142:
25:
256:
490:), where they have little discernible impact on the bearer. Mutations in the introns (or non-coding regions of RNA) have little or no effect on phenotype of an individual if the coding portion of individual's genome does not contain mutations. The Alu insertions that can be detrimental to the human body are inserted into coding regions (
225:
back 65 million years, the AluJ lineage is the oldest and least active in the human genome. The younger AluS lineage is about 30 million years old and still contains some active elements. Finally, the AluY elements are the youngest of the three and have the greatest disposition to move along the human genome. The discovery of
2097:
Nyström-Lahti, Minna; Kristo, Paula; Nicolaides, Nicholas C; Chang, Sheng-Yung; Aaltonen, Lauri A; Moisio, Anu-Liisa; Järvinen, Heikki J; Mecklin, Jukka-Pekka; Kinzler, Kenneth W; Vogelstein, Bert; de la
Chapelle, Albert; Peltomäki, Päivi (1995). "Founding mutations and Alu-mediated recombination in
434:
element insertion if they have a common ancestor. This is because insertion of an Alu element occurs only 100 - 200 times per million years, and no known mechanism of deletion of one has been found. Therefore, individuals with an element likely descended from an ancestor with one—and vice versa, for
188:
elements emerged from a head to tail fusion of two distinct FAMs (fossil antique monomers) over 100 million years ago, hence its dimeric structure of two similar, but distinct monomers (left and right arms) joined by an A-rich linker. Both monomers are thought to have evolved from 7SL, also known as
2348:
Cortese, A.; Simone, R.; Sullivan, R.; Vandrovcova, J.; Tariq, H.; Yau, W. Y.; Humphrey, J.; Jaunmuktane, Z.; Sivakumar, P.; Polke, J.; Ilyas, M.; Tribollet, E.; Tomaselli, P. J.; Devigili, G.; Callegari, I.; Versino, M.; Salpietro, V.; Efthymiou, S.; Kaski, D.; Wood, N. W.; Andrade, N. S.; Buglo,
224:
elements were split in two major subfamilies known as AluJ (named after Jurka) and AluS (named after Smith), and other Alu subfamilies were also independently discovered by several groups. Later on, a sub-subfamily of AluS which included active Alu elements was given the separate name AluY. Dating
238:
B1 elements in rats and mice are similar to Alus in that they also evolved from 7SL RNA, but they only have one left monomer arm. 95% percent of human Alus are also found in chimpanzees, and 50% of B elements in mice are also found in rats. These elements are mostly found in introns and upstream
542:. In the human genome, the most recently active have been the 22 AluY and 6 AluS Transposon Element subfamilies due to their inherited activity to cause various cancers. Thus due to their major heritable damage it is important to understand the causes that affect their transpositional activity.
1849:
Lander, Eric S; Linton, Lauren M; Birren, Bruce; Nusbaum, Chad; Zody, Michael C; Baldwin, Jennifer; Devon, Keri; Dewar, Ken; Doyle, Michael; Fitzhugh, William; Funke, Roel; Gage, Diane; Harris, Katrina; Heaford, Andrew; Howland, John; Kann, Lisa; Lehoczky, Jessica; Levine, Rosie; McEwan, Paul;
184:(SINEs) among the class of repetitive RNA elements. The typical structure is 5' - Part A - A5TACA6 - Part B - PolyA Tail - 3', where Part A and Part B (also known as "left arm" and "right arm") are similar nucleotide sequences. Expressed another way, it is believed modern
242:
The ancestral form of Alu and B1 is the fossil Alu monomer (FAM). Free-floating forms of the left and right arms exist, termed free left Alu monomers (FLAMs) and free right Alu monomers (FRAMs) respectively. A notable FLAM in primates is the
2243:
Fukuda, Shinichi; Varshney, Akhil; Fowler, Benjamin J.; Wang, Shao-bin; Narendran, Siddharth; Ambati, Kameshwari; Yasuma, Tetsuhiro; Magagnoli, Joseph; Leung, Hannah; Hirahara, Shuichiro; Nagasaka, Yosuke (2021-02-09).
229:
subfamilies led to the hypothesis of master/source genes, and provided the definitive link between transposable elements (active elements) and interspersed repetitive DNA (mutated copies of active elements).
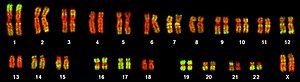
152:(46, XX). Chromosomes were hybridized with a probe for Alu elements (green) and counterstained with TOPRO-3 (red). Alu elements were used as a marker for chromosomes and chromosome bands rich in genes.
1850:
McKernan, Kevin; Meldrim, James; Mesirov, Jill P; Miranda, Cher; Morris, William; Naylor, Jerome; Raymond, Christina; Rosetti, Mark; Santos, Ralph; Sheridan, Andrew; et al. (2001).
89:
elements were thought to be selfish or parasitic DNA, because their sole known function is self reproduction. However, they are likely to play a role in evolution and have been used as
360:
elements are responsible for regulation of tissue-specific genes. They are also involved in the transcription of nearby genes and can sometimes change the way a gene is expressed.
925:
Arcot, Santosh S.; Wang, Zhenyuan; Weber, James L.; Deininger, Prescott L.; Batzer, Mark A. (September 1995). "Alu
Repeats: A Source for the Genesis of Primate Microsatellites".
683:
element is associated with better performance in endurance-oriented events (e.g. triathlons), whereas its absence is associated with strength- and power-oriented performance.
426:
element insertion events have a characteristic signature that is both easy to read and faithfully recorded in the genome from generation to generation. The study of
890:
Kriegs, Jan Ole; Churakov, Gennady; Jurka, Jerzy; Brosius, Jürgen; Schmitz, Jürgen (2007). "Evolutionary history of 7SL RNA-derived SINEs in
Supraprimates".
2536:
2398:
Puthucheary, Zudin; Skipworth, James RA; Rawal, Jai; Loosemore, Mike; Van
Someren, Ken; Montgomery, Hugh E (2011). "The ACE Gene and Human Performance".
305:) RNA example below, functional hexamers are underlined using a solid line, with the non-functional third hexamer denoted using a dotted line:
694:
2246:"Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration"
486:
elements are also a common source of mutations in humans; however, such mutations are often confined to non-coding regions of pre-mRNA (
497:
However, the variation generated can be used in studies of the movement and ancestry of human populations, and the mutagenic effect of
2722:
2709:
181:
501:
and retrotransposons in general has played a major role in the evolution of the human genome. There are also a number of cases where
3025:
2732:
379:
430:
elements (the more recently evolved) thus reveals details of ancestry because individuals will most likely only share a particular
1751:
Norris, J; Fan, D; Aleman, C; Marks, J. R; Futreal, P. A; Wiseman, R. W; Iglehart, J. D; Deininger, P. L; McDonnell, D. P (1995).
1753:"Identification of a new subclass of Alu DNA repeats that can function as estrogen receptor-dependent transcriptional enhancers"
1233:"Alu and B1 Repeats Have Been Selectively Retained in the Upstream and Intronic Regions of Genes of Specific Functional Classes"
2529:
611:
302:
864:
2763:
2609:
2596:
2349:
E.; Rebelo, A.; Rossor, A. M.; Bronstein, A.; Fratta, P.; Marques, W. J.; Züchner, S.; Reilly, M. M.; Houlden, H. (2019).
660:
578:
1020:
Roy-Engel, A. M; Carroll, M. L; Vogel, E; Garber, R. K; Nguyen, S. V; Salem, A. H; Batzer, M. A; Deininger, P. L (2001).
2319:
204:
elements interspersed throughout the human genome, and it is estimated that about 10.7% of the human genome consists of
820:
Kidwell, Margaret G; Lisch, Damon R (2001). "Perspective: Transposable
Elements, Parasitic Dna, and Genome Evolution".
396:
RNA forms a specific RNA:protein complex with a protein heterodimer consisting of SRP9 and SRP14. SRP9/14 facilitates
389:
98:
1645:
Terreros, Maria C.; Alfonso-Sanchez, Miguel A.; Novick; Luis; Lacau; Lowery; Regueiro; Herrera (September 11, 2009).
2880:
2522:
2768:
568:
443:
element insertions can be found in the corresponding positions in the genomes of other primates, but about 7,000
833:
563:
2496:
467:
70:
3030:
2574:
2549:
2514:
1989:
Shen, S; Lin, L; Cai, J. J; Jiang, P; Kenkel, E. J; Stroik, M. R; Sato, S; Davidson, B. L; Xing, Y (2011).
3004:
2932:
2825:
2443:"The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates"
634:
The following disease have been associated with repeat expansion of AAGGG pentamere in Alu element :
618:
378:
elements do not encode for protein products. They are replicated as any other DNA sequence, but depend on
209:
2850:
2800:
1851:
593:
532:
409:
279:
2860:
1131:
Bennett, E. A; Keller, H; Mills, R. E; Schmidt, S; Moran, J. V; Weichenrieder, O; Devine, S. E (2008).
141:
2982:
2900:
1385:"The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element"
1333:"Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data"
1180:
Richard Shen, M; Batzer, Mark A; Deininger, Prescott L (1991). "Evolution of the master Alu gene(s)".
2977:
2890:
2805:
2691:
2553:
2442:
2257:
2002:
1866:
1707:
1603:
1506:
1451:
1396:
1244:
1189:
1082:
604:
78:
2492:
119:
insertions have been implicated in several inherited human diseases and in various forms of cancer.
2755:
2626:
2143:"Computational identification of harmful mutation regions to the activity of transposable elements"
463:
337:
123:
65:
2927:
2835:
2810:
2654:
2472:
2423:
2301:
2123:
1971:
1925:
1782:
1733:
1676:
1627:
1475:
1213:
845:
802:
755:
371:
298:
287:
573:
2905:
2464:
2415:
2380:
2293:
2275:
2225:
2174:
2115:
2079:
2030:
1963:
1917:
1882:
1831:
1774:
1725:
1668:
1619:
1569:
1534:
1467:
1424:
1362:
1313:
1272:
1205:
1162:
1110:
1051:
1002:
950:
942:
907:
837:
794:
747:
599:
538:
471:
2870:
2820:
2815:
2989:
2919:
2845:
2795:
2557:
2454:
2407:
2370:
2362:
2283:
2265:
2215:
2205:
2164:
2154:
2107:
2069:
2061:
2020:
2010:
1955:
1909:
1874:
1821:
1813:
1764:
1715:
1658:
1611:
1561:
1524:
1514:
1459:
1414:
1404:
1352:
1344:
1303:
1262:
1252:
1197:
1152:
1144:
1100:
1090:
1041:
1033:
992:
984:
934:
899:
829:
786:
739:
690:
642:
2840:
2745:
730:
Schmid, Carl W; Deininger, Prescott L (1975). "Sequence organization of the human genome".
2865:
2700:
2672:
2641:
2636:
2351:"Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia"
1331:
Conti, A; Carnevali, D; Bollati, V; Fustinoni, S; Pellegrini, M; Dieci, G (January 2015).
553:
520:
variation acts as markers that segregate with the disease so the presence of a particular
479:
459:
367:
290:, have a similar but stronger promoter structure. Both boxes are located in the left arm.
135:
1946:
Batzer, Mark A; Deininger, Prescott L (2002). "Alu
Repeats and Human Genomic Diversity".
516:
insertions are sometimes disruptive and can result in inherited disorders. However, most
2261:
2006:
1870:
1711:
1607:
1510:
1455:
1400:
1248:
1193:
1086:
679:-D). This variation has been linked to changes in sporting ability: the presence of the
38:
Please help update this article to reflect recent events or newly available information.
2972:
2830:
2787:
2615:
2605:
2375:
2350:
2288:
2245:
2220:
2193:
2169:
2142:
2074:
2049:
2025:
1990:
1592:"Structure and assembly of the Alu domain of the mammalian signal recognition particle"
1357:
1332:
1267:
1232:
1157:
1132:
1046:
1021:
997:
972:
628:
588:
90:
1826:
1801:
1565:
1105:
1070:
422:
elements in primates form a fossil record that is relatively easy to decipher because
3019:
2937:
2677:
2600:
2591:
2583:
2411:
2305:
1975:
1529:
1494:
1419:
1384:
743:
558:
527:
does not mean that the carrier will definitely get the disease. The first report of
439:
element may be a good property to consider when studying human evolution. Most human
294:
110:
2476:
2427:
2127:
1929:
1786:
1217:
849:
806:
759:
2949:
2503:
1737:
1680:
1631:
1479:
244:
217:
82:
341:
endonuclease is 5' ag/ct 3'; that is, the enzyme cuts the DNA segment between the
1257:
1037:
2944:
2855:
1696:"Initial sequence of the chimpanzee genome and comparison with the human genome"
1590:
Weichenrieder, Oliver; Wild, Klemens; Strub, Katharina; Cusack, Stephen (2000).
698:
623:
435:
those without. In genetics, the presence or lack thereof of a recently inserted
213:
1900:
Deininger, Prescott L; Batzer, Mark A (1999). "Alu
Repeats and Human Disease".
1499:
Proceedings of the
National Academy of Sciences of the United States of America
1231:
Tsirigos, Aristotelis; Rigoutsos, Isidore; Stormo, Gary D. (18 December 2009).
535:
causing a prevalent inherited predisposition to cancer was a 1995 report about
392:(SRPs), which aid newly translated proteins to reach their final destinations.
255:
2956:
2773:
2366:
2210:
2159:
1591:
903:
702:
583:
149:
2327:
2279:
1817:
1769:
1752:
1552:
Kramerov, D; Vassetzky, N (2005). "Short
Retroposons in Eukaryotic Genomes".
946:
773:
Szmulewicz, Martin N; Novick, Gabriel E; Herrera, Rene J (1998). "Effects of
2875:
2566:
2270:
2015:
1991:"Widespread establishment and regulatory impact of Alu exons in human genes"
1519:
1409:
1308:
1291:
1095:
790:
177:
145:
127:
2468:
2419:
2384:
2297:
2229:
2178:
2083:
2034:
1967:
1921:
1913:
1886:
1729:
1672:
1623:
1573:
1366:
1317:
1276:
1166:
1055:
1006:
938:
911:
841:
2119:
1835:
1778:
1538:
1471:
1428:
1348:
1209:
1148:
1114:
954:
798:
751:
482:, contributing to up to 30% of the methylation sites in the human genome.
270:
Two main promoter "boxes" are found in Alu: a 5' A box with the consensus
2999:
2885:
2649:
2545:
1647:"Insights on human evolution: an analysis of Alu insertion polymorphisms"
988:
346:
2111:
1720:
1695:
1663:
1646:
505:
insertions or deletions are associated with specific effects in humans:
2895:
2459:
1442:
Ullu E, Tschudi C (1984). "Alu sequences are processed 7SL RNA genes".
1201:
487:
416:'s RNA sequence gets copied into the genome rather than the L1's mRNA.
342:
190:
162:
131:
122:
The study of Alu elements has also been important in elucidating human
94:
1495:"DNA sequence insertion and evolutionary variation in gene regulation"
1022:"Alu insertion polymorphisms for the study of human genomic diversity"
2994:
1878:
1615:
1463:
524:
260:
169:
106:
16:
Mobile genetic element in the primate genome (including human genome)
2065:
645:(Cerebellar Ataxia, Neuropathy & Vestibular Areflexia Syndrome)
329:
GTAGTGCGCTATGCCGATCGGAATAGCCACTGCACTCCAGCCTGGGCAACATAGCGAGACCCCGTCTC
1959:
2740:
687:
491:
401:
263:
254:
166:
388:
element replication and mobilization begins by interactions with
1071:"A fundamental division in the Alu family of repeated sequences"
638:
283:
2518:
301:, with the last one overlapping with the "B box". In this 7SL (
60:
18:
2441:
Dulai, K. S; von Dornum, M; Mollon, J. D; Hunt, D. M (1999).
2320:"SNP in the promoter region of the myeloperoxidase MPO gene"
834:
10.1554/0014-3820(2001)055[0001:ptepda]2.0.co;2
212:(i.e., occurring in more than one form or morph). In 1988,
2050:"The impact of retrotransposons on human genome evolution"
1292:"Alu Monomer Revisited: Recent Generation of Alu Monomers"
1852:"Initial sequencing and analysis of the human genome"
610:
And the following diseases have been associated with
161:
The Alu family is a family of repetitive elements in
2965:
2918:
2786:
2754:
2731:
2708:
2699:
2690:
2665:
2625:
2582:
2573:
2141:Jin, Lingling; McQuillan, Ian; Li, Longhai (2017).
545:The following human diseases have been linked with
1694:Chimpanzee Sequencing Analysis Consortium (2005).
1133:"Active Alu retrotransposons in the human genome"
382:retrotransposons for generation of new elements.
614:in Alu elements affecting transcription levels:
400:'s attachment to ribosomes that capture nascent
2250:Proceedings of the National Academy of Sciences
1995:Proceedings of the National Academy of Sciences
1941:
1939:
1585:
1583:
1389:Proceedings of the National Academy of Sciences
1126:
1124:
1075:Proceedings of the National Academy of Sciences
973:"Alu elements as regulators of gene expression"
297:response element hexamer sites in its internal
109:and originated in the genome of an ancestor of
93:. They are derived from the small cytoplasmic
1378:
1376:
966:
964:
494:) or into mRNA after the process of splicing.
193:. The length of the polyA tail varies between
63:originally characterized by the action of the
2530:
2508:National Center for Biotechnology Information
408:element can take control of the L1 protein's
105:elements are highly conserved within primate
8:
1802:"Does SINE evolution preclude Alu function?"
865:"Functions and Utility of Alu Jumping Genes"
478:elements, these regions serve as a site of
85:, present in excess of one million copies.
2705:
2696:
2579:
2537:
2523:
2515:
310:GCCGGGCGCGGTGGCGCGTGCCTGTAGTCCCAGCTACTCGGG
2495:at the U.S. National Library of Medicine
2458:
2374:
2287:
2269:
2219:
2209:
2168:
2158:
2073:
2048:Cordaux, Richard; Batzer, Mark A (2009).
2024:
2014:
1825:
1768:
1719:
1662:
1528:
1518:
1418:
1408:
1356:
1307:
1266:
1256:
1156:
1104:
1094:
1045:
996:
971:Häsler, Julien; Strub, Katharina (2006).
713:in the evolution of three colour vision.
140:
722:
208:sequences. However, less than 0.5% are
458:elements have been proposed to affect
663:, has 2 common variants, one with an
462:and been found to contain functional
180:long and are therefore classified as
7:
705:(including humans) is flanked by an
274:, and a 3' B box with the consensus
2504:"NCBI Genbank DNA encoding 7SL RNA"
1757:The Journal of Biological Chemistry
1383:Vansant, G; Reynolds, W. F (1995).
370:and look like DNA copies made from
293:Alu elements contain four or fewer
182:short interspersed nuclear elements
2606:Short tandem repeat/Microsatellite
14:
1902:Molecular Genetics and Metabolism
709:element, implicating the role of
470:. Due to the abundant content of
447:insertions are unique to humans.
2412:10.2165/11588720-000000000-00000
1554:International Review of Cytology
1290:Kojima, K. K. (16 August 2010).
612:single-nucleotide DNA variations
335:The recognition sequence of the
23:
1296:Molecular Biology and Evolution
509:Associations with human disease
349:residues (in lowercase above).
77:elements are the most abundant
2610:Trinucleotide repeat disorders
2194:"Alu elements: Know the SINEs"
1182:Journal of Molecular Evolution
777:insertions on gene function".
239:regulatory elements of genes.
1:
2597:Variable number tandem repeat
1566:10.1016/S0074-7696(05)47004-7
661:angiotensin-converting enzyme
579:Familial hypercholesterolemia
2192:Deininger, Prescott (2011).
1258:10.1371/journal.pcbi.1000610
744:10.1016/0092-8674(75)90184-1
390:signal recognition particles
1069:Jurka, J; Smith, T (1988).
286:, which are transcribed by
200:There are over one million
99:signal recognition particle
3047:
2098:hereditary colon cancer".
1237:PLOS Computational Biology
1038:10.1093/genetics/159.1.279
650:Associated human mutations
564:chorioretinal degeneration
2564:
2367:10.1038/s41588-019-0372-4
2211:10.1186/gb-2011-12-12-236
2160:10.1186/s12864-017-4227-z
1651:Journal of Human Genetics
904:10.1016/j.tig.2007.02.002
569:Diabetes mellitus type II
468:steroid hormone receptors
266:and SINEs, including Alu.
32:This article needs to be
3026:Repetitive DNA sequences
2497:Medical Subject Headings
2493:Alu+Repetitive+Sequences
1770:10.1074/jbc.270.39.22777
641:mutation responsible of
537:hereditary nonpolyposis
71:restriction endonuclease
2271:10.1073/pnas.2022751118
2054:Nature Reviews Genetics
2016:10.1073/pnas.1012834108
1948:Nature Reviews Genetics
1520:10.1073/pnas.93.18.9374
1410:10.1073/pnas.92.18.8229
1096:10.1073/pnas.85.13.4775
791:10.1002/elps.1150190806
176:elements are about 300
165:genomes, including the
3005:Protein tandem repeats
2933:Tandemly arrayed genes
1914:10.1006/mgme.1999.2864
1818:10.1093/nar/26.20.4541
1806:Nucleic Acids Research
1493:Britten, R. J (1996).
1337:Nucleic Acids Research
977:Nucleic Acids Research
939:10.1006/geno.1995.1224
267:
153:
59:is a short stretch of
1800:Schmid, C. W (1998).
1309:10.1093/molbev/msq218
1149:10.1101/gr.081737.108
863:Pray, Leslie (2008).
697:in the re-gaining of
671:-I) and one with the
594:mucopolysaccharidosis
410:reverse transcriptase
280:nucleic acid notation
259:Genetic structure of
258:
144:
97:, a component of the
79:transposable elements
2978:Pathogenicity island
605:Macular degeneration
412:, ensuring that the
148:from a female human
2262:2021PNAS..11822751F
2112:10.1038/nm1195-1203
2007:2011PNAS..108.2837S
1871:2001Natur.409..860L
1721:10.1038/nature04072
1712:2005Natur.437...69.
1664:10.1038/jhg.2009.86
1608:2000Natur.408..167W
1511:1996PNAS...93.9374B
1456:1984Natur.312..171U
1401:1995PNAS...92.8229V
1349:10.1093/nar/gku1361
1249:2009PLSCB...5E0610T
1194:1991JMolE..33..311R
1087:1988PNAS...85.4775J
619:Alzheimer's disease
136:evolution of humans
124:population genetics
66:Arthrobacter luteus
2928:Gene amplification
2460:10.1101/gr.9.7.629
2256:(6): e2022751118.
1202:10.1007/bf02102862
989:10.1093/nar/gkl706
892:Trends in Genetics
703:Old World primates
372:RNA polymerase III
288:RNA polymerase III
268:
154:
3013:
3012:
2914:
2913:
2782:
2781:
2686:
2685:
2575:Repeated sequence
2550:repeated sequence
1865:(6822): 860–921.
600:Neurofibromatosis
539:colorectal cancer
472:CpG dinucleotides
251:Sequence features
53:
52:
3038:
2990:Low copy repeats
2983:Symbiosis island
2920:Gene duplication
2706:
2697:
2580:
2558:gene duplication
2539:
2532:
2525:
2516:
2511:
2481:
2480:
2462:
2438:
2432:
2431:
2395:
2389:
2388:
2378:
2345:
2339:
2338:
2336:
2335:
2326:. Archived from
2316:
2310:
2309:
2291:
2273:
2240:
2234:
2233:
2223:
2213:
2189:
2183:
2182:
2172:
2162:
2153:(Suppl 9): 862.
2138:
2132:
2131:
2094:
2088:
2087:
2077:
2045:
2039:
2038:
2028:
2018:
1986:
1980:
1979:
1943:
1934:
1933:
1897:
1891:
1890:
1879:10.1038/35057062
1856:
1846:
1840:
1839:
1829:
1797:
1791:
1790:
1772:
1763:(39): 22777–82.
1748:
1742:
1741:
1723:
1691:
1685:
1684:
1666:
1642:
1636:
1635:
1616:10.1038/35041507
1602:(6809): 167–73.
1587:
1578:
1577:
1549:
1543:
1542:
1532:
1522:
1490:
1484:
1483:
1464:10.1038/312171a0
1439:
1433:
1432:
1422:
1412:
1380:
1371:
1370:
1360:
1328:
1322:
1321:
1311:
1287:
1281:
1280:
1270:
1260:
1243:(12): e1000610.
1228:
1222:
1221:
1177:
1171:
1170:
1160:
1128:
1119:
1118:
1108:
1098:
1066:
1060:
1059:
1049:
1017:
1011:
1010:
1000:
968:
959:
958:
922:
916:
915:
887:
881:
880:
878:
876:
860:
854:
853:
817:
811:
810:
770:
764:
763:
727:
691:gene duplication
451:Impact in humans
368:retrotransposons
331:
330:
277:
273:
234:Related elements
220:discovered that
134:, including the
48:
45:
39:
27:
26:
19:
3046:
3045:
3041:
3040:
3039:
3037:
3036:
3035:
3016:
3015:
3014:
3009:
2961:
2910:
2778:
2750:
2727:
2701:Retrotransposon
2682:
2673:Inverted repeat
2661:
2646:DNA transposon
2642:Retrotransposon
2637:Gene conversion
2628:
2621:
2618:
2569:
2560:
2543:
2502:
2489:
2484:
2447:Genome Research
2440:
2439:
2435:
2400:Sports Medicine
2397:
2396:
2392:
2347:
2346:
2342:
2333:
2331:
2318:
2317:
2313:
2242:
2241:
2237:
2191:
2190:
2186:
2140:
2139:
2135:
2100:Nature Medicine
2096:
2095:
2091:
2066:10.1038/nrg2640
2060:(10): 691–703.
2047:
2046:
2042:
1988:
1987:
1983:
1945:
1944:
1937:
1899:
1898:
1894:
1854:
1848:
1847:
1843:
1812:(20): 4541–50.
1799:
1798:
1794:
1750:
1749:
1745:
1706:(7055): 69–87.
1693:
1692:
1688:
1657:(10): 603–611.
1644:
1643:
1639:
1589:
1588:
1581:
1551:
1550:
1546:
1492:
1491:
1487:
1450:(5990): 171–2.
1441:
1440:
1436:
1395:(18): 8229–33.
1382:
1381:
1374:
1330:
1329:
1325:
1289:
1288:
1284:
1230:
1229:
1225:
1179:
1178:
1174:
1143:(12): 1875–83.
1137:Genome Research
1130:
1129:
1122:
1068:
1067:
1063:
1019:
1018:
1014:
970:
969:
962:
924:
923:
919:
889:
888:
884:
874:
872:
862:
861:
857:
819:
818:
814:
785:(8–9): 1260–4.
779:Electrophoresis
772:
771:
767:
729:
728:
724:
720:
659:gene, encoding
652:
574:Ewing's sarcoma
554:Alport syndrome
511:
460:gene expression
453:
374:-encoded RNAs.
355:
309:
308:
275:
271:
253:
236:
159:
91:genetic markers
49:
43:
40:
37:
28:
24:
17:
12:
11:
5:
3044:
3042:
3034:
3033:
3031:Human genetics
3028:
3018:
3017:
3011:
3010:
3008:
3007:
3002:
2997:
2992:
2987:
2986:
2985:
2980:
2973:Genomic island
2969:
2967:
2963:
2962:
2960:
2959:
2954:
2953:
2952:
2942:
2941:
2940:
2930:
2924:
2922:
2916:
2915:
2912:
2911:
2909:
2908:
2903:
2898:
2893:
2888:
2883:
2878:
2873:
2868:
2863:
2858:
2853:
2848:
2843:
2838:
2833:
2828:
2823:
2818:
2813:
2808:
2803:
2798:
2792:
2790:
2788:DNA transposon
2784:
2783:
2780:
2779:
2777:
2776:
2771:
2766:
2760:
2758:
2752:
2751:
2749:
2748:
2743:
2737:
2735:
2729:
2728:
2726:
2725:
2720:
2714:
2712:
2703:
2694:
2688:
2687:
2684:
2683:
2681:
2680:
2675:
2669:
2667:
2663:
2662:
2660:
2659:
2658:
2657:
2652:
2644:
2639:
2633:
2631:
2623:
2622:
2620:
2619:
2616:Macrosatellite
2613:
2603:
2594:
2588:
2586:
2584:Tandem repeats
2577:
2571:
2570:
2565:
2562:
2561:
2544:
2542:
2541:
2534:
2527:
2519:
2513:
2512:
2500:
2488:
2487:External links
2485:
2483:
2482:
2433:
2390:
2361:(4): 649–658.
2340:
2311:
2235:
2198:Genome Biology
2184:
2133:
2106:(11): 1203–6.
2089:
2040:
2001:(7): 2837–42.
1981:
1960:10.1038/nrg798
1935:
1892:
1841:
1792:
1743:
1686:
1637:
1579:
1544:
1505:(18): 9374–7.
1485:
1434:
1372:
1323:
1282:
1223:
1172:
1120:
1081:(13): 4775–8.
1061:
1012:
983:(19): 5491–7.
960:
933:(1): 136–144.
917:
882:
855:
812:
765:
721:
719:
716:
715:
714:
684:
651:
648:
647:
646:
632:
631:
629:Gastric cancer
626:
621:
608:
607:
602:
597:
591:
589:Leigh syndrome
586:
581:
576:
571:
566:
561:
556:
510:
507:
452:
449:
354:
351:
252:
249:
235:
232:
158:
155:
51:
50:
31:
29:
22:
15:
13:
10:
9:
6:
4:
3:
2:
3043:
3032:
3029:
3027:
3024:
3023:
3021:
3006:
3003:
3001:
2998:
2996:
2993:
2991:
2988:
2984:
2981:
2979:
2976:
2975:
2974:
2971:
2970:
2968:
2964:
2958:
2955:
2951:
2948:
2947:
2946:
2943:
2939:
2938:Ribosomal DNA
2936:
2935:
2934:
2931:
2929:
2926:
2925:
2923:
2921:
2917:
2907:
2904:
2902:
2899:
2897:
2894:
2892:
2889:
2887:
2884:
2882:
2879:
2877:
2874:
2872:
2869:
2867:
2864:
2862:
2859:
2857:
2854:
2852:
2849:
2847:
2844:
2842:
2839:
2837:
2834:
2832:
2829:
2827:
2824:
2822:
2819:
2817:
2814:
2812:
2809:
2807:
2804:
2802:
2799:
2797:
2794:
2793:
2791:
2789:
2785:
2775:
2772:
2770:
2767:
2765:
2762:
2761:
2759:
2757:
2753:
2747:
2744:
2742:
2739:
2738:
2736:
2734:
2730:
2724:
2721:
2719:
2716:
2715:
2713:
2711:
2707:
2704:
2702:
2698:
2695:
2693:
2689:
2679:
2678:Direct repeat
2676:
2674:
2671:
2670:
2668:
2664:
2656:
2653:
2651:
2648:
2647:
2645:
2643:
2640:
2638:
2635:
2634:
2632:
2630:
2624:
2617:
2614:
2611:
2607:
2604:
2602:
2601:Minisatellite
2598:
2595:
2593:
2592:Satellite DNA
2590:
2589:
2587:
2585:
2581:
2578:
2576:
2572:
2568:
2563:
2559:
2555:
2551:
2547:
2540:
2535:
2533:
2528:
2526:
2521:
2520:
2517:
2510:. 2018-05-12.
2509:
2505:
2501:
2498:
2494:
2491:
2490:
2486:
2478:
2474:
2470:
2466:
2461:
2456:
2453:(7): 629–38.
2452:
2448:
2444:
2437:
2434:
2429:
2425:
2421:
2417:
2413:
2409:
2406:(6): 433–48.
2405:
2401:
2394:
2391:
2386:
2382:
2377:
2372:
2368:
2364:
2360:
2356:
2352:
2344:
2341:
2330:on 2010-05-21
2329:
2325:
2321:
2315:
2312:
2307:
2303:
2299:
2295:
2290:
2285:
2281:
2277:
2272:
2267:
2263:
2259:
2255:
2251:
2247:
2239:
2236:
2231:
2227:
2222:
2217:
2212:
2207:
2203:
2199:
2195:
2188:
2185:
2180:
2176:
2171:
2166:
2161:
2156:
2152:
2148:
2144:
2137:
2134:
2129:
2125:
2121:
2117:
2113:
2109:
2105:
2101:
2093:
2090:
2085:
2081:
2076:
2071:
2067:
2063:
2059:
2055:
2051:
2044:
2041:
2036:
2032:
2027:
2022:
2017:
2012:
2008:
2004:
2000:
1996:
1992:
1985:
1982:
1977:
1973:
1969:
1965:
1961:
1957:
1953:
1949:
1942:
1940:
1936:
1931:
1927:
1923:
1919:
1915:
1911:
1908:(3): 183–93.
1907:
1903:
1896:
1893:
1888:
1884:
1880:
1876:
1872:
1868:
1864:
1860:
1853:
1845:
1842:
1837:
1833:
1828:
1823:
1819:
1815:
1811:
1807:
1803:
1796:
1793:
1788:
1784:
1780:
1776:
1771:
1766:
1762:
1758:
1754:
1747:
1744:
1739:
1735:
1731:
1727:
1722:
1717:
1713:
1709:
1705:
1701:
1697:
1690:
1687:
1682:
1678:
1674:
1670:
1665:
1660:
1656:
1652:
1648:
1641:
1638:
1633:
1629:
1625:
1621:
1617:
1613:
1609:
1605:
1601:
1597:
1593:
1586:
1584:
1580:
1575:
1571:
1567:
1563:
1559:
1555:
1548:
1545:
1540:
1536:
1531:
1526:
1521:
1516:
1512:
1508:
1504:
1500:
1496:
1489:
1486:
1481:
1477:
1473:
1469:
1465:
1461:
1457:
1453:
1449:
1445:
1438:
1435:
1430:
1426:
1421:
1416:
1411:
1406:
1402:
1398:
1394:
1390:
1386:
1379:
1377:
1373:
1368:
1364:
1359:
1354:
1350:
1346:
1343:(2): 817–35.
1342:
1338:
1334:
1327:
1324:
1319:
1315:
1310:
1305:
1301:
1297:
1293:
1286:
1283:
1278:
1274:
1269:
1264:
1259:
1254:
1250:
1246:
1242:
1238:
1234:
1227:
1224:
1219:
1215:
1211:
1207:
1203:
1199:
1195:
1191:
1188:(4): 311–20.
1187:
1183:
1176:
1173:
1168:
1164:
1159:
1154:
1150:
1146:
1142:
1138:
1134:
1127:
1125:
1121:
1116:
1112:
1107:
1102:
1097:
1092:
1088:
1084:
1080:
1076:
1072:
1065:
1062:
1057:
1053:
1048:
1043:
1039:
1035:
1032:(1): 279–90.
1031:
1027:
1023:
1016:
1013:
1008:
1004:
999:
994:
990:
986:
982:
978:
974:
967:
965:
961:
956:
952:
948:
944:
940:
936:
932:
928:
921:
918:
913:
909:
905:
901:
898:(4): 158–61.
897:
893:
886:
883:
870:
866:
859:
856:
851:
847:
843:
839:
835:
831:
827:
823:
816:
813:
808:
804:
800:
796:
792:
788:
784:
780:
776:
769:
766:
761:
757:
753:
749:
745:
741:
738:(3): 345–58.
737:
733:
726:
723:
717:
712:
708:
704:
700:
696:
692:
689:
685:
682:
678:
674:
670:
666:
662:
658:
654:
653:
649:
644:
640:
637:
636:
635:
630:
627:
625:
622:
620:
617:
616:
615:
613:
606:
603:
601:
598:
595:
592:
590:
587:
585:
582:
580:
577:
575:
572:
570:
567:
565:
562:
560:
559:Breast cancer
557:
555:
552:
551:
550:
548:
543:
541:
540:
534:
533:recombination
530:
526:
523:
519:
515:
508:
506:
504:
500:
495:
493:
489:
485:
481:
477:
473:
469:
465:
461:
457:
450:
448:
446:
442:
438:
433:
429:
425:
421:
417:
415:
411:
407:
403:
399:
395:
391:
387:
383:
381:
377:
373:
369:
366:elements are
365:
361:
359:
352:
350:
348:
344:
340:
339:
333:
328:
326:
321:
317:
313:
306:
304:
300:
296:
295:Retinoic Acid
291:
289:
285:
281:
265:
262:
257:
250:
248:
246:
240:
233:
231:
228:
223:
219:
215:
211:
207:
203:
198:
196:
192:
187:
183:
179:
175:
171:
168:
164:
156:
151:
147:
143:
139:
137:
133:
129:
125:
120:
118:
114:
112:
111:Supraprimates
108:
104:
100:
96:
92:
88:
84:
80:
76:
72:
69:
67:
62:
58:
47:
44:February 2021
35:
30:
21:
20:
2950:Gene cluster
2718:Alu sequence
2717:
2627:Interspersed
2507:
2450:
2446:
2436:
2403:
2399:
2393:
2358:
2354:
2343:
2332:. Retrieved
2328:the original
2323:
2314:
2253:
2249:
2238:
2201:
2197:
2187:
2150:
2147:BMC Genomics
2146:
2136:
2103:
2099:
2092:
2057:
2053:
2043:
1998:
1994:
1984:
1954:(5): 370–9.
1951:
1947:
1905:
1901:
1895:
1862:
1858:
1844:
1809:
1805:
1795:
1760:
1756:
1746:
1703:
1699:
1689:
1654:
1650:
1640:
1599:
1595:
1557:
1553:
1547:
1502:
1498:
1488:
1447:
1443:
1437:
1392:
1388:
1340:
1336:
1326:
1302:(1): 13–15.
1299:
1295:
1285:
1240:
1236:
1226:
1185:
1181:
1175:
1140:
1136:
1078:
1074:
1064:
1029:
1025:
1015:
980:
976:
930:
926:
920:
895:
891:
885:
873:. Retrieved
869:Scitable.com
868:
858:
825:
821:
815:
782:
778:
774:
768:
735:
731:
725:
710:
706:
680:
676:
672:
668:
664:
656:
633:
609:
549:insertions:
546:
544:
536:
528:
521:
517:
513:
512:
502:
498:
496:
483:
475:
466:regions for
455:
454:
444:
440:
436:
431:
427:
423:
419:
418:
413:
405:
397:
393:
385:
384:
375:
363:
362:
357:
356:
353:Alu elements
336:
334:
324:
323:
319:
315:
311:
307:
292:
269:
245:BC200 lncRNA
241:
237:
226:
221:
218:Temple Smith
205:
201:
199:
194:
185:
173:
160:
121:
116:
115:
102:
86:
83:human genome
74:
64:
56:
54:
41:
33:
2945:Gene family
2856:Tc1/mariner
2811:EnSpm/CACTA
2204:(12): 236.
1560:: 165–221.
828:(1): 1–24.
699:trichromacy
667:insertion (
624:Lung cancer
480:methylation
404:. Thus, an
402:L1 proteins
272:TGGCTCACGCC
214:Jerzy Jurka
210:polymorphic
57:Alu element
3020:Categories
2957:Pseudogene
2774:retroposon
2692:Transposon
2554:transposon
2334:2010-03-14
718:References
584:Hemophilia
531:-mediated
197:families.
178:base pairs
157:Alu family
150:lymphocyte
2876:P element
2826:Harbinger
2567:Repeatome
2355:Nat Genet
2306:231761522
2280:0027-8424
1976:205486422
947:0888-7543
822:Evolution
675:deleted (
474:found in
276:GTTCGAGAC
172:. Modern
146:Karyotype
128:evolution
3000:Telomere
2966:See also
2906:Zisupton
2886:Polinton
2881:PiggyBac
2836:Helitron
2655:Helitron
2650:Polinton
2546:Genetics
2477:10637615
2469:10413401
2428:42531424
2420:21615186
2385:30926972
2298:33526699
2230:22204421
2179:29219079
2128:39468812
2084:19763152
2035:21282640
1968:11988762
1930:15651921
1922:10381326
1887:11237011
1787:45796017
1730:16136131
1673:19745832
1624:11089964
1574:16344113
1367:25550429
1318:20713470
1277:20019790
1218:13091552
1167:18836035
1056:11560904
1026:Genetics
1007:17020921
927:Genomics
912:17307271
871:. Nature
850:25273865
842:11263730
807:45917758
760:42804857
695:resulted
464:promoter
347:cytosine
314:AGGCTGGA
299:promoter
132:primates
126:and the
2896:Transib
2871:Novosib
2851:Kolobok
2821:Ginger2
2816:Ginger1
2801:Crypton
2376:6709527
2324:SNPedia
2289:8017980
2258:Bibcode
2221:3334610
2170:5773891
2120:7584997
2075:2884099
2026:3041063
2003:Bibcode
1867:Bibcode
1836:9753719
1779:7559405
1738:2638825
1708:Bibcode
1681:8153502
1632:4427070
1604:Bibcode
1539:8790336
1507:Bibcode
1480:4328237
1472:6209580
1452:Bibcode
1429:7667273
1397:Bibcode
1358:4333407
1268:2784220
1245:Bibcode
1210:1774786
1190:Bibcode
1158:2593586
1115:3387438
1083:Bibcode
1047:1461783
998:1636486
955:8530063
875:26 June
799:9694261
752:1052772
488:introns
343:guanine
278:(IUPAC
191:SRP RNA
163:primate
107:genomes
95:7SL RNA
81:in the
34:updated
2995:CRISPR
2861:Merlin
2846:ISL2EU
2796:Academ
2629:repeat
2499:(MeSH)
2475:
2467:
2426:
2418:
2383:
2373:
2304:
2296:
2286:
2278:
2228:
2218:
2177:
2167:
2126:
2118:
2082:
2072:
2033:
2023:
1974:
1966:
1928:
1920:
1885:
1859:Nature
1834:
1827:147893
1824:
1785:
1777:
1736:
1728:
1700:Nature
1679:
1671:
1630:
1622:
1596:Nature
1572:
1537:
1527:
1478:
1470:
1444:Nature
1427:
1417:
1365:
1355:
1316:
1275:
1265:
1216:
1208:
1165:
1155:
1113:
1106:280518
1103:
1054:
1044:
1005:
995:
953:
945:
910:
848:
840:
805:
797:
758:
750:
693:which
643:CANVAS
525:allele
325:AGTTCT
320:AGTCCA
316:GGATCG
312:AGGCTG
261:murine
170:genome
2901:Zator
2841:IS3EU
2746:LINE2
2741:LINE1
2733:LINEs
2710:SINEs
2666:Other
2473:S2CID
2424:S2CID
2302:S2CID
2124:S2CID
1972:S2CID
1926:S2CID
1855:(PDF)
1783:S2CID
1734:S2CID
1677:S2CID
1628:S2CID
1530:38434
1476:S2CID
1420:41130
1214:S2CID
846:S2CID
803:S2CID
756:S2CID
688:opsin
492:exons
428:Alu Y
338:Alu I
327:GGGCT
284:tRNAs
264:LINE1
167:human
68:(Alu)
2891:Sola
2866:MuDR
2806:Dada
2769:MER4
2764:HERV
2756:LTRs
2465:PMID
2416:PMID
2381:PMID
2294:PMID
2276:ISSN
2226:PMID
2175:PMID
2116:PMID
2080:PMID
2031:PMID
1964:PMID
1918:PMID
1883:PMID
1832:PMID
1775:PMID
1726:PMID
1669:PMID
1620:PMID
1570:PMID
1535:PMID
1468:PMID
1425:PMID
1363:PMID
1314:PMID
1273:PMID
1206:PMID
1163:PMID
1111:PMID
1052:PMID
1003:PMID
951:PMID
943:ISSN
908:PMID
877:2019
838:PMID
795:PMID
748:PMID
732:Cell
686:The
655:The
639:RFC1
380:LINE
345:and
318:CTTG
216:and
2831:hAT
2723:MIR
2455:doi
2408:doi
2371:PMC
2363:doi
2284:PMC
2266:doi
2254:118
2216:PMC
2206:doi
2165:PMC
2155:doi
2108:doi
2070:PMC
2062:doi
2021:PMC
2011:doi
1999:108
1956:doi
1910:doi
1875:doi
1863:409
1822:PMC
1814:doi
1765:doi
1761:270
1716:doi
1704:437
1659:doi
1612:doi
1600:408
1562:doi
1558:247
1525:PMC
1515:doi
1460:doi
1448:312
1415:PMC
1405:doi
1353:PMC
1345:doi
1304:doi
1263:PMC
1253:doi
1198:doi
1153:PMC
1145:doi
1101:PMC
1091:doi
1042:PMC
1034:doi
1030:159
993:PMC
985:doi
935:doi
900:doi
830:doi
787:doi
775:Alu
740:doi
711:Alu
707:Alu
701:in
681:Alu
677:ACE
673:Alu
669:ACE
665:Alu
657:ACE
596:VII
547:Alu
529:Alu
522:Alu
518:Alu
514:Alu
503:Alu
499:Alu
484:Alu
476:Alu
456:Alu
445:Alu
441:Alu
437:Alu
432:Alu
424:Alu
420:Alu
414:Alu
406:Alu
398:Alu
394:Alu
386:Alu
376:Alu
364:Alu
358:Alu
303:SRP
282:).
227:Alu
222:Alu
206:Alu
202:Alu
195:Alu
186:Alu
174:Alu
130:of
117:Alu
103:Alu
87:Alu
75:Alu
61:DNA
55:An
3022::
2556:,
2552:,
2548::
2506:.
2471:.
2463:.
2449:.
2445:.
2422:.
2414:.
2404:41
2402:.
2379:.
2369:.
2359:51
2357:.
2353:.
2322:.
2300:.
2292:.
2282:.
2274:.
2264:.
2252:.
2248:.
2224:.
2214:.
2202:12
2200:.
2196:.
2173:.
2163:.
2151:18
2149:.
2145:.
2122:.
2114:.
2102:.
2078:.
2068:.
2058:10
2056:.
2052:.
2029:.
2019:.
2009:.
1997:.
1993:.
1970:.
1962:.
1950:.
1938:^
1924:.
1916:.
1906:67
1904:.
1881:.
1873:.
1861:.
1857:.
1830:.
1820:.
1810:26
1808:.
1804:.
1781:.
1773:.
1759:.
1755:.
1732:.
1724:.
1714:.
1702:.
1698:.
1675:.
1667:.
1655:54
1653:.
1649:.
1626:.
1618:.
1610:.
1598:.
1594:.
1582:^
1568:.
1556:.
1533:.
1523:.
1513:.
1503:93
1501:.
1497:.
1474:.
1466:.
1458:.
1446:.
1423:.
1413:.
1403:.
1393:92
1391:.
1387:.
1375:^
1361:.
1351:.
1341:43
1339:.
1335:.
1312:.
1300:28
1298:.
1294:.
1271:.
1261:.
1251:.
1239:.
1235:.
1212:.
1204:.
1196:.
1186:33
1184:.
1161:.
1151:.
1141:18
1139:.
1135:.
1123:^
1109:.
1099:.
1089:.
1079:85
1077:.
1073:.
1050:.
1040:.
1028:.
1024:.
1001:.
991:.
981:34
979:.
975:.
963:^
949:.
941:.
931:29
929:.
906:.
896:23
894:.
867:.
844:.
836:.
826:55
824:.
801:.
793:.
783:19
781:.
754:.
746:.
734:.
332:.
322:GG
247:.
138:.
113:.
101:.
73:.
2612:)
2608:(
2599:/
2538:e
2531:t
2524:v
2479:.
2457::
2451:9
2430:.
2410::
2387:.
2365::
2337:.
2308:.
2268::
2260::
2232:.
2208::
2181:.
2157::
2130:.
2110::
2104:1
2086:.
2064::
2037:.
2013::
2005::
1978:.
1958::
1952:3
1932:.
1912::
1889:.
1877::
1869::
1838:.
1816::
1789:.
1767::
1740:.
1718::
1710::
1683:.
1661::
1634:.
1614::
1606::
1576:.
1564::
1541:.
1517::
1509::
1482:.
1462::
1454::
1431:.
1407::
1399::
1369:.
1347::
1320:.
1306::
1279:.
1255::
1247::
1241:5
1220:.
1200::
1192::
1169:.
1147::
1117:.
1093::
1085::
1058:.
1036::
1009:.
987::
957:.
937::
914:.
902::
879:.
852:.
832::
809:.
789::
762:.
742::
736:6
46:)
42:(
36:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.