788:
receptor subtypes. Also, these modulators have a decreased potential for toxic effects, since modulators with limited co-operativity will have a ceiling level to their effect, irrespective of the administered dose. Another type of pharmacological selectivity that is unique to allosteric modulators is based on co-operativity. An allosteric modulator may display neutral co-operativity with an orthosteric ligand at all subtypes of a given receptor except the subtype of interest, which is termed "absolute subtype selectivity". If an allosteric modulator does not possess appreciable efficacy, it can provide another powerful therapeutic advantage over orthosteric ligands, namely the ability to selectively tune up or down tissue responses only when the endogenous agonist is present. Oligomer-specific small molecule binding sites are drug targets for medically relevant
839:(ASD) provides a central resource for the display, search and analysis of the structure, function and related annotation for allosteric molecules. Currently, ASD contains allosteric proteins from more than 100 species and modulators in three categories (activators, inhibitors, and regulators). Each protein is annotated with detailed description of allostery, biological process and related diseases, and each modulator with binding affinity, physicochemical properties and therapeutic area. Integrating the information of allosteric proteins in ASD should allow the prediction of allostery for unknown proteins, to be followed with experimental validation. In addition, modulators curated in ASD can be used to investigate potential allosteric targets for a query compound, and can help chemists to implement structure modifications for novel allosteric drug design.
852:. Pharmacologically important proteins with difficult-to-target sites may yield to approaches in which one alternatively targets easier-to-reach residues that are capable of allosterically regulating the primary site of interest. These residues can broadly be classified as surface- and interior-allosteric amino acids. Allosteric sites at the surface generally play regulatory roles that are fundamentally distinct from those within the interior; surface residues may serve as receptors or effector sites in allosteric signal transmission, whereas those within the interior may act to transmit such signals.
372:, and then relate specific statistical measurements of allostery to specific energy terms in the energy function (such as an intermolecular salt bridge between two domains). Ensemble models like the ensemble allosteric model and allosteric Ising model assume that each domain of the system can adopt two states similar to the MWC model. The allostery landscape model introduced by Cuendet, Weinstein, and LeVine allows for the domains to have any number of states and the contribution of a specific molecular interaction to a given allosteric coupling can be estimated using a rigorous set of rules.
305:, postulates that enzyme subunits are connected in such a way that a conformational change in one subunit is necessarily conferred to all other subunits. Thus, all subunits must exist in the same conformation. The model further holds that, in the absence of any ligand (substrate or otherwise), the equilibrium favors one of the conformational states, T or R. The equilibrium can be shifted to the R or T state through the binding of one
221:
403:. They can be positive (activating) causing an increase of the enzyme activity or negative (inhibiting) causing a decrease of the enzyme activity. The use of allosteric modulation allows the control of the effects of specific enzyme activities; as a result, allosteric modulators are very effective in pharmacology. In a biological system, allosteric modulation can be difficult to distinguish from modulation by
813:
coupling between several binding sites is in artificial systems usually much larger than in proteins with their usually larger flexibility. The parameter which determines the efficiency (as measured by the ratio of equilibrium constants Krel = KA(E)/KA in presence and absence of an effector E ) is the conformational energy needed to adopt a closed or strained conformation for the binding of a ligand A.
33:
820:
systems direct interaction between bound ligands can occur, which can lead to large cooperativities. Most common is such a direct interaction between ions in receptors for ion-pairs. This cooperativity is often also referred to as allostery, even though conformational changes here are not necessarily
659:
are heterotropic allosteric modulators of hemoglobin. Once again, in IMP/GMP specific 5' nucleotidase, binding of GTP molecule at the dimer interface in the tetrameric enzyme leads to increased affinity for substrate GMP at the active site indicating towards K-type heterotropic allosteric activation.
317:
The sequential model of allosteric regulation holds that subunits are not connected in such a way that a conformational change in one induces a similar change in the others. Thus, all enzyme subunits do not necessitate the same conformation. Moreover, the sequential model dictates that molecules of a
847:
Not all protein residues play equally important roles in allosteric regulation. The identification of residues that are essential to allostery (so-called “allosteric residues”) has been the focus of many studies, especially within the last decade. In part, this growing interest is a result of their
642:
and CO are homotropic allosteric modulators of hemoglobin. Likewise, in IMP/GMP specific 5' nucleotidase, binding of one GMP molecule to a single subunit of the tetrameric enzyme leads to increased affinity for GMP by the subsequent subunits as revealed by sigmoidal substrate versus velocity plots.
398:
binds to an allosteric site (also known as a regulatory site) of an enzyme and alters the enzyme activity. Allosteric modulators are designed to fit the allosteric site to cause a conformational change of the enzyme, in particular a change in the shape of the active site, which then causes a change
393:
is used to alter the activity of molecules and enzymes in biochemistry and pharmacology. For comparison, a typical drug is made to bind to the active site of an enzyme which thus prohibits binding of a substrate to that enzyme causing a decrease in enzyme activity. Allosteric modulation occurs when
207:
Mechanism of Action: Binding to the allosteric site induces a conformational change in the enzyme that can either reduce the affinity of the active site for the substrate or alter the enzyme's catalytic activity. This indirect interference can inhibit the enzyme's function even if the substrate is
787:
to accommodate an endogenous ligand, so are more diverse. Therefore, greater GPCR selectivity may be obtained by targeting allosteric sites. This is particularly useful for GPCRs where selective orthosteric therapy has been difficult because of sequence conservation of the orthosteric site across
680:
A non-regulatory allosteric site is any non-regulatory component of an enzyme (or any protein), that is not itself an amino acid. For instance, many enzymes require sodium binding to ensure proper function. However, the sodium does not necessarily act as a regulatory subunit; the sodium is always
349:
A morpheein is a homo-oligomeric structure that can exist as an ensemble of physiologically significant and functionally different alternate quaternary assemblies. Transitions between alternate morpheein assemblies involve oligomer dissociation, conformational change in the dissociated state, and
322:
protocol. While such an induced fit converts a subunit from the tensed state to relaxed state, it does not propagate the conformational change to adjacent subunits. Instead, substrate-binding at one subunit only slightly alters the structure of other subunits so that their binding sites are more
812:
at a second site, and negative if the affinity isn't highered. Most synthetic allosteric complexes rely on conformational reorganization upon the binding of one effector ligand which then leads to either enhanced or weakened association of second ligand at another binding site. Conformational
834:
Allostery is a direct and efficient means for regulation of biological macromolecule function, produced by the binding of a ligand at an allosteric site topographically distinct from the orthosteric site. Due to the often high receptor selectivity and lower target-based toxicity, allosteric
671:
Some allosteric activators are referred to as "essential", or "obligate" activators, in the sense that in their absence, the activity of their target enzyme activity is very low or negligible, as is the case with N-acetylglutamate's activity on carbamoyl phosphate synthetase I, for example.
293:, the allostery landscape model described by Cuendet, Weinstein, and LeVine, can be used. Allosteric regulation may be facilitated by the evolution of large-scale, low-energy conformational changes, which enables long-range allosteric interaction between distant binding sites.
681:
present and there are no known biological processes to add/remove sodium to regulate enzyme activity. Non-regulatory allostery could comprise any other ions besides sodium (calcium, magnesium, zinc), as well as other chemicals and possibly vitamins.
285:, tensed (T) or relaxed (R), and that relaxed subunits bind substrate more readily than those in the tense state. The two models differ most in their assumptions about subunit interaction and the preexistence of both states. For proteins in which
2510:
Ghosh A, Vishveshwara S (November 2008). "Variations in clique and community patterns in protein structures during allosteric communication: investigation of dynamically equilibrated structures of methionyl tRNA synthetase complexes".
68:, resulting in a conformational change that alters the protein's activity, either enhancing or inhibiting its function. In contrast, substances that bind directly to an enzyme's active site or the binding site of the
473:
oxygen affinity. Another example of allosteric activation is seen in cytosolic IMP-GMP specific 5'-nucleotidase II (cN-II), where the affinity for substrate GMP increases upon GTP binding at the dimer interface.
663:
As has been amply highlighted above, some allosteric proteins can be regulated by both their substrates and other molecules. Such proteins are capable of both homotropic and heterotropic interactions.
194:
Competitive
Inhibition: Most orthosteric inhibitors compete with the substrate for the active site, which means their effectiveness can be reduced if substrate concentration increases.
617:
and maintaining balanced levels of cellular ATP. In this way, ATP serves as a negative allosteric modulator for PFK, despite the fact that it is also a substrate of the enzyme.
651:
A heterotropic allosteric modulator is a regulatory molecule that is not the enzyme's substrate. It may be either an activator or an inhibitor of the enzyme. For example, H, CO
156:), "solid (object)". This is in reference to the fact that the regulatory site of an allosteric protein is physically distinct from its active site. Allostery contrasts with
350:
reassembly to a different oligomer. The required oligomer disassembly step differentiates the morpheein model for allosteric regulation from the classic MWC and KNF models.
211:
Non-Competitive
Inhibition: Allosteric inhibitors often exhibit non-competitive inhibition, meaning their inhibitory effect is not dependent on the substrate concentration.
1984:
425:
that is perfectly suited to adapt to living in the macrophages of humans. The enzyme's sites serve as a communication between different substrates. Specifically between
1998:
Takeuchi M, Ikeda M, Sugasaki A, Shinkai S (November 2001). "Molecular design of artificial molecular and ion recognition systems with allosteric guest responses".
191:
Mechanism of Action: By occupying the active site, these inhibitors prevent the substrate from binding, thereby directly blocking the enzyme's catalytic activity.
2357:
Süel GM, Lockless SW, Wall MA, Ranganathan R (January 2003). "Evolutionarily conserved networks of residues mediate allosteric communication in proteins".
494:
binds to an allosteric site on hemoglobin, the affinity for oxygen of all subunits decreases. This is when a regulator is absent from the binding site.
1096:
Koshland DE, Némethy G, Filmer D (January 1966). "Comparison of experimental binding data and theoretical models in proteins containing subunits".
204:
Binding Site: Allosteric inhibitors bind to a site on the enzyme that is distinct and separate from the active site, known as the allosteric site.
779:. There are a number of advantages in using allosteric modulators as preferred therapeutic agents over classic orthosteric ligands. For example,
469:. The binding of oxygen to one subunit induces a conformational change in that subunit that interacts with the remaining active sites to enhance
804:
binding sites, which exhibit conformational changes upon occupation of one site. Cooperativity between single binding contributions in such
2138:
Badjić JD, Nelson A, Cantrill SJ, Turnbull WB, Stoddart JF (September 2005). "Multivalency and cooperativity in supramolecular chemistry".
1858:
Christopoulos A, May LT, Avlani VA, Sexton PM (November 2004). "G-protein-coupled receptor allosterism: the promise and the problem(s)".
2925:
1037:
1391:"AIM for Allostery: Using the Ising Model to Understand Information Processing and Transmission in Allosteric Biomolecular Systems"
376:
simulations can be used to estimate a system's statistical ensemble so that it can be analyzed with the allostery landscape model.
546:
of the glycine receptor for glycine. Thus, strychnine inhibits the action of an inhibitory transmitter, leading to convulsions.
3081:
1499:"Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis"
542:. Strychnine acts at a separate binding site on the glycine receptor in an allosteric manner; i.e., its binding lowers the
152:
713:
ligand, and can be thought to act like a dimmer switch in an electrical circuit, adjusting the intensity of the response.
638:, as well as a regulatory molecule of the protein's activity. It is typically an activator of the protein. For example, O
2894:
2898:
780:
701:") and enhances or inhibits the effects of the endogenous ligand. Under normal circumstances, it acts by causing a
501:
497:
417:
262:
188:
Binding Site: Orthosteric inhibitors bind directly to the enzyme's active site, where the substrate normally binds.
3066:
775:
Allosteric proteins are involved in, and are central in many diseases, and allosteric sites may represent a novel
3182:
3169:
3156:
3143:
3130:
3117:
3104:
578:
3076:
170:
142:
3209:
3030:
2973:
631:
598:
458:
353:
232:
689:
Allosteric modulation of a receptor results from the binding of allosteric modulators at a different site (a "
2978:
1898:
May LT, Leach K, Sexton PM, Christopoulos A (2007). "Allosteric modulation of G protein-coupled receptors".
725:
656:
594:
543:
491:
426:
61:
449:
enhances the attraction between substrate molecules and other binding sites. An example is the binding of
871:
606:
586:
550:
404:
400:
274:
157:
111:
1563:"Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila"
2918:
1978:
1020:
Bu Z, Callaway DJ (2011). "Proteins move! Protein dynamics and long-range allostery in cell signaling".
706:
702:
694:
390:
306:
88:
3071:
1698:"Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis"
1061:
Monod J, Wyman J, Changeux JP (May 1965). "On the nature of allosteric transitions:A plausible model".
759:
More recent examples of drugs that allosterically modulate their targets include the calcium-mimicking
2846:
2776:"Identifying Allosteric Hotspots with Dynamics: Application to Inter- and Intra-species Conservation"
2728:
2559:
2413:
1817:
1709:
1510:
1402:
1296:
1206:
966:
602:
574:
385:
365:
266:
709:
of the ligand. In this way, an allosteric ligand modulates the receptor's activation by its primary
3035:
1696:
de Cima S, Polo LM, Díez-Fernández C, Martínez AI, Cervera J, Fita I, et al. (November 2015).
1561:
Srinivasan B, Forouhar F, Shukla A, Sampangi C, Kulkarni S, Abashidze M, et al. (March 2014).
876:
753:
733:
590:
554:
430:
910:
Cooper A, Dryden DT (October 1984). "Allostery without conformational change. A plausible model".
736:
regulatory binding sites. These regulatory sites can each produce positive allosteric modulation,
2968:
2836:
2718:
2382:
1807:
1230:
1196:
1002:
935:
395:
373:
1935:"Morpheeins – A New Pathway for Allosteric Drug Discovery~!2010-02-12~!2010-05-21~!2010-06-08~!"
848:
general importance in protein science, but also because allosteric residues may be exploited in
3204:
2874:
2805:
2756:
2687:
2636:
2587:
2528:
2492:
2441:
2374:
2339:
2288:
2239:
2204:
2155:
2120:
2085:
2050:
2015:
1966:
1915:
1911:
1875:
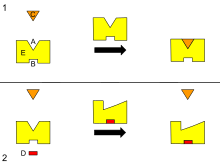
1835:
1776:
1735:
1678:
1627:
1592:
1536:
1479:
1430:
1371:
1322:
1265:
1248:
Jaffe EK (September 2005). "Morpheeins--a new structural paradigm for allosteric regulation".
1222:
1165:
1113:
1078:
1043:
1033:
994:
927:
558:
369:
290:
282:
69:
835:
regulation is also expected to play an increasing role in drug discovery and bioengineering.
3014:
3009:
2983:
2911:
2864:
2854:
2795:
2787:
2746:
2736:
2705:
Negre CF, Morzan UN, Hendrickson HP, Pal R, Lisi GP, Loria JP, et al. (December 2018).
2677:
2667:
2626:
2618:
2577:
2567:
2520:
2482:
2472:
2431:
2421:
2366:
2329:
2319:
2278:
2270:
2231:
2194:
2186:
2147:
2112:
2077:
2042:
2007:
1956:
1946:
1907:
1867:
1825:
1766:
1725:
1717:
1668:
1658:
1619:
1582:
1574:
1526:
1518:
1469:
1461:
1420:
1410:
1361:
1353:
1312:
1304:
1257:
1214:
1155:
1147:
1105:
1070:
1025:
984:
974:
919:
886:
528:
516:
270:
237:
92:
3061:
3045:
2958:
1357:
891:
881:
690:
570:
466:
286:
278:
2103:
Schneider HJ (September 2016). "Efficiency parameters in artificial allosteric systems".
2850:
2732:
2563:
2417:
2068:
Kovbasyuk L, Krämer R (June 2004). "Allosteric supramolecular receptors and catalysts".
1821:
1713:
1623:
1514:
1406:
1300:
1210:
1184:
1024:. Advances in Protein Chemistry and Structural Biology. Vol. 83. pp. 163–221.
970:
3099:
3040:
2869:
2825:"Green function of correlated genes in a minimal mechanical model of protein evolution"
2824:
2800:
2775:
2751:
2706:
2682:
2655:
2631:
2606:
2582:
2547:
2487:
2461:"Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities"
2460:
2436:
2401:
2334:
2307:
2283:
2258:
2199:
2174:
1961:
1934:
1730:
1697:
1673:
1646:
1587:
1562:
1531:
1498:
1474:
1449:
1425:
1390:
1366:
1341:
1317:
1284:
1160:
1135:
1029:
989:
954:
861:
817:
805:
776:
729:
717:
462:
119:
115:
2222:
McConnell AJ, Beer PD (May 2012). "Heteroditopic receptors for ion-pair recognition".
1136:"The Allostery Landscape: Quantifying Thermodynamic Couplings in Biomolecular Systems"
1074:
220:
3198:
3004:
2963:
1234:
866:
309:(the allosteric effector or ligand) to a site that is different from the active site
258:
165:
134:
2607:"Exploring residue component contributions to dynamical network models of allostery"
2386:
939:
593:, causing a change in the enzyme's three-dimensional shape. This change causes its
2953:
1006:
589:
within the cell. When ATP levels are high, ATP will bind to an allosteric site on
504:
of thrombin have been discovered that could potentially be used as anticoagulants.
433:. Sites like these also serve as a sensing mechanism for the enzyme's performance.
45:
41:
710:
609:) at the active site to decrease, and the enzyme is deemed inactive. This causes
2426:
1830:
1795:
1218:
979:
783:(GPCR) allosteric binding sites have not faced the same evolutionary pressure as
3177:
3112:
2948:
1951:
801:
698:
549:
Another instance in which negative allosteric modulation can be seen is between
535:
319:
225:
87:. Allosteric sites allow effectors to bind to the protein, often resulting in a
65:
2829:
Proceedings of the
National Academy of Sciences of the United States of America
2711:
Proceedings of the
National Academy of Sciences of the United States of America
2660:
Proceedings of the
National Academy of Sciences of the United States of America
2552:
Proceedings of the
National Academy of Sciences of the United States of America
2465:
Proceedings of the
National Academy of Sciences of the United States of America
1771:
1754:
1522:
1261:
2897:
introducing a classification system for protein allostery mechanisms from the
2791:
2459:
Gasper PM, Fuglestad B, Komives EA, Markwick PR, McCammon JA (December 2012).
760:
749:
610:
562:
539:
508:
490:
decreases the affinity for substrate at other active sites. For example, when
454:
160:
which requires no conformational change for an enzyme's activation. The term
2324:
1497:
Zhong W, Cui L, Goh BC, Cai Q, Ho P, Chionh YH, et al. (December 2017).
1226:
1151:
808:
systems is positive if occupation of one binding site enhances the affinity Δ
301:
The concerted model of allostery, also referred to as the symmetry model or
3151:
3125:
2859:
2774:
Clarke D, Sethi A, Li S, Kumar S, Chang RW, Chen J, et al. (May 2016).
2741:
2707:"Eigenvector centrality for characterization of protein allosteric pathways"
2672:
2572:
2477:
2308:"Exploiting protein flexibility to predict the location of allosteric sites"
1647:"The N-Acetylglutamate Synthase Family: Structures, Function and Mechanisms"
955:"Allostery: An Overview of Its History, Concepts, Methods, and Applications"
789:
764:
343:
302:
254:
2878:
2809:
2760:
2691:
2640:
2591:
2532:
2496:
2445:
2378:
2343:
2292:
2243:
2235:
2208:
2159:
2124:
2089:
2054:
2046:
2019:
1970:
1919:
1879:
1839:
1780:
1739:
1682:
1596:
1540:
1483:
1434:
1375:
1326:
1269:
1169:
1082:
1047:
998:
2654:
Rivalta I, Sultan MM, Lee NS, Manley GA, Loria JP, Batista VS (May 2012).
2274:
1663:
1117:
931:
364:
Ensemble models of allosteric regulation enumerate an allosteric system's
114:
from upstream substrates. Long-range allostery is especially important in
32:
17:
1631:
741:
422:
107:
2257:
Huang Z, Zhu L, Cao Y, Wu G, Liu X, Chen Y, et al. (January 2011).
1308:
1109:
2116:
1871:
1578:
923:
745:
737:
635:
614:
524:
520:
512:
106:
Allosteric regulations are a natural example of control loops, such as
2622:
2524:
2151:
2081:
2011:
1721:
1465:
1415:
1185:"Colloquium : Proteins: The physics of amorphous evolving matter"
3164:
2934:
2259:"ASD: a comprehensive database of allosteric proteins and modulators"
2190:
532:
487:
450:
446:
242:
123:
57:
2841:
2723:
2370:
1812:
1201:
95:. Effectors that enhance the protein's activity are referred to as
3138:
2033:
Kremer C, Lützen A (May 2013). "Artificial allosteric receptors".
399:
in its activity. In contrast to typical drugs, modulators are not
346:
model of allosteric regulation is a dissociative concerted model.
219:
31:
2605:
Vanwart AT, Eargle J, Luthey-Schulten Z, Amaro RE (August 2012).
2402:"Binding leverage as a molecular basis for allosteric regulation"
1796:"Known allosteric proteins have central roles in genetic disease"
500:
provides an excellent example of negative allosteric modulation.
569:) is an enzyme that catalyses the third step of glycolysis: the
99:, whereas those that decrease the protein's activity are called
2907:
1610:
Edelstein SJ (1975). "Cooperative interactions of hemoglobin".
2656:"Allosteric pathways in imidazole glycerol phosphate synthase"
1450:"Allosteric modulators: an emerging concept in drug discovery"
836:
613:
to cease when ATP levels are high, thus conserving the body's
582:
566:
118:. Allosteric regulation is also particularly important in the
2546:
Sethi A, Eargle J, Black AA, Luthey-Schulten Z (April 2009).
2903:
461:
and the effector. The allosteric, or "other", site is the
705:
in a receptor molecule, which results in a change in the
333:
conformational changes are not propagated to all subunits
748:
at the benzodiazepine regulatory site, and its antidote
246:
This is a diagram of allosteric regulation of an enzyme.
174:) meaning “straight”, “upright”, “right” or “correct”.
2823:
Dutta S, Eckmann JP, Libchaber A, Tlusty T (May 2018).
800:
There are many synthetic compounds containing several
849:
515:
poison, which acts as an allosteric inhibitor of the
1283:
Motlagh HN, Wrabl JO, Li J, Hilser VJ (April 2014).
1134:
Cuendet MA, Weinstein H, LeVine MV (December 2016).
330:
molecules of substrate bind via induced-fit protocol
3090:
3054:
3023:
2992:
2941:
1183:Eckmann JP, Rougemont J, Tlusty T (July 30, 2019).
79:The site to which the effector binds is termed the
585:can be allosterically inhibited by high levels of
565:. Phosphofructokinase (generally referred to as
250:Many allosteric effects can be explained by the
327:subunits need not exist in the same conformation
2400:Mitternacht S, Berezovsky IN (September 2011).
1556:
1554:
1552:
1550:
724:has two active sites that the neurotransmitter
2548:"Dynamical networks in tRNA:protein complexes"
1129:
1127:
482:Negative allosteric modulation (also known as
441:Positive allosteric modulation (also known as
2919:
1983:: CS1 maint: DOI inactive as of March 2024 (
1342:"Structural and energetic basis of allostery"
8:
1900:Annual Review of Pharmacology and Toxicology
1755:"Allostery in disease and in drug discovery"
277:, Nemethy, and Filmer. Both postulate that
56:) is a substance that binds to a site on an
1651:International Journal of Molecular Sciences
1645:Shi D, Allewell NM, Tuchman M (June 2015).
273:(also known as the KNF model) described by
2926:
2912:
2904:
2611:Journal of Chemical Theory and Computation
1140:Journal of Chemical Theory and Computation
415:An example of this model is seen with the
2868:
2858:
2840:
2799:
2750:
2740:
2722:
2681:
2671:
2630:
2581:
2571:
2486:
2476:
2435:
2425:
2333:
2323:
2282:
2198:
1960:
1950:
1912:10.1146/annurev.pharmtox.47.120505.105159
1829:
1811:
1770:
1729:
1672:
1662:
1586:
1530:
1473:
1424:
1414:
1365:
1316:
1200:
1159:
988:
978:
1340:Hilser VJ, Wrabl JO, Motlagh HN (2012).
843:Allosteric residues and their prediction
1939:The Open Conference Proceedings Journal
1794:Abrusan G, Ascher DB, Inouye M (2022).
902:
630:A homotropic allosteric modulator is a
457:, where oxygen is effectively both the
2306:Panjkovich A, Daura X (October 2012).
1976:
323:receptive to substrate. To summarize:
1893:
1891:
1889:
1853:
1851:
1849:
1358:10.1146/annurev-biophys-050511-102319
7:
2105:Organic & Biomolecular Chemistry
2173:Kim SK, Sessler JL (October 2010).
1624:10.1146/annurev.bi.44.070175.001233
1389:LeVine MV, Weinstein H (May 2015).
356:(PBGS) is the prototype morpheein.
1285:"The ensemble nature of allostery"
1030:10.1016/B978-0-12-381262-9.00005-7
36:Allosteric regulation of an enzyme
25:
486:) occurs when the binding of one
445:) occurs when the binding of one
1860:Biochemical Society Transactions
1448:Abdel-Magid AF (February 2015).
771:Allosteric sites as drug targets
1454:ACS Medicinal Chemistry Letters
953:Liu J, Nussinov R (June 2016).
178:Ortho vs. allosteric inhibitors
1250:Trends in Biochemical Sciences
1022:Protein Structure and Diseases
1:
2140:Accounts of Chemical Research
2035:Chemistry: A European Journal
2000:Accounts of Chemical Research
1612:Annual Review of Biochemistry
1075:10.1016/s0022-2836(65)80285-6
746:positive allosteric modulator
27:Regulation of enzyme activity
2427:10.1371/journal.pcbi.1002148
1831:10.1371/journal.pcbi.1009806
1219:10.1103/RevModPhys.91.031001
1063:Journal of Molecular Biology
980:10.1371/journal.pcbi.1004966
796:Synthetic allosteric systems
110:from downstream products or
1955:(inactive March 11, 2024).
1952:10.2174/2210289201001010001
1753:Nussinov R, Tsai C (2013).
1346:Annual Review of Biophysics
912:European Biophysics Journal
821:triggering binding events.
728:(GABA) binds, but also has
3226:
2899:Royal Society of Chemistry
2406:PLOS Computational Biology
2269:(Database issue): D663–9.
1800:PLOS Computational Biology
1772:10.1016/j.cell.2013.03.034
1523:10.1038/s41467-017-02086-y
1262:10.1016/j.tibs.2005.07.003
959:PLOS Computational Biology
781:G protein-coupled receptor
498:Direct thrombin inhibitors
418:Mycobacterium tuberculosis
383:
169:
151:
141:
76:regulators or modulators.
3082:Michaelis–Menten kinetics
2792:10.1016/j.str.2016.03.008
2359:Nature Structural Biology
1189:Reviews of Modern Physics
734:general anaesthetic agent
579:fructose 1,6-bisphosphate
370:potential energy function
72:of a receptor are called
2974:Diffusion-limited enzyme
2325:10.1186/1471-2105-13-273
2179:Chemical Society Reviews
1152:10.1021/acs.jctc.6b00841
676:Non-regulatory allostery
354:Porphobilinogen synthase
2860:10.1073/pnas.1716215115
2742:10.1073/pnas.1810452115
2673:10.1073/pnas.1120536109
2573:10.1073/pnas.0810961106
2478:10.1073/pnas.1218414109
837:The AlloSteric Database
726:gamma-aminobutyric acid
657:2,3-bisphosphoglycerate
289:exist in more than two
2263:Nucleic Acids Research
2236:10.1002/anie.201107244
2047:10.1002/chem.201203814
872:Competitive inhibition
763:and the HIV treatment
740:the activity of GABA.
405:substrate presentation
401:competitive inhibitors
318:substrate bind via an
247:
158:substrate presentation
37:
3067:Eadie–Hofstee diagram
3000:Allosteric regulation
2717:(52): E12201–E12208.
1664:10.3390/ijms160613004
1503:Nature Communications
703:conformational change
502:Allosteric inhibitors
484:allosteric inhibition
443:allosteric activation
391:Allosteric modulation
380:Allosteric modulation
368:as a function of its
223:
101:allosteric inhibitors
97:allosteric activators
89:conformational change
35:
3077:Lineweaver–Burk plot
2175:"Ion pair receptors"
816:In many multivalent
693:") from that of the
667:Essential activators
603:fructose-6-phosphate
575:fructose-6-phosphate
561:loop that regulates
411:Energy sensing model
386:Allosteric modulator
366:statistical ensemble
281:exist in one of two
54:allosteric modulator
50:allosteric regulator
2851:2018PNAS..115E4559D
2835:(20): E4559–E4568.
2733:2018PNAS..11512201N
2564:2009PNAS..106.6620S
2418:2011PLSCB...7E2148M
2275:10.1093/nar/gkq1022
1822:2022PLSCB..18E9806A
1714:2015NatSR...516950D
1515:2017NatCo...8.1986Z
1407:2015Entrp..17.2895L
1309:10.1038/nature13001
1301:2014Natur.508..331M
1211:2019RvMP...91c1001E
1110:10.1021/bi00865a047
971:2016PLSCB..12E4966L
877:Cooperative binding
850:biomedical contexts
830:Allosteric Database
754:receptor antagonist
591:phosphofructokinase
555:phosphofructokinase
507:Another example is
478:Negative modulation
437:Positive modulation
229:B – Allosteric site
91:and/or a change in
3036:Enzyme superfamily
2969:Enzyme promiscuity
2312:BMC Bioinformatics
2117:10.1039/c6ob01303a
1872:10.1042/BST0320873
1702:Scientific Reports
1579:10.1111/febs.12727
924:10.1007/BF00276625
374:Molecular dynamics
248:
122:ability to adjust
64:distinct from the
38:
3192:
3191:
2623:10.1021/ct300377a
2525:10.1021/bi8007559
2519:(44): 11398–407.
2224:Angewandte Chemie
2152:10.1021/ar040223k
2111:(34): 7994–8001.
2082:10.1021/cr030673a
2012:10.1021/ar0000410
1933:Jaffe EK (2010).
1722:10.1038/srep16950
1466:10.1021/ml5005365
1416:10.3390/e17052895
1146:(12): 5758–5767.
785:orthosteric sites
716:For example, the
695:endogenous ligand
559:negative feedback
70:endogenous ligand
40:In the fields of
16:(Redirected from
3217:
3072:Hanes–Woolf plot
3015:Enzyme activator
3010:Enzyme inhibitor
2984:Enzyme catalysis
2928:
2921:
2914:
2905:
2883:
2882:
2872:
2862:
2844:
2820:
2814:
2813:
2803:
2771:
2765:
2764:
2754:
2744:
2726:
2702:
2696:
2695:
2685:
2675:
2666:(22): E1428–36.
2651:
2645:
2644:
2634:
2617:(8): 2949–2961.
2602:
2596:
2595:
2585:
2575:
2543:
2537:
2536:
2507:
2501:
2500:
2490:
2480:
2471:(52): 21216–22.
2456:
2450:
2449:
2439:
2429:
2397:
2391:
2390:
2354:
2348:
2347:
2337:
2327:
2303:
2297:
2296:
2286:
2254:
2248:
2247:
2219:
2213:
2212:
2202:
2191:10.1039/c002694h
2185:(10): 3784–809.
2170:
2164:
2163:
2135:
2129:
2128:
2100:
2094:
2093:
2070:Chemical Reviews
2065:
2059:
2058:
2030:
2024:
2023:
1995:
1989:
1988:
1982:
1974:
1964:
1954:
1930:
1924:
1923:
1895:
1884:
1883:
1855:
1844:
1843:
1833:
1815:
1791:
1785:
1784:
1774:
1750:
1744:
1743:
1733:
1693:
1687:
1686:
1676:
1666:
1642:
1636:
1635:
1607:
1601:
1600:
1590:
1573:(6): 1613–1628.
1567:The FEBS Journal
1558:
1545:
1544:
1534:
1494:
1488:
1487:
1477:
1445:
1439:
1438:
1428:
1418:
1401:(5): 2895–2918.
1386:
1380:
1379:
1369:
1337:
1331:
1330:
1320:
1280:
1274:
1273:
1245:
1239:
1238:
1204:
1180:
1174:
1173:
1163:
1131:
1122:
1121:
1093:
1087:
1086:
1058:
1052:
1051:
1017:
1011:
1010:
992:
982:
950:
944:
943:
907:
887:Protein dynamics
825:Online resources
707:binding affinity
529:neurotransmitter
523:is a major post-
517:glycine receptor
465:of an adjoining
313:Sequential model
279:protein subunits
271:sequential model
173:
155:
146:), "other", and
145:
93:protein dynamics
21:
3225:
3224:
3220:
3219:
3218:
3216:
3215:
3214:
3210:Enzyme kinetics
3195:
3194:
3193:
3188:
3100:Oxidoreductases
3086:
3062:Enzyme kinetics
3050:
3046:List of enzymes
3019:
2988:
2959:Catalytic triad
2937:
2932:
2895:Instant insight
2891:
2886:
2822:
2821:
2817:
2773:
2772:
2768:
2704:
2703:
2699:
2653:
2652:
2648:
2604:
2603:
2599:
2545:
2544:
2540:
2509:
2508:
2504:
2458:
2457:
2453:
2412:(9): e1002148.
2399:
2398:
2394:
2356:
2355:
2351:
2305:
2304:
2300:
2256:
2255:
2251:
2230:(21): 5052–61.
2221:
2220:
2216:
2172:
2171:
2167:
2137:
2136:
2132:
2102:
2101:
2097:
2067:
2066:
2062:
2041:(20): 6162–96.
2032:
2031:
2027:
1997:
1996:
1992:
1975:
1932:
1931:
1927:
1897:
1896:
1887:
1866:(Pt 5): 873–7.
1857:
1856:
1847:
1806:(2): e1009806.
1793:
1792:
1788:
1752:
1751:
1747:
1695:
1694:
1690:
1657:(6): 13004–22.
1644:
1643:
1639:
1609:
1608:
1604:
1560:
1559:
1548:
1496:
1495:
1491:
1447:
1446:
1442:
1388:
1387:
1383:
1339:
1338:
1334:
1295:(7496): 331–9.
1282:
1281:
1277:
1247:
1246:
1242:
1182:
1181:
1177:
1133:
1132:
1125:
1095:
1094:
1090:
1060:
1059:
1055:
1040:
1019:
1018:
1014:
965:(6): e1004966.
952:
951:
947:
909:
908:
904:
900:
892:Receptor theory
882:Enzyme kinetics
858:
845:
832:
827:
798:
773:
721:
691:regulatory site
687:
678:
669:
654:
649:
641:
634:for its target
628:
623:
571:phosphorylation
553:and the enzyme
480:
467:protein subunit
439:
413:
388:
382:
362:
360:Ensemble models
340:
338:Morpheein model
315:
299:
297:Concerted model
245:
240:
235:
230:
228:
218:
201:
185:
180:
164:comes from the
133:comes from the
85:regulatory site
81:allosteric site
28:
23:
22:
15:
12:
11:
5:
3223:
3221:
3213:
3212:
3207:
3197:
3196:
3190:
3189:
3187:
3186:
3173:
3160:
3147:
3134:
3121:
3108:
3094:
3092:
3088:
3087:
3085:
3084:
3079:
3074:
3069:
3064:
3058:
3056:
3052:
3051:
3049:
3048:
3043:
3038:
3033:
3027:
3025:
3024:Classification
3021:
3020:
3018:
3017:
3012:
3007:
3002:
2996:
2994:
2990:
2989:
2987:
2986:
2981:
2976:
2971:
2966:
2961:
2956:
2951:
2945:
2943:
2939:
2938:
2933:
2931:
2930:
2923:
2916:
2908:
2902:
2901:
2890:
2889:External links
2887:
2885:
2884:
2815:
2786:(5): 826–837.
2766:
2697:
2646:
2597:
2558:(16): 6620–5.
2538:
2502:
2451:
2392:
2371:10.1038/nsb881
2349:
2298:
2249:
2214:
2165:
2130:
2095:
2076:(6): 3161–87.
2060:
2025:
2006:(11): 865–73.
1990:
1925:
1885:
1845:
1786:
1765:(2): 293–305.
1745:
1688:
1637:
1602:
1546:
1489:
1440:
1381:
1332:
1275:
1240:
1175:
1123:
1088:
1053:
1038:
1012:
945:
918:(2): 103–109.
901:
899:
896:
895:
894:
889:
884:
879:
874:
869:
864:
857:
854:
844:
841:
831:
828:
826:
823:
818:supramolecular
806:supramolecular
797:
794:
772:
769:
730:benzodiazepine
719:
686:
683:
677:
674:
668:
665:
652:
648:
645:
639:
627:
624:
622:
619:
479:
476:
438:
435:
412:
409:
384:Main article:
381:
378:
361:
358:
339:
336:
335:
334:
331:
328:
314:
311:
298:
295:
217:
214:
213:
212:
209:
205:
200:
197:
196:
195:
192:
189:
184:
181:
179:
176:
116:cell signaling
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3222:
3211:
3208:
3206:
3203:
3202:
3200:
3184:
3180:
3179:
3174:
3171:
3167:
3166:
3161:
3158:
3154:
3153:
3148:
3145:
3141:
3140:
3135:
3132:
3128:
3127:
3122:
3119:
3115:
3114:
3109:
3106:
3102:
3101:
3096:
3095:
3093:
3089:
3083:
3080:
3078:
3075:
3073:
3070:
3068:
3065:
3063:
3060:
3059:
3057:
3053:
3047:
3044:
3042:
3041:Enzyme family
3039:
3037:
3034:
3032:
3029:
3028:
3026:
3022:
3016:
3013:
3011:
3008:
3006:
3005:Cooperativity
3003:
3001:
2998:
2997:
2995:
2991:
2985:
2982:
2980:
2977:
2975:
2972:
2970:
2967:
2965:
2964:Oxyanion hole
2962:
2960:
2957:
2955:
2952:
2950:
2947:
2946:
2944:
2940:
2936:
2929:
2924:
2922:
2917:
2915:
2910:
2909:
2906:
2900:
2896:
2893:
2892:
2888:
2880:
2876:
2871:
2866:
2861:
2856:
2852:
2848:
2843:
2838:
2834:
2830:
2826:
2819:
2816:
2811:
2807:
2802:
2797:
2793:
2789:
2785:
2781:
2777:
2770:
2767:
2762:
2758:
2753:
2748:
2743:
2738:
2734:
2730:
2725:
2720:
2716:
2712:
2708:
2701:
2698:
2693:
2689:
2684:
2679:
2674:
2669:
2665:
2661:
2657:
2650:
2647:
2642:
2638:
2633:
2628:
2624:
2620:
2616:
2612:
2608:
2601:
2598:
2593:
2589:
2584:
2579:
2574:
2569:
2565:
2561:
2557:
2553:
2549:
2542:
2539:
2534:
2530:
2526:
2522:
2518:
2514:
2506:
2503:
2498:
2494:
2489:
2484:
2479:
2474:
2470:
2466:
2462:
2455:
2452:
2447:
2443:
2438:
2433:
2428:
2423:
2419:
2415:
2411:
2407:
2403:
2396:
2393:
2388:
2384:
2380:
2376:
2372:
2368:
2364:
2360:
2353:
2350:
2345:
2341:
2336:
2331:
2326:
2321:
2317:
2313:
2309:
2302:
2299:
2294:
2290:
2285:
2280:
2276:
2272:
2268:
2264:
2260:
2253:
2250:
2245:
2241:
2237:
2233:
2229:
2225:
2218:
2215:
2210:
2206:
2201:
2196:
2192:
2188:
2184:
2180:
2176:
2169:
2166:
2161:
2157:
2153:
2149:
2146:(9): 723–32.
2145:
2141:
2134:
2131:
2126:
2122:
2118:
2114:
2110:
2106:
2099:
2096:
2091:
2087:
2083:
2079:
2075:
2071:
2064:
2061:
2056:
2052:
2048:
2044:
2040:
2036:
2029:
2026:
2021:
2017:
2013:
2009:
2005:
2001:
1994:
1991:
1986:
1980:
1972:
1968:
1963:
1958:
1953:
1948:
1944:
1940:
1936:
1929:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1894:
1892:
1890:
1886:
1881:
1877:
1873:
1869:
1865:
1861:
1854:
1852:
1850:
1846:
1841:
1837:
1832:
1827:
1823:
1819:
1814:
1809:
1805:
1801:
1797:
1790:
1787:
1782:
1778:
1773:
1768:
1764:
1760:
1756:
1749:
1746:
1741:
1737:
1732:
1727:
1723:
1719:
1715:
1711:
1707:
1703:
1699:
1692:
1689:
1684:
1680:
1675:
1670:
1665:
1660:
1656:
1652:
1648:
1641:
1638:
1633:
1629:
1625:
1621:
1617:
1613:
1606:
1603:
1598:
1594:
1589:
1584:
1580:
1576:
1572:
1568:
1564:
1557:
1555:
1553:
1551:
1547:
1542:
1538:
1533:
1528:
1524:
1520:
1516:
1512:
1508:
1504:
1500:
1493:
1490:
1485:
1481:
1476:
1471:
1467:
1463:
1459:
1455:
1451:
1444:
1441:
1436:
1432:
1427:
1422:
1417:
1412:
1408:
1404:
1400:
1396:
1392:
1385:
1382:
1377:
1373:
1368:
1363:
1359:
1355:
1351:
1347:
1343:
1336:
1333:
1328:
1324:
1319:
1314:
1310:
1306:
1302:
1298:
1294:
1290:
1286:
1279:
1276:
1271:
1267:
1263:
1259:
1255:
1251:
1244:
1241:
1236:
1232:
1228:
1224:
1220:
1216:
1212:
1208:
1203:
1198:
1195:(3): 031001.
1194:
1190:
1186:
1179:
1176:
1171:
1167:
1162:
1157:
1153:
1149:
1145:
1141:
1137:
1130:
1128:
1124:
1119:
1115:
1111:
1107:
1104:(1): 365–85.
1103:
1099:
1092:
1089:
1084:
1080:
1076:
1072:
1068:
1064:
1057:
1054:
1049:
1045:
1041:
1039:9780123812629
1035:
1031:
1027:
1023:
1016:
1013:
1008:
1004:
1000:
996:
991:
986:
981:
976:
972:
968:
964:
960:
956:
949:
946:
941:
937:
933:
929:
925:
921:
917:
913:
906:
903:
897:
893:
890:
888:
885:
883:
880:
878:
875:
873:
870:
868:
867:Anharmonicity
865:
863:
860:
859:
855:
853:
851:
842:
840:
838:
829:
824:
822:
819:
814:
811:
807:
803:
795:
793:
791:
786:
782:
778:
770:
768:
766:
762:
757:
755:
751:
747:
743:
739:
735:
731:
727:
723:
714:
712:
708:
704:
700:
696:
692:
684:
682:
675:
673:
666:
664:
661:
658:
646:
644:
637:
633:
625:
620:
618:
616:
612:
608:
604:
600:
596:
592:
588:
584:
580:
576:
572:
568:
564:
560:
556:
552:
547:
545:
541:
537:
534:
530:
526:
522:
518:
514:
510:
505:
503:
499:
495:
493:
489:
485:
477:
475:
472:
468:
464:
460:
456:
453:molecules to
452:
448:
444:
436:
434:
432:
428:
424:
420:
419:
410:
408:
406:
402:
397:
392:
387:
379:
377:
375:
371:
367:
359:
357:
355:
351:
347:
345:
337:
332:
329:
326:
325:
324:
321:
312:
310:
308:
304:
296:
294:
292:
291:conformations
288:
284:
283:conformations
280:
276:
272:
268:
264:
260:
257:put forth by
256:
253:
244:
239:
234:
227:
222:
215:
210:
206:
203:
202:
198:
193:
190:
187:
186:
182:
177:
175:
172:
167:
166:Ancient Greek
163:
159:
154:
149:
144:
139:
136:
135:Ancient Greek
132:
127:
125:
121:
117:
113:
109:
104:
102:
98:
94:
90:
86:
82:
77:
75:
71:
67:
63:
59:
55:
51:
47:
43:
34:
30:
19:
3178:Translocases
3175:
3162:
3149:
3136:
3123:
3113:Transferases
3110:
3097:
2999:
2954:Binding site
2832:
2828:
2818:
2783:
2779:
2769:
2714:
2710:
2700:
2663:
2659:
2649:
2614:
2610:
2600:
2555:
2551:
2541:
2516:
2513:Biochemistry
2512:
2505:
2468:
2464:
2454:
2409:
2405:
2395:
2365:(1): 59–69.
2362:
2358:
2352:
2315:
2311:
2301:
2266:
2262:
2252:
2227:
2223:
2217:
2182:
2178:
2168:
2143:
2139:
2133:
2108:
2104:
2098:
2073:
2069:
2063:
2038:
2034:
2028:
2003:
1999:
1993:
1979:cite journal
1942:
1938:
1928:
1903:
1899:
1863:
1859:
1803:
1799:
1789:
1762:
1758:
1748:
1708:(1): 16950.
1705:
1701:
1691:
1654:
1650:
1640:
1615:
1611:
1605:
1570:
1566:
1506:
1502:
1492:
1460:(2): 104–7.
1457:
1453:
1443:
1398:
1394:
1384:
1349:
1345:
1335:
1292:
1288:
1278:
1256:(9): 490–7.
1253:
1249:
1243:
1192:
1188:
1178:
1143:
1139:
1101:
1098:Biochemistry
1097:
1091:
1066:
1062:
1056:
1021:
1015:
962:
958:
948:
915:
911:
905:
862:ASD database
846:
833:
815:
809:
799:
784:
774:
758:
738:potentiating
715:
688:
685:Pharmacology
679:
670:
662:
650:
647:Heterotropic
629:
548:
506:
496:
483:
481:
470:
442:
440:
416:
414:
389:
363:
352:
348:
341:
316:
300:
269:, or by the
251:
249:
161:
147:
137:
130:
128:
105:
100:
96:
84:
80:
78:
73:
53:
49:
46:pharmacology
42:biochemistry
39:
29:
2949:Active site
1509:(1): 1986.
1352:: 585–609.
802:noncovalent
777:drug target
711:orthosteric
699:active site
557:within the
536:spinal cord
527:inhibitory
463:active site
320:induced fit
226:Active site
183:Orthosteric
112:feedforward
74:orthosteric
66:active site
3199:Categories
3152:Isomerases
3126:Hydrolases
2993:Regulation
2842:1801.03681
2724:1706.02327
1813:2107.04318
1618:: 209–32.
1202:1907.13371
1069:: 88–118.
898:References
790:morpheeins
761:cinacalcet
750:flumazenil
626:Homotropic
611:glycolysis
563:glycolysis
540:brain stem
513:convulsant
509:strychnine
455:hemoglobin
199:Allosteric
162:orthostery
126:activity.
18:Allosteric
3031:EC number
2780:Structure
1235:199001124
1227:0034-6861
765:maraviroc
632:substrate
599:substrate
533:mammalian
459:substrate
423:bacterium
344:morpheein
303:MWC model
255:MWC model
252:concerted
238:Inhibitor
233:Substrate
131:allostery
129:The term
3205:Proteins
3055:Kinetics
2979:Cofactor
2942:Activity
2879:29712824
2810:27066750
2761:30530700
2692:22586084
2641:23139645
2592:19351898
2533:18842003
2497:23197839
2446:21935347
2387:67749580
2379:12483203
2344:23095452
2293:21051350
2244:22419667
2209:20737073
2160:16171315
2125:27431438
2090:15186190
2055:23463705
2020:11714258
1971:21643557
1920:17009927
1906:: 1–51.
1880:15494038
1840:10138267
1781:23582321
1740:26592762
1683:26068232
1597:24456211
1541:29215013
1484:25699154
1435:26594108
1376:22577828
1327:24740064
1270:16023348
1170:27766843
1083:14343300
1048:21570668
999:27253437
940:12591175
856:See also
742:Diazepam
722:receptor
595:affinity
544:affinity
525:synaptic
396:effector
287:subunits
275:Koshland
267:Changeux
208:present.
168:orthós (
108:feedback
62:receptor
3165:Ligases
2935:Enzymes
2870:5960285
2847:Bibcode
2801:4883016
2752:6310864
2729:Bibcode
2683:3365145
2632:3489502
2583:2672494
2560:Bibcode
2488:3535651
2437:3174156
2414:Bibcode
2335:3562710
2318:: 273.
2284:3013650
2200:3016456
1962:3107518
1945:: 1–6.
1818:Bibcode
1731:4655335
1710:Bibcode
1674:4490483
1588:3982195
1532:5719368
1511:Bibcode
1475:4329591
1426:4652859
1403:Bibcode
1395:Entropy
1367:3935618
1318:4224315
1297:Bibcode
1207:Bibcode
1161:5156960
1118:5938952
1007:3610740
990:4890769
967:Bibcode
932:6544679
636:protein
615:glucose
521:Glycine
492:2,3-BPG
153:στερεός
148:stereos
3139:Lyases
2877:
2867:
2808:
2798:
2759:
2749:
2690:
2680:
2639:
2629:
2590:
2580:
2531:
2495:
2485:
2444:
2434:
2385:
2377:
2342:
2332:
2291:
2281:
2242:
2207:
2197:
2158:
2123:
2088:
2053:
2018:
1969:
1959:
1918:
1878:
1838:
1779:
1738:
1728:
1681:
1671:
1632:237460
1630:
1595:
1585:
1539:
1529:
1482:
1472:
1433:
1423:
1374:
1364:
1325:
1315:
1289:Nature
1268:
1233:
1225:
1168:
1158:
1116:
1081:
1046:
1036:
1005:
997:
987:
938:
930:
655:, and
488:ligand
451:oxygen
447:ligand
307:ligand
265:, and
243:Enzyme
216:Models
124:enzyme
120:cell's
58:enzyme
3091:Types
2837:arXiv
2719:arXiv
2383:S2CID
1808:arXiv
1231:S2CID
1197:arXiv
1003:S2CID
936:S2CID
752:is a
744:is a
697:(an "
621:Types
577:into
471:their
263:Wyman
259:Monod
171:ὀρθός
143:ἄλλος
138:allos
3183:list
3176:EC7
3170:list
3163:EC6
3157:list
3150:EC5
3144:list
3137:EC4
3131:list
3124:EC3
3118:list
3111:EC2
3105:list
3098:EC1
2875:PMID
2806:PMID
2757:PMID
2688:PMID
2637:PMID
2588:PMID
2529:PMID
2493:PMID
2442:PMID
2375:PMID
2340:PMID
2289:PMID
2240:PMID
2205:PMID
2156:PMID
2121:PMID
2086:PMID
2051:PMID
2016:PMID
1985:link
1967:PMID
1916:PMID
1876:PMID
1836:PMID
1777:PMID
1759:Cell
1736:PMID
1679:PMID
1628:PMID
1593:PMID
1537:PMID
1480:PMID
1431:PMID
1372:PMID
1323:PMID
1266:PMID
1223:ISSN
1166:PMID
1114:PMID
1079:PMID
1044:PMID
1034:ISBN
995:PMID
928:PMID
732:and
718:GABA
605:and
597:for
538:and
511:, a
429:and
421:, a
342:The
241:E –
236:D –
231:C –
224:A –
52:(or
44:and
2865:PMC
2855:doi
2833:115
2796:PMC
2788:doi
2747:PMC
2737:doi
2715:115
2678:PMC
2668:doi
2664:109
2627:PMC
2619:doi
2578:PMC
2568:doi
2556:106
2521:doi
2483:PMC
2473:doi
2469:109
2432:PMC
2422:doi
2367:doi
2330:PMC
2320:doi
2279:PMC
2271:doi
2232:doi
2195:PMC
2187:doi
2148:doi
2113:doi
2078:doi
2074:104
2043:doi
2008:doi
1957:PMC
1947:doi
1908:doi
1868:doi
1826:doi
1767:doi
1763:153
1726:PMC
1718:doi
1669:PMC
1659:doi
1620:doi
1583:PMC
1575:doi
1571:281
1527:PMC
1519:doi
1470:PMC
1462:doi
1421:PMC
1411:doi
1362:PMC
1354:doi
1313:PMC
1305:doi
1293:508
1258:doi
1215:doi
1156:PMC
1148:doi
1106:doi
1071:doi
1026:doi
985:PMC
975:doi
920:doi
607:ATP
587:ATP
583:PFK
581:.
573:of
567:PFK
551:ATP
531:in
431:G6P
427:AMP
394:an
83:or
60:or
48:an
3201::
2873:.
2863:.
2853:.
2845:.
2831:.
2827:.
2804:.
2794:.
2784:24
2782:.
2778:.
2755:.
2745:.
2735:.
2727:.
2713:.
2709:.
2686:.
2676:.
2662:.
2658:.
2635:.
2625:.
2613:.
2609:.
2586:.
2576:.
2566:.
2554:.
2550:.
2527:.
2517:47
2515:.
2491:.
2481:.
2467:.
2463:.
2440:.
2430:.
2420:.
2408:.
2404:.
2381:.
2373:.
2363:10
2361:.
2338:.
2328:.
2316:13
2314:.
2310:.
2287:.
2277:.
2267:39
2265:.
2261:.
2238:.
2228:51
2226:.
2203:.
2193:.
2183:39
2181:.
2177:.
2154:.
2144:38
2142:.
2119:.
2109:14
2107:.
2084:.
2072:.
2049:.
2039:19
2037:.
2014:.
2004:34
2002:.
1981:}}
1977:{{
1965:.
1941:.
1937:.
1914:.
1904:47
1902:.
1888:^
1874:.
1864:32
1862:.
1848:^
1834:.
1824:.
1816:.
1804:18
1802:.
1798:.
1775:.
1761:.
1757:.
1734:.
1724:.
1716:.
1704:.
1700:.
1677:.
1667:.
1655:16
1653:.
1649:.
1626:.
1616:44
1614:.
1591:.
1581:.
1569:.
1565:.
1549:^
1535:.
1525:.
1517:.
1505:.
1501:.
1478:.
1468:.
1456:.
1452:.
1429:.
1419:.
1409:.
1399:17
1397:.
1393:.
1370:.
1360:.
1350:41
1348:.
1344:.
1321:.
1311:.
1303:.
1291:.
1287:.
1264:.
1254:30
1252:.
1229:.
1221:.
1213:.
1205:.
1193:91
1191:.
1187:.
1164:.
1154:.
1144:12
1142:.
1138:.
1126:^
1112:.
1100:.
1077:.
1067:12
1065:.
1042:.
1032:.
1001:.
993:.
983:.
973:.
963:12
961:.
957:.
934:.
926:.
916:11
914:.
792:.
767:.
756:.
519:.
407:.
261:,
103:.
3185:)
3181:(
3172:)
3168:(
3159:)
3155:(
3146:)
3142:(
3133:)
3129:(
3120:)
3116:(
3107:)
3103:(
2927:e
2920:t
2913:v
2881:.
2857::
2849::
2839::
2812:.
2790::
2763:.
2739::
2731::
2721::
2694:.
2670::
2643:.
2621::
2615:8
2594:.
2570::
2562::
2535:.
2523::
2499:.
2475::
2448:.
2424::
2416::
2410:7
2389:.
2369::
2346:.
2322::
2295:.
2273::
2246:.
2234::
2211:.
2189::
2162:.
2150::
2127:.
2115::
2092:.
2080::
2057:.
2045::
2022:.
2010::
1987:)
1973:.
1949::
1943:1
1922:.
1910::
1882:.
1870::
1842:.
1828::
1820::
1810::
1783:.
1769::
1742:.
1720::
1712::
1706:5
1685:.
1661::
1634:.
1622::
1599:.
1577::
1543:.
1521::
1513::
1507:8
1486:.
1464::
1458:6
1437:.
1413::
1405::
1378:.
1356::
1329:.
1307::
1299::
1272:.
1260::
1237:.
1217::
1209::
1199::
1172:.
1150::
1120:.
1108::
1102:5
1085:.
1073::
1050:.
1028::
1009:.
977::
969::
942:.
922::
810:G
720:A
653:2
640:2
601:(
150:(
140:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.