428:
390:α-Glucosidase hydrolyzes terminal non-reducing (1→4)-linked α-glucose residues to release a single α-glucose molecule. α-Glucosidase is a carbohydrate-hydrolase that releases α-glucose as opposed to β-glucose. β-Glucose residues can be released by glucoamylase, a functionally similar enzyme. The substrate selectivity of α-glucosidase is due to subsite affinities of the enzyme's active site. Two proposed mechanisms include a nucleophilic displacement and an oxocarbenium ion intermediate.
394:
488:, an α-glucosidase inhibitor, competitively and reversibly inhibits α-glucosidase in the intestines. This inhibition lowers the rate of glucose absorption through delayed carbohydrate digestion and extended digestion time. Acarbose may be able to prevent the development of diabetic symptoms. Hence, α-glucosidase inhibitors (like acarbose) are used as anti-diabetic drugs in combination with other anti-diabetic drugs.
240:
2260:
229:
40:
439:
Human lysosomal α-glucosidase has been studied for the significance of the Asp-518 and other residues in proximity of the enzyme's active site. It was found that substituting Asp-513 with Glu-513 interferes with posttranslational modification and intracellular transport of α-glucosidase's precursor.
414:
Trout liver α-glucosidases were extracted and characterized. It was shown that for one of the trout liver α-glucosidases maximum activity of the enzyme was increased by 80% during exercise in comparison to a resting trout. This change was shown to correlate to an activity increase for liver glycogen
443:
Kinetic changes in α-glucosidase have been shown to be induced by denaturants such as guanidinium chloride (GdmCl) and SDS solutions. These denaturants cause loss of activity and conformational change. A loss of enzyme activity occurs at much lower concentrations of denaturant than required for
1169:
Yoshimizu, M.; Tajima, Y; Matsuzawa, F; Aikawa, S; Iwamoto, K; Kobayashi, T; Edmunds, T; Fujishima, K; Tsuji, D; Itoh, K; Ikekita, M; Kawashima, I; Sugawara, K; Ohyanagi, N; Suzuki, T; Togawa, T; Ohno, K; Sakuraba, H (May 2008). "Binding parameters and thermodynamics of the interaction of imino
318:
entry GO:0090599 represents the broad sense of "alpha-glucosidase". It is defined as "catalysis of the hydrolysis of terminal, non-reducing alpha-linked alpha-D-glucose residue with release of alpha-D-glucose." In this sense, "alpha-glucosidase" can encompass a wide range of enzyme activitiess,
475:
exhibit inhibition of the enzyme. It was found that one compound molecule binds to a single enzyme molecule. It was shown that 1-deoxynojirimycin (DNJ) would bind the strongest of the sugars tested and blocked the active site of the enzyme almost entirely. The studies enhanced knowledge of the
512:
Many animal viruses possess an outer envelope composed of viral glycoproteins. These are often required for the viral life cycle and utilize cellular machinery for synthesis. Inhibitors of α-glucosidase show that the enzyme is involved in the pathway for
777:
Group of enzymes whose specificity is directed mainly toward the exohydrolysis of 1,4-alpha-glucosidic linkages, and that hydrolyze oligosaccharides rapidly, relative to polysaccharides, which are hydrolyzed relatively slowly, or not at
1424:
492:
has been found to be a strong inhibitor of α-glucosidase. The compound can inhibit the enzyme up to 36% with a concentration of 0.5 mg/ml. As of 2016, this substance is being tested in rats, mice and
415:
phosphorylase. It is proposed that α-glucosidase in the glucosidic path plays an important part in complementing the phosphorolytic pathway in the liver's metabolic response to energy demands of exercise.
435:α-Glucosidases can be divided, according to primary structure, into two families. The gene coding for human lysosomal α-glucosidase is about 20 kb long and its structure has been cloned and confirmed.
506:
Diagnosis of azoospermia has potential to be aided by measurement of α-glucosidase activity in seminal plasma. Activity in the seminal plasma corresponds to the functionality of the epididymis.
444:
conformational changes. This leads to a conclusion that the enzyme's active site conformation is less stable than the whole enzyme conformation in response to the two denaturants.
1417:
588:
Bruni, C.B.; Sica, V.; Auricchio, F.; Covelli, I. (1970). "Further kinetic and structural characterization of the lysosomal α-D-glucoside glucohydrolase from cattle liver".
1410:
1467:
192:
411:, a blood-sucking insect, forms hemozoin (Hz) during digestion of host hemoglobin. Hemozoin synthesis is dependent on the substrate binding site of α-glucosidase.
211:
319:
differing by the linkage of their terminal (1→3, 1→4, or 1→6), the specific identity of their substrate (sucrose, maltose, or starch), among other aspects.
714:
Sørensen, S.H.; Norén, O.; Sjöström, H.; Danielsen, E.M. (1982). "Amphiphilic pig intestinal microvillus maltase/glucoamylase. Structure and specificity".
1597:
382:. These names are not recommended because they may only refer to a specific activity of the enzyme, or a specific protein having this acvitity.
517:-glycans for viruses such as HIV and human hepatitis B virus (HBV). Inhibition of α-glucosidase can prevent fusion of HIV and secretion of HBV.
1100:
Wu XQ, Xu H, Yue H, Liu KQ, Wang XY (December 2009). "Inhibition kinetics and the aggregation of α-glucosidase by different denaturants".
687:
Sivikami, S.; Radhakrishnan, A.N. (1973). "Purification of rabbit intestinal glucoamylase by affinity chromatography on
Sephadex G-200".
1979:
455:
204:
1861:
1402:
427:
1590:
171:
147:
2135:
1873:
2250:
1170:
sugars with a recombinant human acid α-glucosidase (alglucosidase alfa): insight into the complex formation mechanism".
1720:
1510:
552:
532:
343:
would otherwise be included). Human genes that produce enzymes with activities specified by this EC number include:
440:
Additionally, the Trp-516 and Asp-518 residues have been deemed critical for the enzyme's catalytic functionality.
2120:
2236:
2223:
2210:
2197:
2184:
2171:
2158:
1931:
1619:
1583:
1477:
467:
became the first released treatment for Pompe disease and acts as an analog to α-glucosidase. Further studies of
2130:
1143:
625:"Purification of rat intestinal maltase/glucoamylase and its anomalous dissociation either by heat or by low pH"
165:
2084:
2027:
1610:
575:
328:
292:
253:
58:
418:
Yeast and rat small intestinal α-glucosidases have been shown to be inhibited by several groups of flavonoids.
152:
2032:
1783:
1437:
372:
maltase, glucoinvertase, glucosidosucrase, maltase-glucoamylase, α-glucopyranosidase, glucosidoinvertase, α-
1946:
1691:
1538:
1505:
353:
1281:
Zhen, et al. (November 2017). "Synthesis of novel flavonoid alkaloids as α-glucosidase inhibitors".
216:
2053:
1972:
140:
2125:
1905:
1856:
1559:
1205:
Bischoff H (August 1995). "The mechanism of α-glucosidase inhibition in the management of diabetes".
889:
75:
935:
Mehrani H, Storey KB (October 1993). "Characterization of α-glucosidases from rainbow trout liver".
2089:
1800:
1788:
1623:
1564:
1357:
Mehta, Anand; Zitzmann, Nicole; Rudd, Pauline M; Block, Timothy M; Dwek, Raymond A (23 June 1998).
331:
3.2.1.20 is narrower. It requires the linkage to be 1→4, and the preferred substrate to be smaller
168:
70:
393:
92:
2022:
1844:
1839:
1817:
1805:
1644:
1515:
1462:
1388:
1263:
1125:
749:
527:
468:
464:
2280:
1812:
1686:
1534:
1472:
1380:
1339:
1298:
1255:
1214:
1187:
1117:
1042:
993:
952:
917:
858:
731:
696:
654:
605:
571:
407:
159:
2068:
2063:
2037:
1965:
1822:
1708:
1370:
1329:
1290:
1245:
1179:
1109:
1073:
1032:
1024:
983:
944:
907:
897:
848:
723:
644:
636:
597:
547:
332:
298:
1060:
Hermans, Monique; Marian Kroos; Jos Van
Beeumen; Ben Oostra; Arnold Reuser (25 July 1991).
878:"α-Glucosidase promotes hemozoin formation in a blood-sucking bug: an evolutionary history"
128:
2115:
2099:
2012:
1942:
104:
1061:
893:
790:
295:
256:
63:
2264:
2153:
2094:
1883:
1795:
1632:
1575:
1445:
1037:
1012:
912:
877:
727:
672:
Larner, J.; Lardy, H.; Myrback, K. (1960). "Other glucosidases". In Boyer, P.D. (ed.).
649:
624:
187:
1375:
1358:
1078:
2274:
2058:
2017:
1829:
1758:
1741:
601:
340:
315:
1392:
1129:
2007:
1673:
1441:
1267:
494:
272:
902:
17:
2231:
2166:
2002:
1900:
1866:
1834:
501:
398:
268:
239:
2259:
1011:
Hoefsloot L; M Hoogeveen-Westerveld; A J Reuser; B A Oostra (1 December 1990).
1910:
1878:
1482:
1433:
1334:
1317:
1294:
1183:
1113:
472:
2205:
2179:
1748:
1681:
1658:
1648:
1606:
542:
1302:
1259:
1191:
1121:
997:
948:
921:
1384:
1343:
1218:
1046:
956:
862:
735:
700:
609:
1753:
1525:
676:. Vol. 4 (2nd ed.). New York: Academic Press. pp. 369–378.
489:
485:
480:
357:
232:
1250:
1233:
988:
971:
853:
836:
768:
658:
116:
1736:
1663:
1653:
1640:
808:
243:
135:
1062:"Human Lysosomal a-Glucosidase Characterization of The Catalytic Site"
1028:
640:
497:. Flavonoid analogues have been demonstrated with inhibition activity.
2218:
1988:
1448:
1359:"α-Glucosidase inhibitors as potential broad based anti-viral agents"
336:
199:
111:
99:
87:
463:: a disorder in which α-glucosidase is deficient. In 2006, the drug
228:
350:
is the "maltase-glucoamylase", found on the intestine brush border.
2192:
1915:
1696:
1234:"Inhibition of α-glucosidase and amylase by luteolin, a flavonoid"
426:
392:
1893:
1888:
1849:
1778:
1773:
1768:
1763:
1713:
1701:
363:
347:
123:
1961:
1579:
1406:
39:
1318:"Seminal plasma α-glucosidase activity and male infertility"
1013:"Characterization of the human lysosomal α-glucosidase gene"
750:"alpha-glucosidase activity Gene Ontology Term (GO:0090599)"
1316:
Mahmoud AM, Geslevich J, Kint J, et al. (March 1998).
1957:
970:
Tadera K, Minami Y, Takamatsu K, Matsuoka T (April 2006).
376:-glucosidase, α-glucoside hydrolase, α-1,4-glucosidase, α-
972:"Inhibition of α-glucosidase and α-amylase by flavonoids"
876:
Mury FB, da Silva JR, Ferreira LS, et al. (2009).
837:"Molecular mechanism in α-glucosidase and glucoamylase"
476:
mechanism by which α-glucosidase binds to imino sugars.
2248:
279:
Hydrolysis of terminal, non-reducing (1→4)-linked α-
275:
of the small intestine that acts upon α(1→4) bonds:
2144:
2108:
2077:
2046:
1995:
1930:
1729:
1672:
1631:
1618:
1552:
1524:
1498:
1491:
1455:
210:
198:
186:
181:
158:
146:
134:
122:
110:
98:
86:
81:
69:
57:
52:
32:
1144:"FDA Approves First Treatment for Pompe Disease"
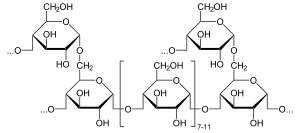
397:Example of an α-glucosidase catalyzed reaction:
431:α-glucosidase in complex with maltose and NAD+
356:is the "acid alpha-glucosidase", found in the
1973:
1591:
1418:
370:Synonyms mentioned by the Commission include
8:
335:(as opposed to larger polysaccharides like
1980:
1966:
1958:
1628:
1598:
1584:
1576:
1495:
1425:
1411:
1403:
178:
38:
1374:
1333:
1249:
1232:Kim JS, Kwon CS, Son KH (November 2000).
1077:
1036:
987:
911:
901:
852:
648:
574:at the U.S. National Library of Medicine
830:
828:
826:
238:
227:
2255:
623:Flanagan, P.R.; Forstner, G.G. (1978).
564:
29:
769:"ENZYME - 3.2.1.20 alpha-glucosidase"
7:
1283:Bioorganic & Medicinal Chemistry
1066:The Journal of Biological Chemistry
728:10.1111/j.1432-1033.1982.tb06817.x
283:-glucose residues with release of
25:
2258:
456:Glycogen storage disease type II
366:, "neutral alpha-glucosidase C".
1862:Alpha-N-acetylgalactosaminidase
327:The definition associated with
758:See: Definition, GO Tree View.
1:
1874:Alpha-N-acetylglucosaminidase
1468:UDP-glucose pyrophosphorylase
1376:10.1016/S0014-5793(98)00525-0
1079:10.1016/S0021-9258(18)92727-4
323:EC 3.2.1.20, the narrow sense
903:10.1371/journal.pone.0006966
602:10.1016/0005-2744(70)90253-6
1238:Biosci. Biotechnol. Biochem
841:Biosci. Biotechnol. Biochem
553:Glycogen debranching enzyme
533:Alpha-glucosidase inhibitor
310:GO:0090599, the broad sense
2297:
689:Indian J. Biochem. Biophys
2136:Michaelis–Menten kinetics
1478:Glycogen branching enzyme
1295:10.1016/j.bmc.2017.07.055
1184:10.1016/j.cca.2008.02.014
1114:10.1007/s10930-009-9213-0
538:Some other glucosidases:
380:-glucoside glucohydrolase
265:-glucoside glucohydrolase
177:
37:
2028:Diffusion-limited enzyme
791:"ExplorEnz: EC 3.2.1.20"
576:Medical Subject Headings
329:Enzyme Commission number
1784:Bacterial neuraminidase
1438:carbohydrate metabolism
1335:10.1093/humrep/13.3.591
976:J. Nutr. Sci. Vitaminol
835:Chiba S (August 1997).
795:www.enzyme-database.org
754:www.informatics.jax.org
291:This is in contrast to
46:Sulfolobus solfataricus
44:α-Glucosidase hexamer,
1947:Oxoguanine glycosylase
1506:Glycogen phosphorylase
949:10.1006/abbi.1993.1499
937:Arch. Biochem. Biophys
590:Biochim. Biophys. Acta
432:
402:
246:
236:
2121:Eadie–Hofstee diagram
2054:Allosteric regulation
430:
396:
242:
231:
2131:Lineweaver–Burk plot
1936:N-Glycosyl compounds
1906:Maltase-glucoamylase
1857:Galactosylceramidase
1624:Glycoside hydrolases
1609:: sugar hydrolases (
1560:Phosphorylase kinase
1789:Viral neuraminidase
1565:Protein phosphatase
1251:10.1271/bbb.64.2458
1072:(21): 13507–13512.
989:10.3177/jnsv.52.149
894:2009PLoSO...4.6966M
854:10.1271/bbb.61.1233
401:+ water → α-glucose
259:, (systematic name
2090:Enzyme superfamily
2023:Enzyme promiscuity
1840:Glucosylceramidase
1721:Debranching enzyme
1645:Sucrase-isomaltase
1516:Phosphoglucomutase
1511:Debranching enzyme
1463:Phosphoglucomutase
572:alpha-Glucosidases
528:Alglucosidase alfa
469:alglucosidase alfa
465:alglucosidase alfa
433:
403:
247:
237:
235:structure segment.
2246:
2245:
1955:
1954:
1926:
1925:
1813:alpha-Mannosidase
1687:Alpha-glucosidase
1573:
1572:
1548:
1547:
1535:Alpha-glucosidase
1473:Glycogen synthase
1029:10.1042/bj2720493
773:enzyme.expasy.org
641:10.1042/bj1730553
510:Antiviral agents:
449:Disease relevance
408:Rhodnius prolixus
379:
375:
286:
282:
264:
226:
225:
222:
221:
141:metabolic pathway
18:Alpha-glucosidase
16:(Redirected from
2288:
2263:
2262:
2254:
2126:Hanes–Woolf plot
2069:Enzyme activator
2064:Enzyme inhibitor
2038:Enzyme catalysis
1982:
1975:
1968:
1959:
1943:DNA glycosylases
1709:Beta-glucosidase
1629:
1600:
1593:
1586:
1577:
1496:
1427:
1420:
1413:
1404:
1397:
1396:
1378:
1354:
1348:
1347:
1337:
1313:
1307:
1306:
1278:
1272:
1271:
1253:
1229:
1223:
1222:
1202:
1196:
1195:
1166:
1160:
1159:
1157:
1155:
1148:FDA News Release
1140:
1134:
1133:
1108:(9–10): 448–56.
1097:
1091:
1090:
1088:
1086:
1081:
1057:
1051:
1050:
1040:
1008:
1002:
1001:
991:
967:
961:
960:
932:
926:
925:
915:
905:
873:
867:
866:
856:
832:
821:
820:
818:
816:
805:
799:
798:
787:
781:
780:
765:
759:
757:
746:
740:
739:
711:
705:
704:
684:
678:
677:
669:
663:
662:
652:
620:
614:
613:
585:
579:
569:
548:Beta-glucosidase
377:
373:
333:oligosaccharides
284:
280:
262:
179:
42:
30:
21:
2296:
2295:
2291:
2290:
2289:
2287:
2286:
2285:
2271:
2270:
2269:
2257:
2249:
2247:
2242:
2154:Oxidoreductases
2140:
2116:Enzyme kinetics
2104:
2100:List of enzymes
2073:
2042:
2013:Catalytic triad
1991:
1986:
1956:
1951:
1935:
1922:
1725:
1668:
1614:
1604:
1574:
1569:
1544:
1520:
1499:extralysosomal:
1487:
1451:
1431:
1401:
1400:
1356:
1355:
1351:
1315:
1314:
1310:
1289:(20): 5355–64.
1280:
1279:
1275:
1244:(11): 2458–61.
1231:
1230:
1226:
1207:Clin Invest Med
1204:
1203:
1199:
1168:
1167:
1163:
1153:
1151:
1142:
1141:
1137:
1099:
1098:
1094:
1084:
1082:
1059:
1058:
1054:
1010:
1009:
1005:
969:
968:
964:
934:
933:
929:
875:
874:
870:
834:
833:
824:
814:
812:
807:
806:
802:
789:
788:
784:
767:
766:
762:
748:
747:
743:
716:Eur. J. Biochem
713:
712:
708:
686:
685:
681:
671:
670:
666:
622:
621:
617:
587:
586:
582:
570:
566:
561:
524:
451:
425:
388:
325:
312:
307:
271:located in the
48:
28:
23:
22:
15:
12:
11:
5:
2294:
2292:
2284:
2283:
2273:
2272:
2268:
2267:
2244:
2243:
2241:
2240:
2227:
2214:
2201:
2188:
2175:
2162:
2148:
2146:
2142:
2141:
2139:
2138:
2133:
2128:
2123:
2118:
2112:
2110:
2106:
2105:
2103:
2102:
2097:
2092:
2087:
2081:
2079:
2078:Classification
2075:
2074:
2072:
2071:
2066:
2061:
2056:
2050:
2048:
2044:
2043:
2041:
2040:
2035:
2030:
2025:
2020:
2015:
2010:
2005:
1999:
1997:
1993:
1992:
1987:
1985:
1984:
1977:
1970:
1962:
1953:
1952:
1950:
1949:
1939:
1937:
1928:
1927:
1924:
1923:
1921:
1920:
1919:
1918:
1908:
1903:
1898:
1897:
1896:
1891:
1884:Hexosaminidase
1881:
1876:
1871:
1870:
1869:
1859:
1854:
1853:
1852:
1847:
1837:
1832:
1827:
1826:
1825:
1815:
1810:
1809:
1808:
1803:
1796:Galactosidases
1793:
1792:
1791:
1786:
1781:
1776:
1771:
1766:
1756:
1751:
1746:
1745:
1744:
1733:
1731:
1727:
1726:
1724:
1723:
1718:
1717:
1716:
1706:
1705:
1704:
1699:
1694:
1684:
1678:
1676:
1670:
1669:
1667:
1666:
1661:
1656:
1651:
1637:
1635:
1633:Disaccharidase
1626:
1616:
1615:
1605:
1603:
1602:
1595:
1588:
1580:
1571:
1570:
1568:
1567:
1562:
1556:
1554:
1550:
1549:
1546:
1545:
1543:
1542:
1531:
1529:
1522:
1521:
1519:
1518:
1513:
1508:
1502:
1500:
1493:
1492:Glycogenolysis
1489:
1488:
1486:
1485:
1480:
1475:
1470:
1465:
1459:
1457:
1453:
1452:
1446:glycogenolysis
1432:
1430:
1429:
1422:
1415:
1407:
1399:
1398:
1369:(1–2): 17–22.
1349:
1308:
1273:
1224:
1197:
1178:(1–2): 68–73.
1172:Clin Chim Acta
1161:
1135:
1092:
1052:
1023:(2): 493–497.
1003:
962:
927:
868:
822:
800:
782:
760:
741:
722:(3): 559–568.
706:
695:(4): 283–284.
679:
664:
635:(2): 553–563.
615:
596:(3): 470–477.
580:
563:
562:
560:
557:
556:
555:
550:
545:
536:
535:
530:
523:
520:
519:
518:
507:
498:
477:
471:revealed that
459:, also called
450:
447:
446:
445:
441:
424:
421:
420:
419:
416:
412:
387:
384:
368:
367:
361:
351:
324:
321:
311:
308:
306:
303:
289:
288:
224:
223:
220:
219:
214:
208:
207:
202:
196:
195:
190:
184:
183:
175:
174:
163:
156:
155:
150:
144:
143:
138:
132:
131:
126:
120:
119:
114:
108:
107:
102:
96:
95:
90:
84:
83:
79:
78:
73:
67:
66:
61:
55:
54:
50:
49:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2293:
2282:
2279:
2278:
2276:
2266:
2261:
2256:
2252:
2238:
2234:
2233:
2228:
2225:
2221:
2220:
2215:
2212:
2208:
2207:
2202:
2199:
2195:
2194:
2189:
2186:
2182:
2181:
2176:
2173:
2169:
2168:
2163:
2160:
2156:
2155:
2150:
2149:
2147:
2143:
2137:
2134:
2132:
2129:
2127:
2124:
2122:
2119:
2117:
2114:
2113:
2111:
2107:
2101:
2098:
2096:
2095:Enzyme family
2093:
2091:
2088:
2086:
2083:
2082:
2080:
2076:
2070:
2067:
2065:
2062:
2060:
2059:Cooperativity
2057:
2055:
2052:
2051:
2049:
2045:
2039:
2036:
2034:
2031:
2029:
2026:
2024:
2021:
2019:
2018:Oxyanion hole
2016:
2014:
2011:
2009:
2006:
2004:
2001:
2000:
1998:
1994:
1990:
1983:
1978:
1976:
1971:
1969:
1964:
1963:
1960:
1948:
1944:
1941:
1940:
1938:
1934:: Hydrolysing
1933:
1929:
1917:
1914:
1913:
1912:
1909:
1907:
1904:
1902:
1899:
1895:
1892:
1890:
1887:
1886:
1885:
1882:
1880:
1877:
1875:
1872:
1868:
1865:
1864:
1863:
1860:
1858:
1855:
1851:
1850:non-lysosomal
1848:
1846:
1843:
1842:
1841:
1838:
1836:
1833:
1831:
1830:Hyaluronidase
1828:
1824:
1821:
1820:
1819:
1818:Glucuronidase
1816:
1814:
1811:
1807:
1804:
1802:
1799:
1798:
1797:
1794:
1790:
1787:
1785:
1782:
1780:
1777:
1775:
1772:
1770:
1767:
1765:
1762:
1761:
1760:
1759:Neuraminidase
1757:
1755:
1752:
1750:
1747:
1743:
1742:Alpha-amylase
1740:
1739:
1738:
1735:
1734:
1732:
1728:
1722:
1719:
1715:
1712:
1711:
1710:
1707:
1703:
1700:
1698:
1695:
1693:
1690:
1689:
1688:
1685:
1683:
1680:
1679:
1677:
1675:
1671:
1665:
1662:
1660:
1657:
1655:
1652:
1650:
1646:
1642:
1639:
1638:
1636:
1634:
1630:
1627:
1625:
1621:
1617:
1612:
1608:
1601:
1596:
1594:
1589:
1587:
1582:
1581:
1578:
1566:
1563:
1561:
1558:
1557:
1555:
1551:
1540:
1536:
1533:
1532:
1530:
1527:
1523:
1517:
1514:
1512:
1509:
1507:
1504:
1503:
1501:
1497:
1494:
1490:
1484:
1481:
1479:
1476:
1474:
1471:
1469:
1466:
1464:
1461:
1460:
1458:
1454:
1450:
1447:
1443:
1439:
1435:
1428:
1423:
1421:
1416:
1414:
1409:
1408:
1405:
1394:
1390:
1386:
1382:
1377:
1372:
1368:
1364:
1360:
1353:
1350:
1345:
1341:
1336:
1331:
1327:
1323:
1319:
1312:
1309:
1304:
1300:
1296:
1292:
1288:
1284:
1277:
1274:
1269:
1265:
1261:
1257:
1252:
1247:
1243:
1239:
1235:
1228:
1225:
1220:
1216:
1213:(4): 303–11.
1212:
1208:
1201:
1198:
1193:
1189:
1185:
1181:
1177:
1173:
1165:
1162:
1149:
1145:
1139:
1136:
1131:
1127:
1123:
1119:
1115:
1111:
1107:
1103:
1096:
1093:
1080:
1075:
1071:
1067:
1063:
1056:
1053:
1048:
1044:
1039:
1034:
1030:
1026:
1022:
1018:
1014:
1007:
1004:
999:
995:
990:
985:
982:(2): 149–53.
981:
977:
973:
966:
963:
958:
954:
950:
946:
943:(1): 188–94.
942:
938:
931:
928:
923:
919:
914:
909:
904:
899:
895:
891:
887:
883:
879:
872:
869:
864:
860:
855:
850:
847:(8): 1233–9.
846:
842:
838:
831:
829:
827:
823:
810:
809:"EC 3.2.1.20"
804:
801:
796:
792:
786:
783:
779:
774:
770:
764:
761:
755:
751:
745:
742:
737:
733:
729:
725:
721:
717:
710:
707:
702:
698:
694:
690:
683:
680:
675:
668:
665:
660:
656:
651:
646:
642:
638:
634:
630:
626:
619:
616:
611:
607:
603:
599:
595:
591:
584:
581:
577:
573:
568:
565:
558:
554:
551:
549:
546:
544:
541:
540:
539:
534:
531:
529:
526:
525:
521:
516:
511:
508:
505:
503:
499:
496:
491:
487:
484:
482:
478:
474:
470:
466:
462:
461:Pompe disease
458:
457:
453:
452:
448:
442:
438:
437:
436:
429:
422:
417:
413:
410:
409:
405:
404:
400:
395:
391:
385:
383:
381:
365:
362:
359:
355:
352:
349:
346:
345:
344:
342:
341:alpha-amylase
338:
334:
330:
322:
320:
317:
316:Gene Ontology
309:
304:
302:
300:
299:β-glucosidase
297:
294:
278:
277:
276:
274:
270:
266:
258:
255:
251:
250:α-Glucosidase
245:
241:
234:
230:
218:
215:
213:
209:
206:
203:
201:
197:
194:
191:
189:
185:
180:
176:
173:
170:
167:
164:
161:
157:
154:
151:
149:
145:
142:
139:
137:
133:
130:
127:
125:
121:
118:
117:NiceZyme view
115:
113:
109:
106:
103:
101:
97:
94:
91:
89:
85:
80:
77:
74:
72:
68:
65:
62:
60:
56:
51:
47:
41:
36:
33:α-Glucosidase
31:
19:
2232:Translocases
2229:
2216:
2203:
2190:
2177:
2167:Transferases
2164:
2151:
2008:Binding site
1674:Glucosidases
1456:Glycogenesis
1442:glycogenesis
1366:
1363:FEBS Letters
1362:
1352:
1328:(3): 591–5.
1325:
1321:
1311:
1286:
1282:
1276:
1241:
1237:
1227:
1210:
1206:
1200:
1175:
1171:
1164:
1152:. Retrieved
1147:
1138:
1105:
1101:
1095:
1083:. Retrieved
1069:
1065:
1055:
1020:
1016:
1006:
979:
975:
965:
940:
936:
930:
888:(9): e6966.
885:
881:
871:
844:
840:
813:. Retrieved
803:
794:
785:
776:
772:
763:
753:
744:
719:
715:
709:
692:
688:
682:
673:
667:
632:
628:
618:
593:
589:
583:
567:
537:
514:
509:
500:
495:cell culture
479:
460:
454:
434:
406:
389:
371:
369:
326:
313:
290:
273:brush border
260:
249:
248:
105:BRENDA entry
45:
2003:Active site
1901:Iduronidase
1835:Pullulanase
1322:Hum. Reprod
674:The Enzymes
502:Azoospermia
473:iminosugars
399:maltotriose
305:Terminology
269:glucosidase
93:IntEnz view
53:Identifiers
2206:Isomerases
2180:Hydrolases
2047:Regulation
1911:Heparanase
1879:Fucosidase
1697:Neutral AB
1553:Regulation
1483:Glycogenin
1434:Metabolism
1017:Biochem. J
629:Biochem. J
559:References
162:structures
129:KEGG entry
76:9001-42-7
2085:EC number
1845:lysosomal
1749:Chitinase
1714:cytosolic
1702:Neutral C
1682:Cellulase
1659:Trehalase
1649:Invertase
1607:Hydrolase
1526:lysosomal
1102:Protein J
543:Cellulase
423:Structure
386:Mechanism
82:Databases
2281:EC 3.2.1
2275:Category
2109:Kinetics
2033:Cofactor
1996:Activity
1754:Lysozyme
1393:25156942
1303:28797772
1260:11193416
1192:18328816
1130:36546023
1122:19921411
998:16802696
922:19742319
882:PLOS ONE
811:. ExPASy
522:See also
490:Luteolin
486:Acarbose
481:Diabetes
358:lysosome
296:3.2.1.21
287:-glucose
257:3.2.1.20
233:Glycogen
217:proteins
205:articles
193:articles
166:RCSB PDB
64:3.2.1.20
2265:Biology
2219:Ligases
1989:Enzymes
1737:Amylase
1664:Lactase
1654:Maltase
1641:Sucrase
1449:enzymes
1385:9678587
1344:9572418
1268:5757649
1219:8549017
1154:1 March
1085:1 March
1047:2268276
1038:1149727
957:8215402
913:2734994
890:Bibcode
863:9301101
815:1 March
736:6814909
701:4792946
650:1185809
610:5466143
267:) is a
244:Maltose
153:profile
136:MetaCyc
71:CAS no.
2251:Portal
2193:Lyases
1823:Klotho
1391:
1383:
1342:
1301:
1266:
1258:
1217:
1190:
1128:
1120:
1068:. 21.
1045:
1035:
996:
955:
920:
910:
861:
734:
699:
657:
647:
608:
578:(MeSH)
337:starch
200:PubMed
182:Search
172:PDBsum
112:ExPASy
100:BRENDA
88:IntEnz
59:EC no.
27:Enzyme
2145:Types
1932:3.2.2
1916:HPSE2
1801:Alpha
1730:Other
1620:3.2.1
1389:S2CID
1264:S2CID
1150:. FDA
1126:S2CID
659:29602
148:PRIAM
2237:list
2230:EC7
2224:list
2217:EC6
2211:list
2204:EC5
2198:list
2191:EC4
2185:list
2178:EC3
2172:list
2165:EC2
2159:list
2152:EC1
1894:HEXB
1889:HEXA
1867:NAGA
1806:Beta
1779:NEU4
1774:NEU3
1769:NEU2
1764:NEU1
1692:Acid
1613:3.2)
1539:Acid
1444:and
1381:PMID
1340:PMID
1299:PMID
1256:PMID
1215:PMID
1188:PMID
1156:2012
1118:PMID
1087:2012
1043:PMID
994:PMID
953:PMID
918:PMID
859:PMID
817:2012
778:all.
732:PMID
697:PMID
655:PMID
606:PMID
364:GANC
348:MGAM
314:The
212:NCBI
169:PDBe
124:KEGG
1371:doi
1367:430
1330:doi
1291:doi
1246:doi
1180:doi
1176:391
1110:doi
1074:doi
1070:266
1033:PMC
1025:doi
1021:272
984:doi
945:doi
941:306
908:PMC
898:doi
849:doi
724:doi
720:126
645:PMC
637:doi
633:173
598:doi
594:212
354:GAA
188:PMC
160:PDB
2277::
1945::
1622::
1611:EC
1440:,
1436::
1387:.
1379:.
1365:.
1361:.
1338:.
1326:13
1324:.
1320:.
1297:.
1287:25
1285:.
1262:.
1254:.
1242:64
1240:.
1236:.
1211:18
1209:.
1186:.
1174:.
1146:.
1124:.
1116:.
1106:28
1104:.
1064:.
1041:.
1031:.
1019:.
1015:.
992:.
980:52
978:.
974:.
951:.
939:.
916:.
906:.
896:.
884:.
880:.
857:.
845:61
843:.
839:.
825:^
793:.
775:.
771:.
752:.
730:.
718:.
693:10
691:.
653:.
643:.
631:.
627:.
604:.
592:.
339::
301:.
293:EC
261:α-
254:EC
2253::
2239:)
2235:(
2226:)
2222:(
2213:)
2209:(
2200:)
2196:(
2187:)
2183:(
2174:)
2170:(
2161:)
2157:(
1981:e
1974:t
1967:v
1647:/
1643:/
1599:e
1592:t
1585:v
1541:)
1537:(
1528::
1426:e
1419:t
1412:v
1395:.
1373::
1346:.
1332::
1305:.
1293::
1270:.
1248::
1221:.
1194:.
1182::
1158:.
1132:.
1112::
1089:.
1076::
1049:.
1027::
1000:.
986::
959:.
947::
924:.
900::
892::
886:4
865:.
851::
819:.
797:.
756:.
738:.
726::
703:.
661:.
639::
612:.
600::
515:N
504::
483::
378:D
374:D
360:.
285:D
281:D
263:D
252:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.