:
2465:
2461:
2454:
2451:
2446:
2442:
2437:
2432:
2428:
2424:
2420:
2413:
2410:
2405:
2401:
2395:
2393:
2391:
2387:
2382:
2378:
2371:
2369:
2365:
2360:
2356:
2351:
2346:
2342:
2338:
2334:
2330:
2326:
2322:
2318:
2311:
2309:
2307:
2305:
2301:
2296:
2292:
2287:
2282:
2278:
2274:
2270:
2266:
2262:
2255:
2252:
2247:
2243:
2238:
2233:
2229:
2225:
2221:
2210:
2207:
2202:
2198:
2193:
2188:
2184:
2180:
2176:
2172:
2168:
2164:
2160:
2149:
2146:
2141:
2137:
2132:
2127:
2122:
2117:
2113:
2109:
2105:
2101:
2097:
2090:
2087:
2082:
2078:
2073:
2068:
2063:
2058:
2054:
2050:
2046:
2039:
2037:
2033:
2028:
2024:
2020:
2016:
2012:
2008:
2001:
1997:
1991:
1988:
1983:
1979:
1974:
1969:
1965:
1961:
1957:
1950:
1948:
1946:
1942:
1937:
1933:
1928:
1923:
1919:
1915:
1912:(1): 63–100.
1911:
1907:
1903:
1896:
1893:
1888:
1884:
1879:
1874:
1870:
1866:
1862:
1855:
1853:
1849:
1844:
1840:
1836:
1832:
1825:
1822:
1817:
1813:
1808:
1803:
1798:
1793:
1789:
1785:
1781:
1774:
1770:
1764:
1761:
1756:
1752:
1748:
1744:
1740:
1736:
1729:
1726:
1721:
1717:
1712:
1707:
1703:
1699:
1695:
1688:
1685:
1680:
1676:
1671:
1666:
1662:
1658:
1654:
1647:
1644:
1639:
1635:
1630:
1625:
1621:
1617:
1613:
1606:
1603:
1597:
1592:
1589:
1587:
1584:
1582:
1579:
1577:
1574:
1572:
1569:
1567:
1564:
1562:
1561:Mitochondrion
1559:
1557:
1554:
1552:
1549:
1547:
1544:
1537:
1536:ATP10 protein
1534:
1533:
1528:
1526:
1524:
1516:
1514:
1512:
1504:
1502:
1500:
1492:
1480:
1472:
1468:
1467:
1462:
1461:
1457:
1455:
1454:
1449:
1448:
1443:
1442:
1437:
1436:
1431:
1430:
1425:
1424:
1419:
1418:
1413:
1412:
1407:
1406:
1401:
1400:
1395:
1394:
1389:
1388:
1383:
1382:
1378:
1376:
1375:
1371:
1369:
1368:
1364:
1363:
1362:
1359:
1357:
1349:
1347:
1345:
1341:
1337:
1329:
1317:
1309:
1307:
1293:
1291:
1288:
1286:
1275:
1270:
1268:
1266:
1262:
1253:
1251:
1186:
1134:
1111:
1107:
1092:
1088:
1087:DNA helicases
1079:
1077:
1073:
1068:
1060:
1058:
1056:
1048:
1044:
1040:
1036:
1032:
1029:In respiring
1027:
1025:
1021:
1018:
1002:
1000:
998:
992:
990:
978:
946:
942:
938:
935:(the ring of
926:
917:
915:
906:
902:
898:
893:
889:
881:
877:
872:
860:
854:Binding model
853:
847:
846:
841:
840:
835:
834:
830:
828:
825:
824:
821:
820:
816:
813:
812:
809:
808:
804:
801:
800:
796:
793:
792:
782:
776:
767:
761:
757:
753:
749:
745:
741:
720:
717:shape with a
716:
712:
695:
684:
677:
675:
674:
670:
668:
665:
664:
660:
658:
657:
653:
651:
648:
647:
643:
641:
640:
636:
634:
631:
630:
627:
625:
624:
620:
618:
615:
614:
611:
609:
608:
603:
602:
598:
596:
593:
592:
589:
587:
586:
581:
580:
576:
574:
571:
570:
566:
563:
560:
559:
549:
537:
535:
527:
511:
509:
471:
467:
458:
451:
438:
400:
393:
391:
385:
369:
367:
357:
353:
348:
346:
342:
341:cyanobacteria
338:
334:
330:
326:
322:
318:
314:
310:
306:
302:
298:
275:
274:
273:
271:
263:
259:
255:
251:
247:
235:
232:
230:
226:
223:
220:
218:
214:
211:
208:
206:
202:
197:
193:
190:
186:
183:
181:
180:Gene Ontology
177:
174:
171:
168:
165:
162:
158:
155:
152:
150:
146:
143:
140:
138:
134:
131:
128:
126:
122:
119:
118:NiceZyme view
116:
114:
110:
107:
104:
102:
98:
95:
92:
90:
86:
81:
78:
75:
73:
69:
66:
63:
61:
57:
52:
47:
41:
36:
31:
19:
18:Atp synthesis
6022:Translocases
6019:
6006:
5993:
5980:
5967:
5957:Transferases
5954:
5941:
5798:Binding site
5535:
5469:
5425:
5403:
5388:Small GTPase
5002:Wilson/ATP7B
4997:Menkes/ATP7A
4843:
4690:ATP synthase
4689:
4497:
4465:
4433:
4381:
4348:ATP synthase
4347:
4328:
4296:
4269:
4247:
4227:Cytochrome c
4210:
4150:
4140:Kynureninase
4127:
4100:
4038:
3460:
3448:ATP synthase
3447:
3313:
3261:
3257:
3251:
3240:
3200:
3196:
3186:
3151:
3147:
3137:
3104:
3100:
3094:
3053:
3049:
3043:
3006:
3002:
2992:
2957:
2953:
2943:
2918:
2914:
2908:
2873:
2869:
2855:
2828:
2824:
2814:
2789:
2785:
2741:
2737:
2692:(1–2): 1–4.
2689:
2686:FEBS Letters
2685:
2675:
2640:
2637:FEBS Letters
2636:
2618:
2577:
2573:
2519:
2515:
2467:
2463:
2453:
2426:
2422:
2412:
2403:
2324:
2320:
2271:(1): 43–50.
2268:
2264:
2254:
2227:
2223:
2209:
2166:
2162:
2148:
2103:
2099:
2089:
2052:
2048:
2010:
2006:
1990:
1963:
1959:
1909:
1905:
1895:
1868:
1864:
1834:
1824:
1787:
1783:
1763:
1738:
1734:
1728:
1701:
1697:
1687:
1660:
1656:
1646:
1619:
1615:
1605:
1556:Flavoprotein
1525:, the LUCA.
1520:
1508:
1476:
1464:
1458:
1451:
1445:
1439:
1433:
1427:
1421:
1415:
1409:
1403:
1397:
1391:
1385:
1379:
1372:
1365:
1360:
1355:
1353:
1340:Calvin cycle
1316:chloroplasts
1313:
1297:
1289:
1280:
1279:
1257:
1204:
1135:
1080:
1064:
1047:mitochondria
1028:
1006:
993:
918:
885:
876:chemiosmotic
843:
837:
831:
817:
805:
768:
704:
671:
654:
637:
621:
605:
599:
583:
577:
538:
519:
463:
449:
373:
370:Nomenclature
349:
315:, while in
294:
256:(ATP) using
246:ATP synthase
245:
244:
106:BRENDA entry
33:ATP Synthase
5793:Active site
5712:Mitofusin-1
5687:3.6.5.5-6:
5656:Prokaryotic
4640:Complex III
3900:3.A.3.8.8:
3794:3.A.3.1.2:
3742:3.A.3.1.1:
3311:Nick Lane:
1663:: 631–657.
1576:Proton pump
1546:Chloroplast
1491:cardiolipin
797:Human Gene
709:is a water
556:– Subunits
526:hydrophilic
337:chloroplast
331:, which in
94:IntEnz view
54:Identifiers
6065:Categories
5996:Isomerases
5970:Hydrolases
5837:Regulation
5678:Eukaryotic
5311:Transducin
5148:(3.6.3.10)
4887:Hydrolases
4660:Complex IV
4467:urea cycle
4155:metabolism
4131:metabolism
4129:tryptophan
3574:H (V-type)
3470:H (F-type)
3452:TC 3A2-3A3
3279:2292/55176
3203:: e51179.
2375:Crofts A.
2055:: 785741.
1790:: e10180.
1598:References
1479:euglenozoa
1356:Bos taurus
1261:oligomycin
1254:Inhibitors
1185:particle.
1091:Rho factor
937:c-subunits
888:Paul Boyer
715:tetrameric
564:Human Gene
384:oligomycin
163:structures
130:KEGG entry
77:9000-83-3
5875:EC number
5646:3.6.5.3:
5386:3.6.5.2:
5328:Gustducin
5260:3.6.5.1:
5094:(3.6.3.9)
5014:(3.6.3.8)
4917:Inorganic
4844:see also
4590:Complex I
4398:Aconitase
4039:see also
4006:type 13:
3824:3.A.3.2:
3440:ion pumps
3296:234747849
3258:BioEssays
2003:;
1966:: 40–48.
1776:;
1328:thylakoid
1302:, eight F
1217:helicase/
1104:-powered
1067:evolution
1061:Evolution
905:Cambridge
711:insoluble
352:F-ATPases
345:cytoplasm
284:⇌ ATP + H
262:phosphate
83:Databases
6086:EC 3.6.3
5899:Kinetics
5823:Cofactor
5786:Activity
4922:Thiamine
4544:Frataxin
4418:Fumarase
4084:proteins
3902:flippase
3718:A-ATPase
3701:ATP6V0E1
3691:ATP6V0D2
3686:ATP6V0D1
3671:ATP6V0A4
3666:ATP6V0A2
3661:ATP6V0A1
3651:ATP6V1G3
3646:ATP6V1G2
3641:ATP6V1G1
3631:ATP6V1E2
3626:ATP6V1E1
3616:ATP6V1C2
3611:ATP6V1C1
3606:ATP6V1B2
3601:ATP6V1B1
3396:Archived
3382:-ATPase.
3288:33998015
3229:31738165
3170:11509182
3129:23229994
3121:11062563
3035:29748256
2984:25759169
2935:10576729
2900:17082766
2847:10838056
2806:26671611
2768:19052322
2716:25800744
2708:15473999
2667:32559858
2602:12511957
2556:26984495
2486:12859904
2445:16449553
2295:18515057
2201:29074581
2140:30760595
2081:22312470
2027:15327958
1982:24878343
1936:14195622
1887:10838057
1816:26439008
1679:25839341
1638:21524994
1591:V-ATPase
1571:P-ATPase
1529:See also
1511:V-ATPase
1276:Bacteria
1076:V-ATPase
1072:V-ATPase
1031:bacteria
1024:flagella
1020:gradient
997:affinity
925:subunits
766:region.
468:and the
234:proteins
222:articles
210:articles
167:RCSB PDB
6071:Enzymes
6055:Biology
6009:Ligases
5779:Enzymes
5733:Tubulin
5702:Dynamin
5554:other:
5234:Katanin
5224:Kinesin
5199:ATP13A3
5194:ATP13A2
4929:Apyrase
4704:MT-ATP8
4699:MT-ATP6
4619:MT-ND4L
4028:ATP13A5
4023:ATP13A4
4018:ATP13A3
4013:ATP13A2
4008:ATP13A1
3736:(3.A.3)
3696:ATP6V0E
3681:ATP6V0C
3676:ATP6V0B
3656:ATP6V1H
3636:ATP6V1F
3621:ATP6V1D
3596:ATP6V1A
3591:ATP6AP2
3586:ATP6AP1
3444:ATPases
3220:6930080
3178:1266814
3086:4275221
3078:8065448
3058:Bibcode
3026:7116070
3003:Science
2975:4422255
2915:Science
2891:1636620
2866:ATPase"
2759:2593570
2659:2136729
2610:4333137
2582:Bibcode
2547:4822572
2524:Bibcode
2494:5765103
2359:8599633
2350:1236464
2329:Bibcode
2286:2581510
2246:6214554
2192:6402782
2171:Bibcode
2163:Science
2131:6410833
2108:Bibcode
2072:3268026
1927:2106494
1807:4718723
1755:9874753
1720:4223640
1505:Archaea
1466:MT-ATP8
1460:MT-ATP6
1283:E. coli
941:rotates
807:MT-ATP6
794:Subunit
775:cristae
650:epsilon
561:Subunit
450:E. coli
339:and in
297:protons
276:ADP + P
189:QuickGO
154:profile
137:MetaCyc
72:CAS no.
65:7.1.2.2
6041:Portal
5983:Lyases
5610:ARL13B
5470:RhoBTB
5364:α12/13
5252:GTPase
5229:Myosin
5219:Dynein
5189:ATP12A
5184:ATP11B
5179:ATP10A
5174:ATP8B1
5164:Other
5136:ATP1B4
5131:ATP1B3
5126:ATP1B2
5121:ATP1B1
5116:ATP1A4
5111:ATP1A3
5106:ATP1A2
5101:ATP1A1
5092:Na+/K+
5080:ATP2C2
5075:ATP2C1
5063:ATP2B4
5058:ATP2B3
5053:ATP2B2
5048:ATP2B1
5036:ATP2A3
5031:ATP2A2
5026:ATP2A1
4975:ATPase
4809:MT-TS2
4804:MT-TS1
4774:MT-TL2
4769:MT-TL1
4679:MT-CO3
4674:MT-CO2
4669:MT-CO1
4649:MT-CYB
4629:MT-ND6
4624:MT-ND5
4614:MT-ND4
4609:MT-ND3
4604:MT-ND2
4599:MT-ND1
4372:Matrix
4002:ATP11C
3997:ATP11B
3992:ATP11A
3986:ATP10D
3981:ATP10B
3976:ATP10A
3959:ATP8B4
3954:ATP8B3
3949:ATP8B2
3944:ATP8B1
3939:ATP8A1
3906:ATP8A2
3881:ATP2C1
3876:ATP2B4
3871:ATP2B3
3866:ATP2B2
3861:ATP2B1
3856:ATP2A3
3851:ATP2A2
3846:ATP2A1
3818:ATP12A
3788:ATP1G1
3783:ATP1B4
3778:ATP1B3
3773:ATP1B2
3768:ATP1B1
3763:ATP1A4
3758:ATP1A3
3753:ATP1A2
3748:ATP1A1
3708:TCIRG1
3554:ATP5L2
3544:ATP5J2
3524:ATP5G3
3519:ATP5G2
3514:ATP5G1
3509:ATP5F1
3494:ATP5C2
3489:ATP5C1
3479:ATP5A1
3321:
3294:
3286:
3227:
3217:
3176:
3168:
3127:
3119:
3084:
3076:
3050:Nature
3033:
3023:
2982:
2972:
2933:
2898:
2888:
2845:
2804:
2766:
2756:
2714:
2706:
2665:
2657:
2608:
2600:
2574:Nature
2554:
2544:
2492:
2484:
2443:
2357:
2347:
2293:
2283:
2244:
2199:
2189:
2138:
2128:
2079:
2069:
2025:
1980:
1934:
1924:
1885:
1814:
1804:
1753:
1718:
1677:
1636:
1441:ATP5J2
1417:ATP5G3
1411:ATP5G2
1405:ATP5G1
1399:ATP5F1
1381:ATP5C1
1367:ATP5A1
1350:Mammal
1336:stroma
1017:proton
945:c-ring
845:ATP5G3
839:ATP5G2
833:ATP5G1
819:ATP5PB
758:, and
689:region
623:ATP5C1
607:ATPAF1
585:ATPAF2
579:ATP5A1
516:region
430:stator
356:ATPase
333:plants
288:O + 2H
250:enzyme
248:is an
217:PubMed
199:Search
185:AmiGO
173:PDBsum
113:ExPASy
101:BRENDA
89:IntEnz
60:EC no.
27:Enzyme
5935:Types
5666:EF-Tu
5605:SAR1B
5588:RAB27
5583:RAB23
5536:RhoDF
5426:RhoUV
5409:CDC42
5404:Cdc42
5390:>
5375:GNA13
5370:GNA12
5355:GNA11
5344:αq/11
5333:GNAT3
5321:GNAT2
5316:GNAT1
5306:GNAI3
5301:GNAI2
5296:GNAI1
5248:3.6.5
5212:3.6.4
5155:ATP4A
5146:H+/K+
5021:SERCA
4983:3.6.3
4967:3.6.3
4944:3.6.2
4904:3.6.1
4829:MT-TY
4824:MT-TW
4819:MT-TV
4814:MT-TT
4799:MT-TR
4794:MT-TQ
4789:MT-TP
4784:MT-TN
4779:MT-TM
4764:MT-TK
4759:MT-TI
4754:MT-TH
4749:MT-TG
4744:MT-TF
4739:MT-TE
4734:MT-TD
4729:MT-TC
4724:MT-TA
4520:PMPCB
4507:ALDH2
4329:other
3970:ATP9B
3965:ATP9A
3894:ATP7B
3889:ATP7A
3830:SERCA
3810:ATP4B
3805:ATP4A
3564:ATP5S
3559:ATP5O
3549:ATP5L
3539:ATP5J
3534:ATP5I
3529:ATP5H
3504:ATP5E
3499:ATP5D
3484:ATP5B
3292:S2CID
3241:eLife
3197:eLife
3174:S2CID
3125:S2CID
3082:S2CID
2712:S2CID
2663:S2CID
2606:S2CID
2490:S2CID
1784:eLife
1453:ATP5O
1447:ATP5L
1435:ATP5J
1429:ATP5I
1423:ATP5H
1393:ATP5E
1387:ATP5D
1374:ATP5B
1310:Plant
1294:Yeast
1081:The F
673:ATP5O
656:ATP5E
639:ATP5D
633:delta
617:gamma
601:ATP5B
573:alpha
567:Note
520:The F
476:and F
374:The F
362:and F
149:PRIAM
6027:list
6020:EC7
6014:list
6007:EC6
6001:list
5994:EC5
5988:list
5981:EC4
5975:list
5968:EC3
5962:list
5955:EC2
5949:list
5942:EC1
5726:OPA1
5719:and
5671:EF-G
5661:IF-2
5627:Rheb
5615:ARL6
5600:ARF6
5571:NRAS
5566:KRAS
5561:HRAS
5546:RhoD
5541:RhoF
5487:RhoH
5463:RhoG
5448:Rac1
5436:RhoV
5431:RhoU
5414:TC10
5350:GNAQ
5070:SPCA
4897:3.6)
4715:tRNA
3933:ATP3
3918:Na/K
3840:) /
3838:SPCA
3834:PMCA
3319:ISBN
3284:PMID
3225:PMID
3166:PMID
3148:Cell
3117:PMID
3074:PMID
3031:PMID
2980:PMID
2931:PMID
2896:PMID
2843:PMID
2829:1458
2802:PMID
2764:PMID
2704:PMID
2655:PMID
2598:PMID
2552:PMID
2482:PMID
2464:Cell
2441:PMID
2355:PMID
2291:PMID
2242:PMID
2197:PMID
2136:PMID
2077:PMID
2053:2011
2023:PMID
2000:1VZS
1978:PMID
1932:PMID
1883:PMID
1869:1458
1812:PMID
1773:5ARA
1751:PMID
1716:PMID
1675:PMID
1634:PMID
1265:DCCD
1263:and
1136:The
1106:T3SS
1065:The
983:to F
967:beta
955:of F
951:beta
892:UCLA
890:, a
667:OSCP
595:beta
426:axle
280:+ 2H
229:NCBI
170:PDBe
125:KEGG
5721:MX2
5717:MX1
5637:RGK
5632:Rap
5622:Ran
5595:Arf
5578:Rab
5556:Ras
5514:Rnd
5492:Rho
5443:Rac
5419:TCL
5280:olf
5012:Ca+
3796:H/K
3274:hdl
3266:doi
3215:PMC
3205:doi
3156:doi
3152:106
3109:doi
3066:doi
3054:370
3021:PMC
3011:doi
3007:360
2970:PMC
2962:doi
2958:468
2923:doi
2919:286
2886:PMC
2878:doi
2833:doi
2794:doi
2754:PMC
2746:doi
2694:doi
2690:576
2645:doi
2641:259
2590:doi
2578:421
2542:PMC
2532:doi
2520:113
2472:doi
2468:114
2431:doi
2427:209
2345:PMC
2337:doi
2281:PMC
2273:doi
2269:476
2232:doi
2228:257
2187:PMC
2179:doi
2167:358
2126:PMC
2116:doi
2104:116
2067:PMC
2057:doi
2015:doi
2011:342
1996:PDB
1968:doi
1922:PMC
1914:doi
1873:doi
1839:doi
1802:PMC
1792:doi
1769:PDB
1743:doi
1739:172
1706:doi
1702:241
1665:doi
1624:doi
1620:149
1318:(CF
1108:or
903:in
480:. F
406:, F
282:out
205:PMC
161:PDB
6067::
5402::
5290:αi
5272:αs
5250::
4973::
4895:EC
4889::
3931::
3920:–
3904::
3844::
3836:,
3832:,
3826:Ca
3803::
3746::
3584::
3446:/
3442:,
3438::
3371:,
3362:Na
3290:.
3282:.
3272:.
3262:43
3260:.
3239:.
3223:.
3213:.
3199:.
3195:.
3172:.
3164:.
3150:.
3146:.
3123:.
3115:.
3103:.
3080:.
3072:.
3064:.
3052:.
3029:.
3019:.
3005:.
3001:.
2978:.
2968:.
2956:.
2952:.
2929:.
2917:.
2894:.
2884:.
2874:25
2872:.
2868:.
2841:.
2827:.
2823:.
2800:.
2790:41
2788:.
2776:^
2762:.
2752:.
2742:72
2740:.
2736:.
2724:^
2710:.
2702:.
2688:.
2684:.
2661:.
2653:.
2639:.
2635:.
2604:.
2596:.
2588:.
2576:.
2564:^
2550:.
2540:.
2530:.
2518:.
2514:.
2502:^
2488:.
2480:.
2466:.
2462:.
2439:.
2425:.
2421:.
2402:.
2389:^
2367:^
2353:.
2343:.
2335:.
2325:69
2323:.
2319:.
2303:^
2289:.
2279:.
2267:.
2263:.
2240:.
2226:.
2222:.
2195:.
2185:.
2177:.
2165:.
2161:.
2134:.
2124:.
2114:.
2102:.
2098:.
2075:.
2065:.
2051:.
2047:.
2035:^
2021:.
2009:.
1998::
1976:.
1964:25
1962:.
1958:.
1944:^
1930:.
1920:.
1910:22
1908:.
1904:.
1881:.
1867:.
1863:.
1851:^
1837:.
1833:.
1810:.
1800:.
1786:.
1782:.
1771::
1749:.
1737:.
1714:.
1700:.
1696:.
1673:.
1661:84
1659:.
1655:.
1632:.
1618:.
1614:.
1501:.
1463:,
1450:,
1444:,
1438:,
1432:,
1426:,
1420:,
1414:,
1408:,
1402:,
1396:,
1390:,
1384:,
1267:.
939:)
916:.
842:,
836:,
756:F6
754:,
750:,
746:,
742:,
604:,
582:,
428:,
424:,
417:,
347:.
290:in
264:(P
187:/
6043::
6029:)
6025:(
6016:)
6012:(
6003:)
5999:(
5990:)
5986:(
5977:)
5973:(
5964:)
5960:(
5951:)
5947:(
5771:e
5764:t
5757:v
5529:3
5524:2
5519:1
5507:C
5502:B
5497:A
5480:2
5475:1
5458:3
5453:2
5362:G
5342:G
5288:G
5278:G
5270:G
4971:4
4969:-
4893:(
4879:e
4872:t
4865:v
4074:e
4067:t
4060:v
3922:H
3828:(
3454:)
3450:(
3428:e
3421:t
3414:v
3373:K
3349:.
3298:.
3276::
3268::
3231:.
3207::
3201:8
3180:.
3158::
3131:.
3111::
3105:7
3088:.
3068::
3060::
3037:.
3013::
2986:.
2964::
2937:.
2925::
2902:.
2880::
2864:1
2849:.
2835::
2808:.
2796::
2770:.
2748::
2718:.
2696::
2669:.
2647::
2631:1
2629:F
2627:O
2612:.
2592::
2584::
2558:.
2534::
2526::
2496:.
2474::
2447:.
2433::
2406:.
2383:.
2361:.
2339::
2331::
2297:.
2275::
2248:.
2234::
2218:1
2203:.
2181::
2173::
2157:O
2142:.
2118::
2110::
2083:.
2059::
2029:.
2017::
1984:.
1970::
1938:.
1916::
1889:.
1875::
1845:.
1841::
1818:.
1794::
1788:4
1757:.
1745::
1722:.
1708::
1681:.
1667::
1640:.
1626::
1540:O
1495:1
1487:O
1483:1
1332:1
1324:O
1322:F
1320:1
1304:O
1300:1
1241:H
1230:H
1219:H
1207:H
1183:1
1179:O
1175:O
1164:H
1153:H
1149:O
1138:H
1131:3
1129:β
1127:3
1122:1
1118:3
1116:β
1114:3
1095:H
1083:1
1051:1
1013:O
1011:F
1009:1
985:O
981:1
973:O
969:3
965:3
961:1
957:1
953:3
949:3
933:O
929:O
921:1
909:1
882:.
864:i
827:c
814:b
802:a
787:O
785:F
779:O
771:O
764:1
760:8
752:g
748:f
744:e
740:d
736:1
734:F
731:O
727:1
723:O
707:O
705:F
699:O
697:F
687:O
685:F
554:1
552:F
546:1
544:F
541:1
530:1
522:1
514:1
512:F
506:1
504:F
502:O
498:O
494:1
490:1
486:1
482:O
478:1
474:O
446:1
444:F
442:O
432:.
421:1
419:F
414:O
412:F
408:1
404:O
388:O
380:O
376:1
364:1
360:O
286:2
278:i
266:i
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.