643:
the high-affinity sites and subsequently activates the enzyme, while UTP and CTP binding leads to inhibition of activity. UTP can bind to the allosteric site, but inhibition of ATCase by UTP is possible only in combination with CTP. With CTP present, UTP binding is enhanced and preferentially directed to the low-affinity sites. On the converse, UTP binding leads to enhanced affinity for CTP at the high-affinity sites and together they inhibit enzyme activity by up to 95%, while CTP binding alone inhibits activity to 50% to 70%. Comparison of the crystal structures of the T and R forms of ATCase show that it swells in size during the allosteric transition, and that the catalytic subunits condense during this process. The two catalytic trimers move apart along the threefold axis by 12 Å, and they rotate about this axis by 5° each, ultimately leading to a reorientation of the regulatory subunits around their twofold axis by 15°. This
634:
Ser52, Thr53, Arg54, Thr55, Arg105, His134, Gln137, Arg167, Arg229, Glu231, and Ser80 and Lys84 from an adjacent catalytic chain. The active site is a highly positively charged pocket. One of the most critical side-chains is from Arg54, which interacts with a terminal oxygen and the anhydride oxygen of carbamoyl phosphate, stabilizing the negative charge of the leaving phosphate group. Arg105, His134, and Thr55 help to increase the electrophilicity of the carbonyl carbon by interacting with the carbonyl oxygen. In general, the rate enhancement of ATCase is achieved by orientation and stabilization of substrates, intermediates, and products rather than by direct involvement of amino acid residues in the catalytic mechanism.
29:
521:
510:
2959:
671:
Located close to the 240s loop and the active site, the loop region encompassing residues 160–166 plays a role in both the internal architecture of the enzyme and its regulatory properties. In particular, the residue Asp162 interacts with Gln231 (known to be involved in aspartate binding), and binds
642:
The allosteric site in the allosteric domain of the R chains of the ATCase complex binds to the nucleotides ATP, CTP and/or UTP. There is one site with high affinity for ATP and CTP and one with 10- to 20-fold lower affinity for these nucleotides in each regulatory dimer. ATP binds predominantly to
633:
of the substrates. Additionally, crystal structures of ATCase bound to carbamoylphosphate and succinate have been obtained. These studies, in addition to investigations using site-directed mutagenesis of specific amino acids, have identified several residues that are crucial for catalysis, such as
696:
The regulatory and catalytic subunits exist as fused protein homologs, providing strong evidence that they would interact together. Two catalytic trimers and two regulatory dimers assemble to form an intermediate of aspartate carbamoyltransferase consisting of 6 catalytic subunits and 4 regulatory
628:
The catalytic site of ATCase is located at the interface between two neighboring catalytic chains in the same trimer and incorporates amino acid side-chains from both of these subunits. Insight into the mode of binding of substrates to the catalytic center of ATCase was first made possible by the
619:
stabilizing contacts between amino acid residues. Each catalytic chain is in contact with three other catalytic chains and two regulatory chains. Each regulatory monomer is in contact with one other regulatory chain and two catalytic chains. In the unliganded enzyme, the two catalytic trimers are
667:
During this structural transition, some interactions between side-chains are lost and some others are established. Studies have confirmed that the position of the 240s loop directly affects substrate binding in the corresponding active site. Earlier studies using site-directed mutagenesis of the
647:
change is associated with alterations in inter-subunit and inter-domain interactions. The interaction between subunits C1-C4 and R1 is extensively modified during this conversion. In particular, there is large movement of amino acid residues 230–254, known collectively as the 240s loop. These
1271:
Fetler L, Vachette P, Hervé G, Ladjimi MM (Dec 1995). "Unlike the quaternary structure transition, the tertiary structure change of the 240s loop in allosteric aspartate transcarbamylase requires active site saturation by substrate for completion".
463:. Instead, it lies between its low-activity, low-affinity "tense" and its high-activity, high-affinity "relaxed" states. The binding of substrate to the catalytic subunits results in an equilibrium shift towards the R state, whereas binding of
668:
240s loop showed that interactions between Asp271 and Tyr240, and between Glu239 of C1 and Tyr165 of C4 would stabilize the T-state, while interactions between Glu239 of C1 and both Lys164 and Tyr165 of C4 would stabilize the R-state.
684:. It was suggested that the change in the overall structure caused by the introduction of this residue affects other residues in the R1-C1, R1-C4 and C1-C4 interfaces, which are involved in the
629:
binding of a bisubstrate analogue, N-(phosphonoacetyl)-L-aspartate (PALA). This compound is a strong inhibitor of ATCase and has a structure that is thought to be very close to that of the
656:
domains at the C1-C4 interface. The overall outcome of these structural changes is that the two domains of each catalytic chain come closer together, ensuring a better contact with the
1683:
1592:
1403:"Replacement of Asp-162 by Ala prevents the cooperative transition by the substrates while enhancing the effect of the allosteric activator ATP on E. coli aspartate transcarbamoylase"
562:
is made of two catalytic trimers that are in contact and held together by three regulatory dimers, so the native form of the enzyme contains six chains of each type, with a total
2139:
467:
to the regulatory subunits results in an equilibrium shift towards the T state. Binding of ATP to the regulatory subunits results in an equilibrium shift towards the R state.
772:
Macol CP, Tsuruta H, Stec B, Kantrowitz ER (May 2001). "Direct structural evidence for a concerted allosteric transition in
Escherichia coli aspartate transcarbamoylase".
2289:
2490:
1366:
Newton CJ, Stevens RC, Kantrowitz ER (Mar 1992). "Importance of a conserved residue, aspartate-162, for the function of
Escherichia coli aspartate transcarbamoylase".
922:
1083:
Krause KL, Volz KW, Lipscomb WN (Feb 1987). "2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate".
491:. The catalysis by ATCase serves as the rate limiting step in pyrimidine biosynthesis because it alters its catalytic velocity in response to cellular levels of both
209:
2031:
1792:
2444:
266:
228:
2345:
2508:
2414:
1668:
1585:
524:
Schematic diagram of ATCase structure, depicting spatial arrangement of green regulatory (R) and blue catalytic (C) subunits. Redrawn and modified from Ke
2609:
573:
Each of the catalytic domains is composed of two structural domains, the aspartate domain, which contains most of the residues responsible for binding
456:
behaviour with respect to its substrates. The enzyme is an archetypal example of allosteric modulation of fine control of metabolic enzyme reactions.
1758:
2388:
1678:
612:
1236:
Kantrowitz ER, Lipscomb WN (Feb 1990). "Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition".
452:
of regulatory subunits (17 kDa). The particular arrangement of catalytic and regulatory subunits in this enzyme affords the complex with strongly
1901:
715:"Mammalian dihydroorotase: nucleotide sequence, peptide sequences, and evolution of the dihydroorotase domain of the multifunctional protein CAD"
2619:
2335:
1578:
28:
2282:
885:
608:
that is not involved in any catalytic property, but has been shown to be essential for the association of regulatory and catalytic subunits.
532:
The discussion of structure, catalytic center, and allosteric site that follows is based on the prokaryotic version of ATCase, specifically
2979:
2573:
2645:
2365:
2361:
2224:
2024:
290:
2117:
2678:
1797:
1016:
1040:
Kantrowitz ER, Lipscomb WN (Aug 1988). "Escherichia coli aspartate transcarbamylase: the relation between structure and function".
2529:
2275:
1905:
1658:
278:
38:
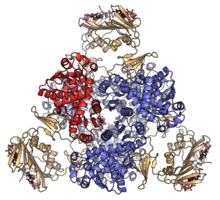
aspartate carbamoyltransferase heterododecamer with catalytic subunits coloured red and blue, and regulatory subunits in orange.
221:
2604:
2485:
2182:
1938:
1871:
1818:
410:
1309:"Importance of the loop at residues 230–245 in the allosteric interactions of Escherichia coli aspartate carbamoyltransferase"
509:
2594:
2513:
2393:
2159:
2132:
2110:
2095:
2060:
2017:
1688:
1663:
271:
148:
2834:
475:
ATCase is a highly regulated enzyme that catalyses the first committed step in pyrimidine biosynthesis, the condensation of
460:
172:
2370:
2330:
1983:
1673:
820:"Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase"
2949:
2599:
2439:
2105:
1896:
554:
but do not have regulatory properties, while the regulatory subunits do not have any catalytic activity but contain the
2614:
2819:
2935:
2922:
2909:
2896:
2883:
2870:
2857:
2633:
2548:
2458:
2424:
2340:
2311:
1712:
2829:
2783:
2726:
2477:
2302:
2209:
1978:
1564:
657:
414:
331:
166:
59:
520:
484:
2731:
2241:
2219:
2199:
1998:
1742:
1701:
153:
1570:
1402:
2534:
2204:
2100:
1717:
1560:
1179:"Three-dimensional structure of carbamoyl phosphate and succinate bound to aspartate carbamoyltransferase"
681:
598:
504:
283:
2752:
2671:
2434:
1952:
1847:
500:
464:
233:
2824:
141:
2429:
1993:
1512:
1320:
1190:
1131:
1049:
726:
685:
644:
348:
76:
2788:
2398:
2256:
2246:
2214:
1970:
1943:
1813:
649:
578:
488:
480:
71:
1003:. Advances in Enzymology – and Related Areas of Molecular Biology. Vol. 68. pp. 67–151.
169:
2721:
2251:
1784:
1643:
1607:
1120:"Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase"
999:
Lipscomb WN (1994). "Aspartate transcarbamylase from
Escherichia coli: activity and regulation".
903:
797:
661:
586:
445:
93:
2267:
581:. Each regulatory domain is also composed of two domains, the allosteric domain, which has the
2984:
2500:
2315:
1823:
1753:
1625:
1540:
1481:
1432:
1383:
1348:
1289:
1253:
1218:
1159:
1100:
1065:
1022:
1012:
976:
891:
881:
849:
789:
754:
160:
40:
546:
chains, which have different roles. The catalytic subunits catalyze the carbamylation of the
2767:
2762:
2736:
2664:
1828:
1530:
1520:
1471:
1463:
1422:
1414:
1375:
1338:
1328:
1281:
1245:
1208:
1198:
1149:
1139:
1092:
1057:
1004:
966:
956:
839:
831:
781:
744:
734:
630:
611:
The three-dimensional arrangement of the catalytic and regulatory subunits involves several
343:
34:
2814:
2798:
2711:
2552:
2044:
1450:
Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA (Apr 2013).
915:
555:
430:
372:
336:
129:
672:
the same residues in both the T and R states. A mutant that had this residue mutated to
352:
1516:
1324:
1194:
1135:
1053:
730:
436:
complex composed of 12 subunits (300 kDa in total). The composition of the subunits is C
105:
2963:
2852:
2793:
2192:
2075:
2052:
1988:
1918:
1881:
1476:
1451:
1427:
1154:
1119:
577:, and the carbamoyl phosphate domain, which contains most of the residues that bind to
563:
204:
64:
1535:
1500:
1343:
1308:
1213:
1178:
971:
940:
713:
Simmer JP, Kelly RE, Rinker AG, Zimmermann BH, Scully JL, Kim H, Evans DR (Jan 1990).
676:
showed a huge reduction in specific activity, a two-fold decrease in the affinity for
184:
2973:
2757:
2716:
2462:
2087:
2070:
1833:
1768:
1727:
1647:
1452:"Protein complexes are under evolutionary selection to assemble via ordered pathways"
1249:
1096:
844:
819:
749:
714:
449:
179:
801:
2706:
2380:
835:
582:
248:
Human carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase
2009:
307:
2930:
2865:
2701:
2298:
2065:
616:
543:
507:, the end-product of the parallel purine pathway, increases catalytic velocity.
476:
382:
188:
2958:
1505:
Proceedings of the
National Academy of Sciences of the United States of America
1467:
1313:
Proceedings of the
National Academy of Sciences of the United States of America
1183:
Proceedings of the
National Academy of Sciences of the United States of America
1124:
Proceedings of the
National Academy of Sciences of the United States of America
949:
Proceedings of the
National Academy of Sciences of the United States of America
719:
Proceedings of the
National Academy of Sciences of the United States of America
2177:
2154:
1866:
1612:
1603:
1008:
895:
559:
492:
453:
314:
2904:
2878:
2558:
1401:
Fetler L, Tauc P, Baker DP, Macol CP, Kantrowitz ER, Vachette P (May 2002).
1333:
1203:
1144:
1061:
961:
677:
653:
574:
551:
1525:
1501:"An intermediate complex in the dissociation of aspartate transcarbamylase"
1485:
1436:
1163:
853:
793:
1544:
1387:
1352:
1293:
1257:
1222:
1104:
1069:
1026:
980:
758:
739:
542:
Early studies demonstrated that ATCase consists of two different kinds of
594:
406:
319:
1379:
1285:
2637:
1418:
673:
534:
433:
425:
417:
136:
117:
44:
2917:
2687:
2467:
2419:
2149:
2144:
2040:
1615:
496:
302:
216:
112:
100:
88:
2891:
2234:
2229:
785:
680:, a loss of homotropic cooperativity, and decreased activation by
547:
2127:
2122:
1956:
1947:
605:
602:
590:
295:
124:
2660:
2271:
2013:
1574:
567:
1001:
Advances in
Enzymology and Related Areas of Molecular Biology
597:
residues clustered in its C-terminal region. These residues
1118:
Wang J, Stieglitz KA, Cardia JP, Kantrowitz ER (Jun 2005).
2656:
941:"Structure of unligated aspartate carbamoyltransferase of
1684:
Phosphoribosylaminoimidazolesuccinocarboxamide synthase
2947:
2140:
Branched-chain alpha-keto acid dehydrogenase complex
1499:
Evans DR, Pastra-Landis SC, Lipscomb WN (Apr 1974).
2843:
2807:
2776:
2745:
2694:
2632:
2582:
2566:
2547:
2522:
2499:
2476:
2457:
2407:
2379:
2354:
2323:
2310:
2242:
Phosphoenolpyruvate sugar phosphotransferase system
2170:
2086:
2051:
1969:
1931:
1855:
1846:
1806:
1783:
1738:
1697:
1640:
1633:
1624:
378:
368:
363:
342:
330:
325:
313:
301:
289:
277:
265:
257:
252:
247:
227:
215:
203:
198:
178:
159:
147:
135:
123:
111:
99:
87:
82:
70:
58:
53:
21:
2491:3-methyl-2-oxobutanoate hydroxymethyltransferase
818:Helmstaedt K, Krappmann S, Braus GH (Sep 2001).
2061:Photosynthetic reaction center complex proteins
866:Biochemistry, by Campbell and Farrel, Chapter 7
1793:Hypoxanthine-guanine phosphoribosyltransferase
813:
811:
648:residues are located at the cleft between the
499:. The end-product of the pyrimidine pathway,
2672:
2445:Cyclopropane-fatty-acyl-phospholipid synthase
2283:
2025:
1586:
994:
992:
990:
939:Ke HM, Honzatko RB, Lipscomb WN (July 1984).
934:
932:
8:
2346:Phosphatidylethanolamine N-methyltransferase
2509:Phosphoribosylglycinamide formyltransferase
2415:Phosphatidyl ethanolamine methyltransferase
1669:Phosphoribosylglycinamide formyltransferase
2679:
2665:
2657:
2610:3-hydroxymethylcephem carbamoyltransferase
2563:
2473:
2320:
2290:
2276:
2268:
2032:
2018:
2010:
1859:
1852:
1637:
1630:
1593:
1579:
1571:
824:Microbiology and Molecular Biology Reviews
360:
195:
27:
1563:at the U.S. National Library of Medicine
1534:
1524:
1475:
1426:
1342:
1332:
1212:
1202:
1153:
1143:
970:
960:
921:CS1 maint: multiple names: authors list (
843:
748:
738:
1679:Phosphoribosylaminoimidazole carboxylase
1307:Middleton SA, Kantrowitz ER (Aug 1986).
519:
503:, decreases catalytic velocity, whereas
2954:
705:
2620:N-acetylornithine carbamoyltransferase
2389:Betaine-homocysteine methyltransferase
2336:Phenylethanolamine N-methyltransferase
911:
901:
244:
18:
448:of catalytic subunits (34 kDa) and 3
7:
2574:methylmalonyl-CoA carboxytransferase
1902:Orotidine 5'-phosphate decarboxylase
2646:Arginine:glycine amidinotransferase
2366:Acetylserotonin O-methyltransferase
2362:5-hydroxyindole-O-methyltransferase
2225:Mitochondrial trifunctional protein
1177:Gouaux JE, Lipscomb WN (Jun 1988).
558:for effector binding. The ATCase
14:
1798:Adenine phosphoribosyltransferase
2957:
2530:Glutamate formimidoyltransferase
1906:Uridine monophosphate synthetase
1659:Ribose-phosphate diphosphokinase
830:(3): 404–21, table of contents.
508:
2605:Putrescine carbamoyltransferase
2486:Serine hydroxymethyltransferase
2183:Carbamoyl phosphate synthase II
1939:Dihydropyrimidine dehydrogenase
1872:Carbamoyl phosphate synthase II
1819:Purine nucleoside phosphorylase
411:pyrimidine biosynthetic pathway
2595:Ornithine carbamoyltransferase
2590:Aspartate carbamoyltransferase
2514:Inosine monophosphate synthase
2394:Homocysteine methyltransferase
2188:Aspartate carbamoyltransferase
2096:Pyruvate dehydrogenase complex
1877:Aspartate carbamoyltransferase
1664:Amidophosphoribosyltransferase
1561:Aspartate+carbamoyltransferase
1238:Trends in Biochemical Sciences
836:10.1128/MMBR.65.3.404-421.2001
395:Aspartate carbamoyltransferase
22:Aspartate carbamoyltransferase
1:
2371:Catechol-O-methyl transferase
2331:Histamine N-methyltransferase
2220:Glycine decarboxylase complex
2215:Fatty acid synthetase complex
1984:Nucleoside-diphosphate kinase
1674:AIR synthetase (FGAM cyclase)
878:Molecular biology of the cell
2600:Oxamate carbamoyltransferase
2440:Thiopurine methyltransferase
1897:Dihydroorotate dehydrogenase
1250:10.1016/0968-0004(90)90176-C
1097:10.1016/0022-2836(87)90265-8
1085:Journal of Molecular Biology
2980:Genes on human chromosome 2
2615:Lysine carbamoyltransferase
2301:: one carbon transferases (
593:domain, consisting of four
399:aspartate transcarbamoylase
3001:
2252:Sucrase-isomaltase complex
2118:Oxoglutarate dehydrogenase
1468:10.1016/j.cell.2013.02.044
2835:Michaelis–Menten kinetics
2425:Histone methyltransferase
2341:Amine N-methyltransferase
1914:
1892:
1862:
1713:Adenylosuccinate synthase
1009:10.1002/9780470123140.ch3
774:Nature Structural Biology
461:Michaelis–Menten kinetics
359:
194:
26:
2727:Diffusion-limited enzyme
2478:Hydroxymethyltransferase
2210:Electron transport chain
1979:Ribonucleotide reductase
1565:Medical Subject Headings
876:Alberts, Bruce, author.
429:, the enzyme is a multi-
2200:P450-containing systems
1999:Dihydrofolate reductase
1334:10.1073/pnas.83.16.5866
1204:10.1073/pnas.85.12.4205
1145:10.1073/pnas.0503742102
1062:10.1126/science.3041592
962:10.1073/pnas.81.13.4037
692:Assembly of the complex
485:N-carbamoyl-L-aspartate
459:ATCase does not follow
2535:Aminomethyltransferase
2205:Cytochrome b6f complex
1718:Adenylosuccinate lyase
1526:10.1073/pnas.71.4.1351
529:
409:the first step in the
2820:Eadie–Hofstee diagram
2753:Allosteric regulation
2435:DNA methyltransferase
2045:multienzyme complexes
1953:Beta-ureidopropionase
1848:Pyrimidine metabolism
1608:amino acid metabolism
740:10.1073/pnas.87.1.174
523:
2830:Lineweaver–Burk plot
2430:Thymidylate synthase
1994:Thymidylate synthase
1971:Deoxyribonucleotides
945:at 2.6-Å resolution"
686:quaternary structure
645:quaternary structure
2399:Methionine synthase
2257:Tryptophan synthase
2247:Polyketide synthase
1944:Dihydropyrimidinase
1814:Adenosine deaminase
1517:1974PNAS...71.1351E
1380:10.1021/bi00126a026
1325:1986PNAS...83.5866M
1286:10.1021/bi00048a008
1195:1988PNAS...85.4205G
1136:2005PNAS..102.8881W
1054:1988Sci...241..669K
731:1990PNAS...87..174S
650:carbamoyl phosphate
585:for the nucleotide
579:carbamoyl phosphate
489:inorganic phosphate
481:carbamoyl phosphate
2789:Enzyme superfamily
2722:Enzyme promiscuity
1785:Nucleotide salvage
1419:10.1110/ps.4500102
914:has generic name (
530:
2945:
2944:
2654:
2653:
2628:
2627:
2543:
2542:
2501:Formyltransferase
2453:
2452:
2265:
2264:
2007:
2006:
1965:
1964:
1927:
1926:
1842:
1841:
1824:Guanine deaminase
1779:
1778:
1754:IMP dehydrogenase
1626:Purine metabolism
887:978-1-315-73536-8
620:also in contact.
392:
391:
388:
387:
243:
242:
239:
238:
142:metabolic pathway
2992:
2962:
2961:
2953:
2825:Hanes–Woolf plot
2768:Enzyme activator
2763:Enzyme inhibitor
2737:Enzyme catalysis
2681:
2674:
2667:
2658:
2564:
2474:
2321:
2292:
2285:
2278:
2269:
2034:
2027:
2020:
2011:
1860:
1853:
1829:Xanthine oxidase
1747:
1706:
1652:
1638:
1631:
1595:
1588:
1581:
1572:
1549:
1548:
1538:
1528:
1496:
1490:
1489:
1479:
1447:
1441:
1440:
1430:
1398:
1392:
1391:
1363:
1357:
1356:
1346:
1336:
1304:
1298:
1297:
1280:(48): 15654–60.
1268:
1262:
1261:
1233:
1227:
1226:
1216:
1206:
1174:
1168:
1167:
1157:
1147:
1115:
1109:
1108:
1080:
1074:
1073:
1048:(4866): 669–74.
1037:
1031:
1030:
996:
985:
984:
974:
964:
943:Escherichia coli
936:
927:
926:
919:
913:
909:
907:
899:
873:
867:
864:
858:
857:
847:
815:
806:
805:
769:
763:
762:
752:
742:
710:
631:transition state
624:Catalytic center
564:molecular weight
556:regulatory sites
512:
361:
245:
196:
47:
35:Escherichia coli
31:
19:
3000:
2999:
2995:
2994:
2993:
2991:
2990:
2989:
2970:
2969:
2968:
2956:
2948:
2946:
2941:
2853:Oxidoreductases
2839:
2815:Enzyme kinetics
2803:
2799:List of enzymes
2772:
2741:
2712:Catalytic triad
2690:
2685:
2655:
2650:
2624:
2578:
2556:
2539:
2518:
2495:
2466:
2449:
2403:
2375:
2350:
2306:
2296:
2266:
2261:
2166:
2082:
2047:
2038:
2008:
2003:
1961:
1923:
1910:
1888:
1838:
1802:
1775:
1739:
1734:
1698:
1693:
1641:
1620:
1599:
1557:
1552:
1498:
1497:
1493:
1449:
1448:
1444:
1407:Protein Science
1400:
1399:
1395:
1374:(11): 3026–32.
1365:
1364:
1360:
1319:(16): 5866–70.
1306:
1305:
1301:
1270:
1269:
1265:
1235:
1234:
1230:
1176:
1175:
1171:
1117:
1116:
1112:
1082:
1081:
1077:
1039:
1038:
1034:
1019:
998:
997:
988:
955:(13): 4037–40.
938:
937:
930:
920:
910:
900:
888:
875:
874:
870:
865:
861:
817:
816:
809:
771:
770:
766:
712:
711:
707:
703:
694:
640:
638:Allosteric site
626:
518:
473:
443:
439:
397:(also known as
49:
39:
17:
12:
11:
5:
2998:
2996:
2988:
2987:
2982:
2972:
2971:
2967:
2966:
2943:
2942:
2940:
2939:
2926:
2913:
2900:
2887:
2874:
2861:
2847:
2845:
2841:
2840:
2838:
2837:
2832:
2827:
2822:
2817:
2811:
2809:
2805:
2804:
2802:
2801:
2796:
2791:
2786:
2780:
2778:
2777:Classification
2774:
2773:
2771:
2770:
2765:
2760:
2755:
2749:
2747:
2743:
2742:
2740:
2739:
2734:
2729:
2724:
2719:
2714:
2709:
2704:
2698:
2696:
2692:
2691:
2686:
2684:
2683:
2676:
2669:
2661:
2652:
2651:
2649:
2648:
2642:
2640:
2630:
2629:
2626:
2625:
2623:
2622:
2617:
2612:
2607:
2602:
2597:
2592:
2586:
2584:
2580:
2579:
2577:
2576:
2570:
2568:
2561:
2545:
2544:
2541:
2540:
2538:
2537:
2532:
2526:
2524:
2520:
2519:
2517:
2516:
2511:
2505:
2503:
2497:
2496:
2494:
2493:
2488:
2482:
2480:
2471:
2455:
2454:
2451:
2450:
2448:
2447:
2442:
2437:
2432:
2427:
2422:
2417:
2411:
2409:
2405:
2404:
2402:
2401:
2396:
2391:
2385:
2383:
2377:
2376:
2374:
2373:
2368:
2358:
2356:
2352:
2351:
2349:
2348:
2343:
2338:
2333:
2327:
2325:
2318:
2308:
2307:
2297:
2295:
2294:
2287:
2280:
2272:
2263:
2262:
2260:
2259:
2254:
2249:
2244:
2239:
2238:
2237:
2232:
2222:
2217:
2212:
2207:
2202:
2197:
2196:
2195:
2193:Dihydroorotase
2190:
2185:
2174:
2172:
2168:
2167:
2165:
2164:
2163:
2162:
2157:
2152:
2147:
2137:
2136:
2135:
2130:
2125:
2115:
2114:
2113:
2108:
2103:
2092:
2090:
2084:
2083:
2081:
2080:
2079:
2078:
2073:
2063:
2057:
2055:
2053:Photosynthesis
2049:
2048:
2039:
2037:
2036:
2029:
2022:
2014:
2005:
2004:
2002:
2001:
1996:
1991:
1989:DCMP deaminase
1986:
1981:
1975:
1973:
1967:
1966:
1963:
1962:
1960:
1959:
1950:
1941:
1935:
1933:
1929:
1928:
1925:
1924:
1922:
1921:
1919:CTP synthetase
1915:
1912:
1911:
1909:
1908:
1899:
1893:
1890:
1889:
1887:
1886:
1885:
1884:
1882:Dihydroorotase
1879:
1874:
1863:
1857:
1850:
1844:
1843:
1840:
1839:
1837:
1836:
1831:
1826:
1821:
1816:
1810:
1808:
1804:
1803:
1801:
1800:
1795:
1789:
1787:
1781:
1780:
1777:
1776:
1774:
1773:
1772:
1771:
1761:
1756:
1750:
1748:
1736:
1735:
1733:
1732:
1731:
1730:
1720:
1715:
1709:
1707:
1695:
1694:
1692:
1691:
1686:
1681:
1676:
1671:
1666:
1661:
1655:
1653:
1635:
1628:
1622:
1621:
1619:
1618:
1610:
1600:
1598:
1597:
1590:
1583:
1575:
1569:
1568:
1556:
1555:External links
1553:
1551:
1550:
1491:
1462:(2): 461–470.
1442:
1413:(5): 1074–81.
1393:
1358:
1299:
1263:
1228:
1189:(12): 4205–8.
1169:
1130:(25): 8881–6.
1110:
1075:
1032:
1017:
986:
928:
886:
868:
859:
807:
764:
704:
702:
699:
693:
690:
639:
636:
625:
622:
517:
514:
472:
469:
441:
437:
390:
389:
386:
385:
380:
376:
375:
370:
366:
365:
357:
356:
346:
340:
339:
334:
328:
327:
323:
322:
317:
311:
310:
305:
299:
298:
293:
287:
286:
281:
275:
274:
269:
263:
262:
259:
255:
254:
250:
249:
241:
240:
237:
236:
231:
225:
224:
219:
213:
212:
207:
201:
200:
192:
191:
182:
176:
175:
164:
157:
156:
151:
145:
144:
139:
133:
132:
127:
121:
120:
115:
109:
108:
103:
97:
96:
91:
85:
84:
80:
79:
74:
68:
67:
62:
56:
55:
51:
50:
32:
24:
23:
16:Protein family
15:
13:
10:
9:
6:
4:
3:
2:
2997:
2986:
2983:
2981:
2978:
2977:
2975:
2965:
2960:
2955:
2951:
2937:
2933:
2932:
2927:
2924:
2920:
2919:
2914:
2911:
2907:
2906:
2901:
2898:
2894:
2893:
2888:
2885:
2881:
2880:
2875:
2872:
2868:
2867:
2862:
2859:
2855:
2854:
2849:
2848:
2846:
2842:
2836:
2833:
2831:
2828:
2826:
2823:
2821:
2818:
2816:
2813:
2812:
2810:
2806:
2800:
2797:
2795:
2794:Enzyme family
2792:
2790:
2787:
2785:
2782:
2781:
2779:
2775:
2769:
2766:
2764:
2761:
2759:
2758:Cooperativity
2756:
2754:
2751:
2750:
2748:
2744:
2738:
2735:
2733:
2730:
2728:
2725:
2723:
2720:
2718:
2717:Oxyanion hole
2715:
2713:
2710:
2708:
2705:
2703:
2700:
2699:
2697:
2693:
2689:
2682:
2677:
2675:
2670:
2668:
2663:
2662:
2659:
2647:
2644:
2643:
2641:
2639:
2635:
2631:
2621:
2618:
2616:
2613:
2611:
2608:
2606:
2603:
2601:
2598:
2596:
2593:
2591:
2588:
2587:
2585:
2581:
2575:
2572:
2571:
2569:
2565:
2562:
2560:
2554:
2550:
2546:
2536:
2533:
2531:
2528:
2527:
2525:
2521:
2515:
2512:
2510:
2507:
2506:
2504:
2502:
2498:
2492:
2489:
2487:
2484:
2483:
2481:
2479:
2475:
2472:
2470:- and Related
2469:
2464:
2463:Hydroxymethyl
2460:
2456:
2446:
2443:
2441:
2438:
2436:
2433:
2431:
2428:
2426:
2423:
2421:
2418:
2416:
2413:
2412:
2410:
2406:
2400:
2397:
2395:
2392:
2390:
2387:
2386:
2384:
2382:
2378:
2372:
2369:
2367:
2363:
2360:
2359:
2357:
2353:
2347:
2344:
2342:
2339:
2337:
2334:
2332:
2329:
2328:
2326:
2322:
2319:
2317:
2313:
2309:
2304:
2300:
2293:
2288:
2286:
2281:
2279:
2274:
2273:
2270:
2258:
2255:
2253:
2250:
2248:
2245:
2243:
2240:
2236:
2233:
2231:
2228:
2227:
2226:
2223:
2221:
2218:
2216:
2213:
2211:
2208:
2206:
2203:
2201:
2198:
2194:
2191:
2189:
2186:
2184:
2181:
2180:
2179:
2176:
2175:
2173:
2169:
2161:
2158:
2156:
2153:
2151:
2148:
2146:
2143:
2142:
2141:
2138:
2134:
2131:
2129:
2126:
2124:
2121:
2120:
2119:
2116:
2112:
2109:
2107:
2104:
2102:
2099:
2098:
2097:
2094:
2093:
2091:
2089:
2088:Dehydrogenase
2085:
2077:
2074:
2072:
2069:
2068:
2067:
2064:
2062:
2059:
2058:
2056:
2054:
2050:
2046:
2042:
2035:
2030:
2028:
2023:
2021:
2016:
2015:
2012:
2000:
1997:
1995:
1992:
1990:
1987:
1985:
1982:
1980:
1977:
1976:
1974:
1972:
1968:
1958:
1954:
1951:
1949:
1945:
1942:
1940:
1937:
1936:
1934:
1930:
1920:
1917:
1916:
1913:
1907:
1903:
1900:
1898:
1895:
1894:
1891:
1883:
1880:
1878:
1875:
1873:
1870:
1869:
1868:
1865:
1864:
1861:
1858:
1854:
1851:
1849:
1845:
1835:
1834:Urate oxidase
1832:
1830:
1827:
1825:
1822:
1820:
1817:
1815:
1812:
1811:
1809:
1805:
1799:
1796:
1794:
1791:
1790:
1788:
1786:
1782:
1770:
1769:GMP reductase
1767:
1766:
1765:
1762:
1760:
1757:
1755:
1752:
1751:
1749:
1746:
1744:
1737:
1729:
1728:AMP deaminase
1726:
1725:
1724:
1721:
1719:
1716:
1714:
1711:
1710:
1708:
1705:
1703:
1696:
1690:
1687:
1685:
1682:
1680:
1677:
1675:
1672:
1670:
1667:
1665:
1662:
1660:
1657:
1656:
1654:
1651:
1649:
1645:
1639:
1636:
1632:
1629:
1627:
1623:
1617:
1614:
1611:
1609:
1605:
1602:
1601:
1596:
1591:
1589:
1584:
1582:
1577:
1576:
1573:
1566:
1562:
1559:
1558:
1554:
1546:
1542:
1537:
1532:
1527:
1522:
1518:
1514:
1511:(4): 1351–5.
1510:
1506:
1502:
1495:
1492:
1487:
1483:
1478:
1473:
1469:
1465:
1461:
1457:
1453:
1446:
1443:
1438:
1434:
1429:
1424:
1420:
1416:
1412:
1408:
1404:
1397:
1394:
1389:
1385:
1381:
1377:
1373:
1369:
1362:
1359:
1354:
1350:
1345:
1340:
1335:
1330:
1326:
1322:
1318:
1314:
1310:
1303:
1300:
1295:
1291:
1287:
1283:
1279:
1275:
1267:
1264:
1259:
1255:
1251:
1247:
1243:
1239:
1232:
1229:
1224:
1220:
1215:
1210:
1205:
1200:
1196:
1192:
1188:
1184:
1180:
1173:
1170:
1165:
1161:
1156:
1151:
1146:
1141:
1137:
1133:
1129:
1125:
1121:
1114:
1111:
1106:
1102:
1098:
1094:
1091:(3): 527–53.
1090:
1086:
1079:
1076:
1071:
1067:
1063:
1059:
1055:
1051:
1047:
1043:
1036:
1033:
1028:
1024:
1020:
1018:9780470123140
1014:
1010:
1006:
1002:
995:
993:
991:
987:
982:
978:
973:
968:
963:
958:
954:
950:
946:
944:
935:
933:
929:
924:
917:
905:
897:
893:
889:
883:
879:
872:
869:
863:
860:
855:
851:
846:
841:
837:
833:
829:
825:
821:
814:
812:
808:
803:
799:
795:
791:
787:
786:10.1038/87582
783:
779:
775:
768:
765:
760:
756:
751:
746:
741:
736:
732:
728:
724:
720:
716:
709:
706:
700:
698:
691:
689:
687:
683:
679:
675:
669:
665:
663:
659:
655:
651:
646:
637:
635:
632:
623:
621:
618:
614:
609:
607:
604:
600:
596:
592:
588:
584:
580:
576:
571:
569:
565:
561:
557:
553:
549:
545:
540:
538:
536:
527:
522:
515:
513:
511:
506:
502:
498:
494:
490:
486:
482:
478:
470:
468:
466:
462:
457:
455:
451:
447:
435:
432:
428:
427:
421:
419:
416:
412:
408:
404:
400:
396:
384:
381:
377:
374:
371:
367:
362:
358:
355:
354:
350:
347:
345:
341:
338:
335:
333:
329:
324:
321:
318:
316:
312:
309:
306:
304:
300:
297:
294:
292:
288:
285:
282:
280:
276:
273:
270:
268:
264:
260:
256:
251:
246:
235:
232:
230:
226:
223:
220:
218:
214:
211:
208:
206:
202:
197:
193:
190:
186:
183:
181:
180:Gene Ontology
177:
174:
171:
168:
165:
162:
158:
155:
152:
150:
146:
143:
140:
138:
134:
131:
128:
126:
122:
119:
118:NiceZyme view
116:
114:
110:
107:
104:
102:
98:
95:
92:
90:
86:
81:
78:
75:
73:
69:
66:
63:
61:
57:
52:
46:
42:
37:
36:
30:
25:
20:
2931:Translocases
2928:
2915:
2902:
2889:
2876:
2866:Transferases
2863:
2850:
2707:Binding site
2589:
2381:Homocysteine
2187:
1876:
1763:
1759:GMP synthase
1740:
1722:
1699:
1689:IMP synthase
1642:
1508:
1504:
1494:
1459:
1455:
1445:
1410:
1406:
1396:
1371:
1368:Biochemistry
1367:
1361:
1316:
1312:
1302:
1277:
1274:Biochemistry
1273:
1266:
1241:
1237:
1231:
1186:
1182:
1172:
1127:
1123:
1113:
1088:
1084:
1078:
1045:
1041:
1035:
1000:
952:
948:
942:
877:
871:
862:
827:
823:
780:(5): 423–6.
777:
773:
767:
725:(1): 174–8.
722:
718:
708:
695:
688:transition.
670:
666:
641:
627:
610:
583:binding site
572:
541:
533:
531:
525:
474:
458:
444:, forming 2
424:
422:
402:
398:
394:
393:
351:
106:BRENDA entry
33:
2702:Active site
2299:Transferase
2066:Photosystem
1244:(2): 53–9.
912:|last=
617:hydrophobic
544:polypeptide
493:pyrimidines
477:L-aspartate
373:Swiss-model
253:Identifiers
94:IntEnz view
54:Identifiers
2974:Categories
2905:Isomerases
2879:Hydrolases
2746:Regulation
1932:Catabolism
1807:Catabolism
1613:nucleotide
1604:Metabolism
896:1082214404
701:References
697:subunits.
658:substrates
599:coordinate
589:, and the
560:holoenzyme
454:allosteric
369:Structures
364:Search for
326:Other data
163:structures
130:KEGG entry
77:9012-49-1
2784:EC number
2583:Carbamoyl
2559:Carbamoyl
1856:Anabolism
1634:Anabolism
904:cite book
678:aspartate
662:analogues
660:or their
654:aspartate
587:effectors
575:aspartate
552:aspartate
550:group of
516:Structure
407:catalyzes
332:EC number
308:NM_004341
267:NCBI gene
83:Databases
2985:EC 2.1.3
2808:Kinetics
2732:Cofactor
2695:Activity
1486:23582331
1437:11967364
1164:15951418
854:11528003
802:35403933
794:11323717
595:cysteine
483:to form
471:Reaction
383:InterPro
234:proteins
222:articles
210:articles
167:RCSB PDB
2964:Biology
2918:Ligases
2688:Enzymes
2638:Amidine
2567:Carboxy
2553:Carboxy
2316:Methyl-
2041:Enzymes
1764:reverse
1723:reverse
1616:enzymes
1545:4598300
1513:Bibcode
1477:4009401
1428:2373563
1388:1550826
1353:3526342
1321:Bibcode
1294:7495794
1258:2186515
1223:3380787
1191:Bibcode
1155:1157055
1132:Bibcode
1105:3586030
1070:3041592
1050:Bibcode
1042:Science
1027:8154326
981:6377306
759:1967494
727:Bibcode
674:alanine
566:of 310
535:E. coli
528:, 1984.
497:purines
446:trimers
434:protein
431:subunit
426:E. coli
418:2.1.3.2
379:Domains
353:p22-p21
337:2.1.3.2
315:UniProt
189:QuickGO
154:profile
137:MetaCyc
72:CAS no.
65:2.1.3.2
48:
2950:Portal
2892:Lyases
2468:Formyl
2420:DNMT3B
2150:BCKDHB
2145:BCKDHA
1567:(MeSH)
1543:
1536:388226
1533:
1484:
1474:
1435:
1425:
1386:
1351:
1344:386397
1341:
1292:
1256:
1221:
1214:280395
1211:
1162:
1152:
1103:
1068:
1025:
1015:
979:
972:345363
969:
894:
884:
852:
842:
800:
792:
757:
747:
526:et al.
450:dimers
403:ATCase
349:Chr. 2
320:P27708
303:RefSeq
296:114010
258:Symbol
217:PubMed
199:Search
185:AmiGO
173:PDBsum
113:ExPASy
101:BRENDA
89:IntEnz
60:EC no.
2844:Types
2634:2.1.4
2549:2.1.3
2523:Other
2459:2.1.2
2408:Other
2312:2.1.1
2235:HADHB
2230:HADHA
2171:Other
845:99034
798:S2CID
750:53223
613:ionic
548:amino
344:Locus
149:PRIAM
2936:list
2929:EC7
2923:list
2916:EC6
2910:list
2903:EC5
2897:list
2890:EC4
2884:list
2877:EC3
2871:list
2864:EC2
2858:list
2851:EC1
2557:and
2305:2.1)
2128:DLST
2123:OGDH
1957:UPB1
1948:DPYS
1741:IMP→
1700:IMP→
1541:PMID
1482:PMID
1456:Cell
1433:PMID
1384:PMID
1349:PMID
1290:PMID
1254:PMID
1219:PMID
1160:PMID
1101:PMID
1066:PMID
1023:PMID
1013:ISBN
977:PMID
923:link
916:help
892:OCLC
882:ISBN
850:PMID
790:PMID
755:PMID
652:and
615:and
606:atom
603:zinc
591:zinc
495:and
487:and
479:and
291:OMIM
284:1424
279:HGNC
229:NCBI
170:PDBe
125:KEGG
45:4FYY
2178:CAD
2160:DLD
2155:DBT
2133:DLD
1867:CAD
1743:GMP
1702:AMP
1648:IMP
1644:R5P
1531:PMC
1521:doi
1472:PMC
1464:doi
1460:153
1423:PMC
1415:doi
1376:doi
1339:PMC
1329:doi
1282:doi
1246:doi
1209:PMC
1199:doi
1150:PMC
1140:doi
1128:102
1093:doi
1089:193
1058:doi
1046:241
1005:doi
967:PMC
957:doi
840:PMC
832:doi
782:doi
745:PMC
735:doi
682:ATP
568:kDa
539:s.
505:ATP
501:CTP
465:CTP
423:In
420:).
401:or
272:790
261:CAD
205:PMC
161:PDB
41:PDB
2976::
2636::
2551::
2465:-,
2461::
2355:O-
2324:N-
2314::
2303:EC
2111:E3
2106:E2
2101:E1
2076:II
2043::
1606::
1539:.
1529:.
1519:.
1509:71
1507:.
1503:.
1480:.
1470:.
1458:.
1454:.
1431:.
1421:.
1411:11
1409:.
1405:.
1382:.
1372:31
1370:.
1347:.
1337:.
1327:.
1317:83
1315:.
1311:.
1288:.
1278:34
1276:.
1252:.
1242:15
1240:.
1217:.
1207:.
1197:.
1187:85
1185:.
1181:.
1158:.
1148:.
1138:.
1126:.
1122:.
1099:.
1087:.
1064:.
1056:.
1044:.
1021:.
1011:.
989:^
975:.
965:.
953:81
951:.
947:.
931:^
908::
906:}}
902:{{
890:.
880:.
848:.
838:.
828:65
826:.
822:.
810:^
796:.
788:.
776:.
753:.
743:.
733:.
723:87
721:.
717:.
664:.
601:a
570:.
415:EC
405:)
187:/
43::
2952::
2938:)
2934:(
2925:)
2921:(
2912:)
2908:(
2899:)
2895:(
2886:)
2882:(
2873:)
2869:(
2860:)
2856:(
2680:e
2673:t
2666:v
2555:-
2364:/
2291:e
2284:t
2277:v
2071:I
2033:e
2026:t
2019:v
1955:/
1946:/
1904:/
1745::
1704::
1650::
1646:→
1594:e
1587:t
1580:v
1547:.
1523::
1515::
1488:.
1466::
1439:.
1417::
1390:.
1378::
1355:.
1331::
1323::
1296:.
1284::
1260:.
1248::
1225:.
1201::
1193::
1166:.
1142::
1134::
1107:.
1095::
1072:.
1060::
1052::
1029:.
1007::
983:.
959::
925:)
918:)
898:.
856:.
834::
804:.
784::
778:8
761:.
737::
729::
537:'
442:6
440:R
438:6
413:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.