409:
24:
491:
van Luenen, Henri G.A.M.; Farris, Carol; Jan, Sabrina; Genest, Paul-Andre; Tripathi, Pankaj; Velds, Arno; Kerkhoven, Ron M.; Nieuwland, Marja; Haydock, Andrew; Ramasamy, Gowthaman; Vainio, Saara; Heidebrecht, Tatjana; Perrakis, Anastassis; Pagie, Ludo; van
Steensel, Bas; Myler, Peter J.;
132:
246:
220:
InChI=1S/C11H16N2O8/c14-2-5-6(15)7(16)8(17)10(21-5)20-3-4-1-12-11(19)13-9(4)18/h1,5-8,10,14-17H,2-3H2,(H2,12,13,18,19)/t5-,6-,7+,8-,10-/m1/s1
230:
InChI=1/C11H16N2O8/c14-2-5-6(15)7(16)8(17)10(21-5)20-3-4-1-12-11(19)13-9(4)18/h1,5-8,10,14-17H,2-3H2,(H2,12,13,18,19)/t5-,6-,7+,8-,10-/m1/s1
211:
386:
being accompanied by a massive read-through at RNA polymerase II termination sites, which ultimately proves lethal to the cell.
372:
and was the first hypermodified nucleobase found in eukaryotic DNA; it has since been found in other kinetoplastids, including
188:
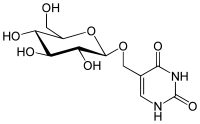
408:
324:
605:
54:
36:
600:
456:, Piet; Sabatini, Robert (October 2008). "Base J: Discovery, Biosynthesis, and Possible Functions".
610:
402:
98:
23:
498:"Glucosylated Hydroxymethyluracil, DNA Base J, Prevents Transcriptional Readthrough in Leishmania"
368:
576:
527:
473:
379:
152:
566:
558:
517:
509:
465:
269:
469:
197:
108:
571:
546:
522:
497:
356:
318:
177:
594:
398:
390:
383:
363:
513:
562:
493:
453:
374:
348:
303:
143:
394:
580:
531:
477:
360:
164:
317:
Except where otherwise noted, data are given for materials in their
407:
131:
121:
352:
545:
Hazelbaker, Dane Z.; Buratowski, Stephen (November 2012).
366:. It was discovered in 1993, in the trypanosome
176:
107:
382:transcription terminator, with its removal in
8:
378:. Within these organisms Base J acts as a
151:
15:
570:
521:
196:
448:
446:
442:
251:
216:
547:"Transcription: Base J Blocks the Way"
470:10.1146/annurev.micro.62.081307.162750
223:Key: SUDBRAWXUGTELR-HPFNVAMJSA-N
7:
233:Key: SUDBRAWXUGTELR-HPFNVAMJBD
167:
254:c1c(c(=O)c(=O)1)CO2((((O2)CO)O)O)O
14:
389:Base J is formed by the initial
287:
281:
22:
321:(at 25 °C , 100 kPa).
80:-Glucosyl-5-hydroxymethyluracil
59:5-({oxy}methyl)pyrimidine-2,4(1
341:-Glucopyranosyloxymethyluracil
293:
275:
1:
458:Annual Review of Microbiology
86:-Glucosyl-hydroxymethyluracil
412:The biosynthesis of base J.
420:: beta-glucosyltranferase;
627:
514:10.1016/j.cell.2012.07.030
401:by an as yet unidentified
563:10.1016/j.cub.2012.10.010
424:: dT (desoxy thymidine);
416:: thymidine hydroxylase;
315:
262:
242:
207:
91:
73:
53:
35:
30:
21:
496:, Piet (August 2012).
433:
411:
55:Systematic IUPAC name
359:including the human
403:glycosyltransferase
397:and the subsequent
347:is a hypermodified
311: g·mol
18:
434:
369:Trypanosoma brucei
325:Infobox references
40:5-pyrimidine-2,4(1
16:
557:(22): R960–R962.
380:RNA polymerase II
340:
333:Chemical compound
331:
330:
133:Interactive image
85:
79:
618:
606:Pyrimidinediones
585:
584:
574:
542:
536:
535:
525:
488:
482:
481:
450:
338:
310:
295:
289:
283:
277:
270:Chemical formula
200:
180:
169:
155:
135:
111:
83:
77:
26:
19:
626:
625:
621:
620:
619:
617:
616:
615:
591:
590:
589:
588:
551:Current Biology
544:
543:
539:
490:
489:
485:
452:
451:
444:
439:
334:
327:
322:
308:
298:
292:
286:
280:
272:
258:
255:
250:
249:
238:
235:
234:
231:
225:
224:
221:
215:
214:
203:
183:
170:
158:
138:
125:
114:
101:
87:
81:
69:
68:
49:
12:
11:
5:
624:
622:
614:
613:
608:
603:
593:
592:
587:
586:
537:
508:(5): 909–921.
483:
464:(1): 235–251.
441:
440:
438:
435:
384:knockout cells
357:kinetoplastids
332:
329:
328:
323:
319:standard state
316:
313:
312:
306:
300:
299:
296:
290:
284:
278:
273:
268:
265:
264:
260:
259:
257:
256:
253:
245:
244:
243:
240:
239:
237:
236:
232:
229:
228:
226:
222:
219:
218:
210:
209:
208:
205:
204:
202:
201:
193:
191:
185:
184:
182:
181:
173:
171:
163:
160:
159:
157:
156:
148:
146:
140:
139:
137:
136:
128:
126:
119:
116:
115:
113:
112:
104:
102:
97:
94:
93:
89:
88:
75:
71:
70:
58:
57:
51:
50:
39:
33:
32:
28:
27:
13:
10:
9:
6:
4:
3:
2:
623:
612:
609:
607:
604:
602:
599:
598:
596:
582:
578:
573:
568:
564:
560:
556:
552:
548:
541:
538:
533:
529:
524:
519:
515:
511:
507:
503:
499:
495:
487:
484:
479:
475:
471:
467:
463:
459:
455:
449:
447:
443:
436:
431:
427:
423:
419:
415:
410:
406:
404:
400:
399:glycosylation
396:
392:
391:hydroxylation
387:
385:
381:
377:
376:
371:
370:
365:
362:
358:
354:
351:found in the
350:
346:
342:
326:
320:
314:
307:
305:
302:
301:
274:
271:
267:
266:
261:
252:
248:
241:
227:
217:
213:
206:
199:
195:
194:
192:
190:
187:
186:
179:
175:
174:
172:
166:
162:
161:
154:
150:
149:
147:
145:
142:
141:
134:
130:
129:
127:
123:
118:
117:
110:
106:
105:
103:
100:
96:
95:
90:
72:
66:
62:
56:
52:
47:
43:
38:
34:
29:
25:
20:
554:
550:
540:
505:
501:
486:
461:
457:
429:
425:
421:
417:
413:
388:
373:
367:
364:trypanosomes
344:
336:
335:
92:Identifiers
74:Other names
64:
60:
45:
41:
601:Nucleobases
428:: HOCH3dU;
263:Properties
611:Glucosides
595:Categories
437:References
375:Leishmania
361:pathogenic
349:nucleobase
304:Molar mass
198:979DWB8FWS
144:ChemSpider
120:3D model (
109:53910-89-7
99:CAS Number
37:IUPAC name
395:thymidine
178:129852278
581:23174300
532:22939620
478:18729733
153:32742278
572:3648658
523:3684241
309:304.255
165:PubChem
67:)-dione
48:)-dione
17:Base J
579:
569:
530:
520:
476:
345:base J
247:SMILES
31:Names
494:Borst
454:Borst
212:InChI
122:JSmol
577:PMID
528:PMID
502:Cell
474:PMID
432:: dJ
189:UNII
567:PMC
559:doi
518:PMC
510:doi
506:150
466:doi
393:of
355:of
353:DNA
343:or
168:CID
597::
575:.
565:.
555:22
553:.
549:.
526:.
516:.
504:.
500:.
472:.
462:62
460:.
445:^
405:.
337:β-
285:16
279:11
82:β-
76:β-
63:,3
44:,3
583:.
561::
534:.
512::
480:.
468::
430:3
426:2
422:1
418:B
414:A
339:D
297:8
294:O
291:2
288:N
282:H
276:C
124:)
84:D
78:D
65:H
61:H
46:H
42:H
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.