44:
1019:
943:
1041:
3404:
910:-acetamido oxygen atom on carbon 1 of the substrate). The aspartate residue stabilizes the positive charge on the nitrogen atom in the oxazolinium ion intermediate. Following the formation of the oxazolinium ion intermediate, water attacks the electrophillic acetal carbon. Glutamate acts as a base by deprotonating the water leading to the formation of the product complex and the G
905:
residue leads to the formation of an oxazolinium ion intermediate. A glutamate residue (α Glu-323/β Glu-355) works as an acid by donating its hydrogen to the glycosidic oxygen atom on the GalNAc residue. An aspartate residue (α Asp-322/β Asp-354) positions the C2-acetamindo group so that it can be
997:
NAG-thiazoline, NGT, acts as mechanism based inhibitor of hexosaminidase A. In patients with Tay–Sachs disease (misfolded hexosaminidase A), NGT acts as a molecular chaperone by binding in the active site of hexosaminidase A which helps create a properly folded hexosaminidase A. The stable dimer
923:
2219:
1034: The light green outline surrounding NGT represents the Van der Waals surface of NGT. The critical amino acids in the active site that are able to hydrogen bond with NGT include Arginine 178 and Glutamate 462.
1056: The light blue outline surrounding NGT represents the Van der Waals surface of NGT. The critical amino acids in the active site that are able to hydrogen bond with NGT include Glutamate 491 and Aspartate 452.
994:. Tay–Sachs causes cerebral degeneration and blindness. Patients also experience flaccid extremities and seizures. At present there has been no cure or effective treatment of Tay–Sachs disease.
983:
The most common mutation, which occurs in over 80 percent of Tay–Sachs patients, results from a four base pair addition (TATC) in exon 11 of the Hex A gene. This insertion leads to an early stop
2212:
2569:
2205:
378:β-hexosaminidase enzymes are dimeric in structure. Three isoenzymes are produced through the combination of α and β subunits to form any one of three active dimers:
209:
1672:
1725:
Knapp S, Vocadlo D, Gao Z, Kirk B, Lou J, Withers SG (1996). "NAG-thiazoline, an N-acetylbeta-hexosaminidase inhibitor that implicates acetamido participation".
2562:
1302:
1143:
680:
527:
228:
2024:"Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain"
980:
gangliosides in the brain than the unaffected person. Over 100 different mutations have been discovered just in infantile cases of Tay–Sachs disease alone.
2708:
1690:
Hou Y, Tse R, Mahuran DJ (April 1996). "Direct determination of the substrate specificity of the alpha-active site in heterodimeric beta-hexosaminidase".
2741:
2555:
2065:"Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer"
43:
3439:
3434:
3424:
972:
occurs when hexosaminidase A loses its ability to function. People with Tay–Sachs disease are unable to remove the GalNAc residue from the G
3429:
968:
There are numerous mutations that lead to hexosaminidase deficiency including gene deletions, nonsense mutations, and missense mutations.
1098:
activities. NCOAT is also known as hexosaminidase C and has distinct substrate specificities compared to lysosomal hexosaminidase A. A
1167:
704:
551:
817:
Even though the α and β subunits of lysosomal hexosaminidase can both cleave GalNAc residues, only the α subunit is able to hydrolyze G
1816:"Crystallographic Structure of Human β-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of GM2 Ganglioside Hydrolysis"
350:
Elevated levels of hexosaminidase in blood and/or urine have been proposed as a biomarker of relapse in the treatment of alcoholism.
3123:
2362:
1875:
825:-424, and a loop structure that forms from the amino acid sequence in the alpha subunit. The loop in the α subunit, consisting of
1314:
1155:
692:
539:
221:
1307:
3005:
2357:
1866:
Ozand PT, Nyhan WL, Barshop BA (2005). "Part
Thirteen Lipid Storage Disorders: Tay-Sachs disease/hexosaminidase A deficiency".
1148:
685:
532:
2734:
2478:
2470:
1099:
172:
148:
3279:
3017:
2521:
2531:
2864:
1754:"Crystal Structure of Human β-Hexosaminidase B: Understanding the Molecular Basis of Sandhoff and Tay–Sachs Disease"
3394:
3264:
3380:
3367:
3354:
3341:
3328:
3315:
3302:
3075:
2763:
2727:
2613:
1095:
3274:
166:
3228:
3171:
2754:
2189:
2183:
1355:
1208:
1103:
745:
592:
249:
59:
2547:
153:
3176:
2927:
2663:
2582:
2483:
3090:
2835:
1647:
1319:
1934:"The histone acetyltransferase NCOAT contains a zinc finger-like motif involved in substrate recognition"
1160:
969:
697:
544:
494:
354:
233:
3197:
3116:
1372:
1225:
999:
609:
353:
Hereditary inability to form functional hexosaminidase enzymes are the cause of lipid storage disorders
141:
3269:
3049:
3000:
2384:
2076:
762:
76:
2063:
Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA (August 2006).
3233:
2944:
2932:
2767:
2699:
2332:
2265:
2197:
169:
71:
1893:"The molecular basis of HEXA mRNA deficiency caused by the most common Tay–Sachs disease mutation"
93:
3166:
2988:
2983:
2961:
2949:
2788:
2634:
2456:
2342:
2337:
2312:
2297:
1229:
990:
Children born with Tay–Sachs usually die between two and four years of age from aspiration and
3444:
2956:
2830:
2651:
2257:
2153:
2104:
2045:
2004:
1955:
1914:
1871:
1845:
1783:
1707:
1628:
1583:
1536:
1479:
1048:
1026:
160:
3212:
3207:
3181:
3109:
2966:
2852:
2656:
2270:
2232:
2179:
2143:
2135:
2094:
2084:
2035:
1994:
1986:
1945:
1904:
1835:
1827:
1773:
1765:
1734:
1699:
1618:
1575:
1526:
1518:
1469:
1461:
1376:
1367:
1220:
841:-283 which is absent in the β subunit, serves as an ideal structure for the binding of the G
757:
613:
604:
474:
358:
1360:
1213:
766:
750:
597:
129:
3259:
3243:
3156:
3086:
2603:
2379:
1396:
1249:
1109:
A fourth mammalian hexosaminidase polypeptide which has been designated hexosaminidase D (
786:
633:
105:
2080:
1018:
252:
64:
3408:
3297:
3238:
2939:
2776:
2719:
2148:
2123:
2099:
2064:
1999:
1974:
1909:
1892:
1840:
1815:
1778:
1753:
1603:"Studies on the glycosidases of jack bean meal. 3. Crystallization and properties of β-
1531:
1498:
1474:
1437:
204:
1769:
1623:
1602:
184:
3418:
3202:
3161:
2973:
2902:
2885:
2629:
2351:
2307:
894:
834:
179:
17:
1046:
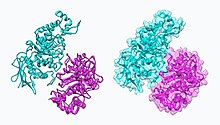
The β subunit active site shown bound to NAG-thiazoline (NGT) in β-hexosaminidase.
942:
3151:
2817:
2586:
2247:
1024:
The α subunit active site shown bound to NAG-thiazoline (NGT) in β-hexosaminidase.
462:
1975:"Studies on human N-acetyl-Beta-d-hexosaminidase C separated from neonatal brain"
1331:
1184:
721:
568:
3375:
3310:
3146:
3044:
3010:
2978:
2673:
2645:
2395:
2291:
1406:
1259:
796:
643:
188:
3403:
2175:
GeneReviews/NCBI/NIH/UW entry on hexosaminidase A deficiency, Tay–Sachs disease
1814:
Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN (June 2006).
1438:"Some comments on the type references of the official nomenclature (IUB) for β-
1040:
869:
to hexosaminidase, so a functional hexosaminidase enzyme is able to hydrolyze G
3054:
3022:
2668:
2578:
2403:
2236:
2228:
2174:
1831:
1338:
1191:
922:
728:
575:
486:
336:
324:
3349:
3323:
2892:
2825:
2802:
2792:
2750:
2373:
2321:
2089:
991:
902:
478:
2157:
2108:
2049:
2040:
2023:
1959:
1950:
1933:
1849:
1787:
976:
ganglioside, and as a result, they end up storing 100 to 1000 times more G
1918:
1711:
1632:
1587:
1483:
2897:
2513:
2281:
2122:
Gutternigg M, Rendić D, Voglauer R, Iskratsch T, Wilson IB (April 2009).
2008:
1752:
Mark BL, Mahuran DJ, Cherney MM, Zhao D, Knapp S, James MN (April 2003).
1343:
1196:
1003:
822:
733:
580:
375:
2192:
1579:
1540:
117:
2880:
2807:
2797:
2784:
2689:
2139:
1465:
838:
826:
136:
1990:
1738:
1703:
1522:
1052:
1030:
3362:
3132:
2589:
2525:
2438:
2433:
2239:
1326:
1179:
830:
716:
563:
320:
216:
112:
100:
88:
1010:
gangliosides. The two subunits of hexosaminidase A are shown below:
2124:"Mammalian cells contain a second nucleocytoplasmic hexosaminidase"
3336:
3059:
2840:
2428:
2423:
2418:
2413:
2408:
1136:
1090:
984:
866:
3037:
3032:
2993:
2922:
2917:
2912:
2907:
2857:
2845:
2608:
2535:
2503:
2498:
2493:
2488:
2327:
2302:
1172:
709:
673:
556:
520:
451:
446:
440:
124:
3105:
2723:
2551:
2201:
998:
conformation of hexosaminidase A has the ability to leave the
2022:
Gao Y, Wells L, Comer FI, Parker GJ, Hart GW (March 2001).
1646:
Allen JP, Sillanaukee P, Strid N, Litten RZ (August 2004).
3101:
952:
ganglioside and removal of a GalNAc residue to produce G
1554:
Frohwein YZ, Gatt S (September 1967). "Isolation of β-
432:
exists in tissues but no known physiological function
421:
exists in tissues but no known physiological function
3392:
1932:
Toleman CA, Paterson AJ, Kudlow JE (February 2006).
438:
The α and β subunits are encoded by separate genes,
3288:
3252:
3221:
3190:
3139:
3074:
2873:
2816:
2775:
2762:
2698:
2682:
2622:
2596:
2512:
2469:
2449:
2393:
2279:
2255:
2246:
1402:
1392:
1387:
1366:
1354:
1349:
1337:
1325:
1313:
1301:
1293:
1285:
1280:
1275:
1255:
1245:
1240:
1219:
1207:
1202:
1190:
1178:
1166:
1154:
1142:
1132:
1127:
1122:
792:
782:
777:
756:
744:
739:
727:
715:
703:
691:
679:
669:
664:
659:
639:
629:
624:
603:
591:
586:
574:
562:
550:
538:
526:
516:
511:
506:
380:
227:
215:
203:
198:
178:
159:
147:
135:
123:
111:
99:
87:
82:
70:
58:
53:
32:
1652:National Institute on Alcohol Abuse and Alcoholism
1497:Calvo P, Reglero A, Cabezas JA (November 1978).
450:respectively. β-Hexosaminidase and the cofactor
849:AP), and arginine is essential for binding the
3117:
2735:
2563:
2213:
964:Gene mutations resulting in Tay–Sachs disease
8:
881:-acetylgalactosamine (GalNAc) residue from G
2709:N-acetylglucosamine-1-phosphate transferase
1870:. London: Hodder Arnold. pp. 539–546.
1566:-acetylgalactosaminidase from calf brain".
1102:in the human O-GlcNAcase gene is linked to
3124:
3110:
3102:
2772:
2742:
2728:
2720:
2570:
2556:
2548:
2252:
2220:
2206:
2198:
1809:
1807:
1805:
1803:
1801:
1799:
1797:
1384:
1237:
936:ganglioside catalyzed by hexosaminidase A.
774:
621:
195:
42:
2182:at the U.S. National Library of Medicine
2147:
2098:
2088:
2039:
1998:
1949:
1908:
1839:
1777:
1622:
1530:
1473:
1006:where it can perform the degradation of G
493:gangliosides, which is the main cause of
1861:
1859:
1673:"Tay-Sachs Disease and Sandhoff Disease"
465:and other molecules containing terminal
3399:
1503:-acetylhexosaminidase from the mollusc
1428:
1094:gene possesses both hexosaminidase and
948:The mechanism of the hydrolysis of a G
821:gangliosides because of a key residue,
469:-acetyl hexosamines. Gene mutations in
1973:Besley GT, Broadhead DM (April 1976).
1272:
1119:
656:
503:
29:
987:, which causes the Hex A deficiency.
7:
893:A Michaelis complex consisting of a
853:-acetyl-neuraminic acid residue of G
1088:ransferase) that is encoded by the
403:only isoenzyme that can hydrolyze G
1499:"Purification and properties of β-
897:residue, a GalNAc residue on the G
25:
2363:Sphingomyelin phosphodiesterase 1
1891:Boles DJ, Proia RL (March 1995).
458:catalyze the degradation of the G
3402:
1113:) has recently been identified.
1068:The bifunctional protein NCOAT (
1039:
1017:
941:
921:
370:Lysosomal A, B, and S isoenzymes
3006:Alpha-N-acetylgalactosaminidase
2358:Sphingomyelin phosphodiesterase
2479:Palmitoyl protein thioesterase
1648:"Biomarkers of Heavy Drinking"
1562:-acetylglucosaminidase, and β-
1100:single-nucleotide polymorphism
865:gangliosides and presents the
861:activator protein transports G
1:
3018:Alpha-N-acetylglucosaminidase
2522:Serine C-palmitoyltransferase
1770:10.1016/S0022-2836(03)00216-X
1624:10.1016/S0021-9258(18)62830-3
1601:Li SC, Li YT (October 1970).
906:attacked by the nucleophile (
877:gangliosides by removing the
3440:Genes on human chromosome 17
3435:Genes on human chromosome 10
3425:Genes on human chromosome 15
2532:Ceramide glucosyltransferase
2069:Proc. Natl. Acad. Sci. U.S.A
1446:-acetylhexosaminidase and β-
431:
428:
425:
420:
417:
414:
402:
399:
396:
257:β-acetylaminodeoxyhexosidase
3430:Genes on human chromosome 5
1868:Atlas of metabolic diseases
3461:
1442:-acetylglucosaminidase, β-
1436:Cabezas JA (August 1989).
1064:Cytosolic C and D isozymes
660:β-hexosaminidase subunit β
507:β-hexosaminidase subunit α
3280:Michaelis–Menten kinetics
2614:Oligosaccharyltransferase
1832:10.1016/j.jmb.2006.04.004
1558:-acetylhexosaminidase, β-
1450:-acetylgalactosaminidase"
1383:
1236:
1096:histone acetyltransferase
773:
620:
301:β-N-acetylglucosaminidase
269:N-acetyl-β-hexosaminidase
194:
41:
3172:Diffusion-limited enzyme
2184:Medical Subject Headings
1104:diabetes mellitus type 2
281:β-acetylhexosaminidinase
48:Hexosaminidase A (Hex A)
2928:Bacterial neuraminidase
2664:Aspartylglucosaminidase
2583:carbohydrate metabolism
2484:Tripeptidyl peptidase I
2090:10.1073/pnas.0601931103
1002:and is directed to the
289:-N-acetylhexosaminidase
273:N-acetyl hexosaminidase
3091:Oxoguanine glycosylase
2041:10.1074/jbc.M010420200
1951:10.1074/jbc.M510485200
1607:-acetylhexosaminidase"
309:N-acetylhexosaminidase
3265:Eadie–Hofstee diagram
3198:Allosteric regulation
1000:endoplasmic reticulum
37:-Acetylhexosaminidase
18:Beta-hexosaminidase A
3275:Lineweaver–Burk plot
3080:N-Glycosyl compounds
3050:Maltase-glucoamylase
3001:Galactosylceramidase
2768:Glycoside hydrolases
2753:: sugar hydrolases (
2385:Galactosylceramidase
1505:Helicella ericetorum
901:ganglioside, and an
857:gangliosides. The G
845:activator protein (G
389:subunit composition
365:Isoenzymes and genes
2933:Viral neuraminidase
2333:Alpha-galactosidase
2266:Glycosyltransferase
2081:2006PNAS..10311952F
1580:10.1021/bi00861a018
889:Mechanism of action
873:gangliosides into G
3234:Enzyme superfamily
3167:Enzyme promiscuity
2984:Glucosylceramidase
2865:Debranching enzyme
2789:Sucrase-isomaltase
2635:Beta-galactosidase
2514:Ceramide synthesis
2457:Sphingosine kinase
2343:Glucocerebrosidase
2338:Beta-galactosidase
2313:Glucocerebrosidase
2298:Beta-galactosidase
2140:10.1042/BJ20081630
1466:10.1042/bj2611059b
3390:
3389:
3099:
3098:
3070:
3069:
2957:alpha-Mannosidase
2831:Alpha-glucosidase
2717:
2716:
2652:alpha-Mannosidase
2545:
2544:
2465:
2464:
2258:glycosphingolipid
1991:10.1042/bj1550205
1897:Am. J. Hum. Genet
1739:10.1021/ja960826u
1733:(28): 6804–6805.
1704:10.1021/bi9524575
1523:10.1042/bj1750743
1420:
1419:
1416:
1415:
1412:
1411:
1269:
1268:
1265:
1264:
970:Tay–Sachs disease
810:
809:
806:
805:
802:
801:
653:
652:
649:
648:
495:Tay–Sachs disease
456:activator protein
436:
435:
355:Tay-Sachs disease
346:
334:
316:
296:
288:
264:
243:
242:
239:
238:
142:metabolic pathway
16:(Redirected from
3452:
3407:
3406:
3398:
3270:Hanes–Woolf plot
3213:Enzyme activator
3208:Enzyme inhibitor
3182:Enzyme catalysis
3126:
3119:
3112:
3103:
3087:DNA glycosylases
2853:Beta-glucosidase
2773:
2744:
2737:
2730:
2721:
2657:beta-mannosidase
2572:
2565:
2558:
2549:
2376:
2354:
2328:Hexosaminidase B
2324:
2303:Hexosaminidase A
2294:
2271:Sulfotransferase
2253:
2233:lipid metabolism
2222:
2215:
2208:
2199:
2180:hexosaminidase A
2162:
2161:
2151:
2119:
2113:
2112:
2102:
2092:
2060:
2054:
2053:
2043:
2019:
2013:
2012:
2002:
1970:
1964:
1963:
1953:
1929:
1923:
1922:
1912:
1888:
1882:
1881:
1863:
1854:
1853:
1843:
1811:
1792:
1791:
1781:
1749:
1743:
1742:
1727:J. Am. Chem. Soc
1722:
1716:
1715:
1687:
1681:
1680:
1668:
1662:
1661:
1659:
1658:
1643:
1637:
1636:
1626:
1598:
1592:
1591:
1551:
1545:
1544:
1534:
1494:
1488:
1487:
1477:
1433:
1385:
1276:hexosaminidase D
1273:
1238:
1123:hexosaminidase C
1120:
1116:
1115:
1055:
1043:
1033:
1021:
945:
932:ganglioside to G
925:
775:
657:
622:
504:
500:
499:
475:Sandhoff disease
473:often result in
400:α/β heterodimer
381:
359:Sandhoff disease
347:-hexosaminides.
344:
332:
323:involved in the
314:
305:hexosaminidase A
294:
286:
277:β-hexosaminidase
262:
196:
46:
30:
27:Class of enzymes
21:
3460:
3459:
3455:
3454:
3453:
3451:
3450:
3449:
3415:
3414:
3413:
3401:
3393:
3391:
3386:
3298:Oxidoreductases
3284:
3260:Enzyme kinetics
3248:
3244:List of enzymes
3217:
3186:
3157:Catalytic triad
3135:
3130:
3100:
3095:
3079:
3066:
2869:
2812:
2758:
2748:
2718:
2713:
2694:
2678:
2618:
2604:Dolichol kinase
2592:
2576:
2546:
2541:
2508:
2461:
2445:
2389:
2380:Arylsulfatase A
2371:
2349:
2319:
2289:
2275:
2242:
2226:
2171:
2166:
2165:
2121:
2120:
2116:
2075:(32): 11952–7.
2062:
2061:
2057:
2034:(13): 9838–45.
2021:
2020:
2016:
1972:
1971:
1967:
1931:
1930:
1926:
1890:
1889:
1885:
1878:
1865:
1864:
1857:
1813:
1812:
1795:
1764:(5): 1093–109.
1751:
1750:
1746:
1724:
1723:
1719:
1689:
1688:
1684:
1671:Demczko, Matt.
1670:
1669:
1665:
1656:
1654:
1645:
1644:
1640:
1617:(19): 5153–60.
1600:
1599:
1595:
1553:
1552:
1548:
1496:
1495:
1491:
1435:
1434:
1430:
1425:
1080:-GlcNAcase and
1066:
1061:
1060:
1059:
1058:
1057:
1047:
1044:
1036:
1035:
1025:
1022:
1009:
979:
975:
966:
961:
960:
959:
958:
957:
955:
951:
946:
938:
937:
935:
931:
928:Hydrolysis of G
926:
913:
900:
891:
884:
876:
872:
864:
860:
856:
848:
844:
820:
815:
492:
461:
455:
406:
385:
372:
367:
317:-hexosaminidase
297:-hexosaminidase
265:-hexosaminidase
49:
28:
23:
22:
15:
12:
11:
5:
3458:
3456:
3448:
3447:
3442:
3437:
3432:
3427:
3417:
3416:
3412:
3411:
3388:
3387:
3385:
3384:
3371:
3358:
3345:
3332:
3319:
3306:
3292:
3290:
3286:
3285:
3283:
3282:
3277:
3272:
3267:
3262:
3256:
3254:
3250:
3249:
3247:
3246:
3241:
3236:
3231:
3225:
3223:
3222:Classification
3219:
3218:
3216:
3215:
3210:
3205:
3200:
3194:
3192:
3188:
3187:
3185:
3184:
3179:
3174:
3169:
3164:
3159:
3154:
3149:
3143:
3141:
3137:
3136:
3131:
3129:
3128:
3121:
3114:
3106:
3097:
3096:
3094:
3093:
3083:
3081:
3072:
3071:
3068:
3067:
3065:
3064:
3063:
3062:
3052:
3047:
3042:
3041:
3040:
3035:
3028:Hexosaminidase
3025:
3020:
3015:
3014:
3013:
3003:
2998:
2997:
2996:
2991:
2981:
2976:
2971:
2970:
2969:
2959:
2954:
2953:
2952:
2947:
2940:Galactosidases
2937:
2936:
2935:
2930:
2925:
2920:
2915:
2910:
2900:
2895:
2890:
2889:
2888:
2877:
2875:
2871:
2870:
2868:
2867:
2862:
2861:
2860:
2850:
2849:
2848:
2843:
2838:
2828:
2822:
2820:
2814:
2813:
2811:
2810:
2805:
2800:
2795:
2781:
2779:
2777:Disaccharidase
2770:
2760:
2759:
2749:
2747:
2746:
2739:
2732:
2724:
2715:
2714:
2712:
2711:
2705:
2703:
2696:
2695:
2693:
2692:
2686:
2684:
2680:
2679:
2677:
2676:
2671:
2666:
2661:
2660:
2659:
2654:
2642:
2640:Hexosaminidase
2637:
2632:
2626:
2624:
2620:
2619:
2617:
2616:
2611:
2606:
2600:
2598:
2594:
2593:
2577:
2575:
2574:
2567:
2560:
2552:
2543:
2542:
2540:
2539:
2529:
2518:
2516:
2510:
2509:
2507:
2506:
2501:
2496:
2491:
2486:
2481:
2475:
2473:
2467:
2466:
2463:
2462:
2460:
2459:
2453:
2451:
2447:
2446:
2444:
2443:
2442:
2441:
2436:
2431:
2426:
2421:
2416:
2411:
2400:
2398:
2391:
2390:
2388:
2387:
2382:
2377:
2368:
2367:
2366:
2365:
2355:
2346:
2345:
2340:
2335:
2330:
2325:
2316:
2315:
2310:
2305:
2300:
2295:
2286:
2284:
2277:
2276:
2274:
2273:
2268:
2262:
2260:
2250:
2244:
2243:
2227:
2225:
2224:
2217:
2210:
2202:
2196:
2195:
2187:
2177:
2170:
2169:External links
2167:
2164:
2163:
2114:
2055:
2014:
1965:
1944:(7): 3918–25.
1924:
1883:
1876:
1855:
1793:
1744:
1717:
1698:(13): 3963–9.
1682:
1663:
1638:
1593:
1574:(9): 2775–82.
1546:
1489:
1460:(3): 1059–60.
1427:
1426:
1424:
1421:
1418:
1417:
1414:
1413:
1410:
1409:
1404:
1400:
1399:
1394:
1390:
1389:
1381:
1380:
1370:
1364:
1363:
1358:
1352:
1351:
1347:
1346:
1341:
1335:
1334:
1329:
1323:
1322:
1317:
1311:
1310:
1305:
1299:
1298:
1295:
1291:
1290:
1287:
1283:
1282:
1278:
1277:
1270:
1267:
1266:
1263:
1262:
1257:
1253:
1252:
1247:
1243:
1242:
1234:
1233:
1223:
1217:
1216:
1211:
1205:
1204:
1200:
1199:
1194:
1188:
1187:
1182:
1176:
1175:
1170:
1164:
1163:
1158:
1152:
1151:
1146:
1140:
1139:
1134:
1130:
1129:
1125:
1124:
1065:
1062:
1045:
1038:
1037:
1023:
1016:
1015:
1014:
1013:
1012:
1007:
977:
973:
965:
962:
953:
949:
947:
940:
939:
933:
929:
927:
920:
919:
918:
917:
916:
911:
898:
890:
887:
885:gangliosides.
882:
874:
870:
862:
858:
854:
846:
842:
818:
814:
811:
808:
807:
804:
803:
800:
799:
794:
790:
789:
784:
780:
779:
771:
770:
760:
754:
753:
748:
742:
741:
737:
736:
731:
725:
724:
719:
713:
712:
707:
701:
700:
695:
689:
688:
683:
677:
676:
671:
667:
666:
662:
661:
654:
651:
650:
647:
646:
641:
637:
636:
631:
627:
626:
618:
617:
607:
601:
600:
595:
589:
588:
584:
583:
578:
572:
571:
566:
560:
559:
554:
548:
547:
542:
536:
535:
530:
524:
523:
518:
514:
513:
509:
508:
490:
459:
453:
434:
433:
430:
429:α/α homodimer
427:
423:
422:
419:
418:β/β homodimer
416:
412:
411:
404:
401:
398:
394:
393:
390:
387:
384:hexosaminidase
371:
368:
366:
363:
246:Hexosaminidase
241:
240:
237:
236:
231:
225:
224:
219:
213:
212:
207:
201:
200:
192:
191:
182:
176:
175:
164:
157:
156:
151:
145:
144:
139:
133:
132:
127:
121:
120:
115:
109:
108:
103:
97:
96:
91:
85:
84:
80:
79:
74:
68:
67:
62:
56:
55:
51:
50:
47:
39:
38:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3457:
3446:
3443:
3441:
3438:
3436:
3433:
3431:
3428:
3426:
3423:
3422:
3420:
3410:
3405:
3400:
3396:
3382:
3378:
3377:
3372:
3369:
3365:
3364:
3359:
3356:
3352:
3351:
3346:
3343:
3339:
3338:
3333:
3330:
3326:
3325:
3320:
3317:
3313:
3312:
3307:
3304:
3300:
3299:
3294:
3293:
3291:
3287:
3281:
3278:
3276:
3273:
3271:
3268:
3266:
3263:
3261:
3258:
3257:
3255:
3251:
3245:
3242:
3240:
3239:Enzyme family
3237:
3235:
3232:
3230:
3227:
3226:
3224:
3220:
3214:
3211:
3209:
3206:
3204:
3203:Cooperativity
3201:
3199:
3196:
3195:
3193:
3189:
3183:
3180:
3178:
3175:
3173:
3170:
3168:
3165:
3163:
3162:Oxyanion hole
3160:
3158:
3155:
3153:
3150:
3148:
3145:
3144:
3142:
3138:
3134:
3127:
3122:
3120:
3115:
3113:
3108:
3107:
3104:
3092:
3088:
3085:
3084:
3082:
3078:: Hydrolysing
3077:
3073:
3061:
3058:
3057:
3056:
3053:
3051:
3048:
3046:
3043:
3039:
3036:
3034:
3031:
3030:
3029:
3026:
3024:
3021:
3019:
3016:
3012:
3009:
3008:
3007:
3004:
3002:
2999:
2995:
2994:non-lysosomal
2992:
2990:
2987:
2986:
2985:
2982:
2980:
2977:
2975:
2974:Hyaluronidase
2972:
2968:
2965:
2964:
2963:
2962:Glucuronidase
2960:
2958:
2955:
2951:
2948:
2946:
2943:
2942:
2941:
2938:
2934:
2931:
2929:
2926:
2924:
2921:
2919:
2916:
2914:
2911:
2909:
2906:
2905:
2904:
2903:Neuraminidase
2901:
2899:
2896:
2894:
2891:
2887:
2886:Alpha-amylase
2884:
2883:
2882:
2879:
2878:
2876:
2872:
2866:
2863:
2859:
2856:
2855:
2854:
2851:
2847:
2844:
2842:
2839:
2837:
2834:
2833:
2832:
2829:
2827:
2824:
2823:
2821:
2819:
2815:
2809:
2806:
2804:
2801:
2799:
2796:
2794:
2790:
2786:
2783:
2782:
2780:
2778:
2774:
2771:
2769:
2765:
2761:
2756:
2752:
2745:
2740:
2738:
2733:
2731:
2726:
2725:
2722:
2710:
2707:
2706:
2704:
2701:
2697:
2691:
2688:
2687:
2685:
2681:
2675:
2672:
2670:
2667:
2665:
2662:
2658:
2655:
2653:
2650:
2649:
2648:
2647:
2643:
2641:
2638:
2636:
2633:
2631:
2630:Neuraminidase
2628:
2627:
2625:
2621:
2615:
2612:
2610:
2607:
2605:
2602:
2601:
2599:
2595:
2591:
2588:
2584:
2580:
2573:
2568:
2566:
2561:
2559:
2554:
2553:
2550:
2537:
2533:
2530:
2527:
2523:
2520:
2519:
2517:
2515:
2511:
2505:
2502:
2500:
2497:
2495:
2492:
2490:
2487:
2485:
2482:
2480:
2477:
2476:
2474:
2472:
2468:
2458:
2455:
2454:
2452:
2448:
2440:
2437:
2435:
2432:
2430:
2427:
2425:
2422:
2420:
2417:
2415:
2412:
2410:
2407:
2406:
2405:
2402:
2401:
2399:
2397:
2392:
2386:
2383:
2381:
2378:
2375:
2370:
2369:
2364:
2361:
2360:
2359:
2356:
2353:
2352:sphingomyelin
2348:
2347:
2344:
2341:
2339:
2336:
2334:
2331:
2329:
2326:
2323:
2318:
2317:
2314:
2311:
2309:
2308:Neuraminidase
2306:
2304:
2301:
2299:
2296:
2293:
2288:
2287:
2285:
2283:
2278:
2272:
2269:
2267:
2264:
2263:
2261:
2259:
2254:
2251:
2249:
2245:
2241:
2238:
2234:
2230:
2223:
2218:
2216:
2211:
2209:
2204:
2203:
2200:
2194:
2191:
2188:
2185:
2181:
2178:
2176:
2173:
2172:
2168:
2159:
2155:
2150:
2145:
2141:
2137:
2133:
2129:
2125:
2118:
2115:
2110:
2106:
2101:
2096:
2091:
2086:
2082:
2078:
2074:
2070:
2066:
2059:
2056:
2051:
2047:
2042:
2037:
2033:
2029:
2028:J. Biol. Chem
2025:
2018:
2015:
2010:
2006:
2001:
1996:
1992:
1988:
1984:
1980:
1976:
1969:
1966:
1961:
1957:
1952:
1947:
1943:
1939:
1938:J. Biol. Chem
1935:
1928:
1925:
1920:
1916:
1911:
1906:
1903:(3): 716–24.
1902:
1898:
1894:
1887:
1884:
1879:
1877:0-340-80970-1
1873:
1869:
1862:
1860:
1856:
1851:
1847:
1842:
1837:
1833:
1829:
1826:(4): 913–29.
1825:
1821:
1817:
1810:
1808:
1806:
1804:
1802:
1800:
1798:
1794:
1789:
1785:
1780:
1775:
1771:
1767:
1763:
1759:
1755:
1748:
1745:
1740:
1736:
1732:
1728:
1721:
1718:
1713:
1709:
1705:
1701:
1697:
1693:
1686:
1683:
1678:
1674:
1667:
1664:
1653:
1649:
1642:
1639:
1634:
1630:
1625:
1620:
1616:
1612:
1611:J. Biol. Chem
1608:
1606:
1597:
1594:
1589:
1585:
1581:
1577:
1573:
1569:
1565:
1561:
1557:
1550:
1547:
1542:
1538:
1533:
1528:
1524:
1520:
1517:(2): 743–50.
1516:
1512:
1508:
1506:
1502:
1493:
1490:
1485:
1481:
1476:
1471:
1467:
1463:
1459:
1455:
1451:
1449:
1445:
1441:
1432:
1429:
1422:
1408:
1405:
1401:
1398:
1395:
1391:
1386:
1382:
1379:
1378:
1374:
1371:
1369:
1365:
1362:
1359:
1357:
1353:
1348:
1345:
1342:
1340:
1336:
1333:
1330:
1328:
1324:
1321:
1318:
1316:
1312:
1309:
1306:
1304:
1300:
1296:
1292:
1288:
1284:
1279:
1274:
1271:
1261:
1258:
1254:
1251:
1248:
1244:
1239:
1235:
1232:
1231:
1227:
1224:
1222:
1218:
1215:
1212:
1210:
1206:
1201:
1198:
1195:
1193:
1189:
1186:
1183:
1181:
1177:
1174:
1171:
1169:
1165:
1162:
1159:
1157:
1153:
1150:
1147:
1145:
1141:
1138:
1135:
1131:
1126:
1121:
1118:
1117:
1114:
1112:
1107:
1105:
1101:
1097:
1093:
1092:
1087:
1083:
1079:
1075:
1071:
1063:
1054:
1050:
1042:
1032:
1028:
1020:
1011:
1005:
1001:
995:
993:
988:
986:
981:
971:
963:
944:
924:
915:
914:ganglioside.
909:
904:
896:
888:
886:
880:
868:
852:
840:
836:
832:
828:
824:
812:
798:
795:
791:
788:
785:
781:
776:
772:
769:
768:
764:
761:
759:
755:
752:
749:
747:
743:
738:
735:
732:
730:
726:
723:
720:
718:
714:
711:
708:
706:
702:
699:
696:
694:
690:
687:
684:
682:
678:
675:
672:
668:
663:
658:
655:
645:
642:
638:
635:
632:
628:
623:
619:
616:
615:
611:
608:
606:
602:
599:
596:
594:
590:
585:
582:
579:
577:
573:
570:
567:
565:
561:
558:
555:
553:
549:
546:
543:
541:
537:
534:
531:
529:
525:
522:
519:
515:
510:
505:
502:
501:
498:
496:
488:
485:decrease the
484:
480:
476:
472:
468:
464:
457:
449:
448:
443:
442:
424:
413:
410:
395:
391:
388:
383:
382:
379:
377:
369:
364:
362:
360:
356:
351:
348:
342:
338:
330:
326:
322:
318:
310:
306:
302:
298:
290:
282:
278:
274:
270:
266:
258:
254:
251:
247:
235:
232:
230:
226:
223:
220:
218:
214:
211:
208:
206:
202:
197:
193:
190:
186:
183:
181:
180:Gene Ontology
177:
174:
171:
168:
165:
162:
158:
155:
152:
150:
146:
143:
140:
138:
134:
131:
128:
126:
122:
119:
118:NiceZyme view
116:
114:
110:
107:
104:
102:
98:
95:
92:
90:
86:
81:
78:
75:
73:
69:
66:
63:
61:
57:
52:
45:
40:
36:
31:
19:
3376:Translocases
3373:
3360:
3347:
3334:
3321:
3311:Transferases
3308:
3295:
3152:Binding site
3027:
2818:Glucosidases
2644:
2639:
2587:glycoprotein
2248:Sphingolipid
2134:(1): 83–90.
2131:
2127:
2117:
2072:
2068:
2058:
2031:
2027:
2017:
1985:(1): 205–8.
1982:
1978:
1968:
1941:
1937:
1927:
1900:
1896:
1886:
1867:
1823:
1820:J. Mol. Biol
1819:
1761:
1758:J. Mol. Biol
1757:
1747:
1730:
1726:
1720:
1695:
1692:Biochemistry
1691:
1685:
1676:
1666:
1655:. Retrieved
1651:
1641:
1614:
1610:
1604:
1596:
1571:
1568:Biochemistry
1567:
1563:
1559:
1555:
1549:
1514:
1510:
1504:
1500:
1492:
1457:
1453:
1447:
1443:
1439:
1431:
1375:
1294:Alt. symbols
1228:
1110:
1108:
1089:
1085:
1081:
1077:
1073:
1069:
1067:
996:
989:
982:
967:
956:ganglioside.
907:
892:
878:
850:
816:
765:
612:
482:
470:
466:
463:gangliosides
445:
439:
437:
408:
407:ganglioside
373:
352:
349:
340:
339:residues in
328:
327:of terminal
312:
308:
304:
300:
292:
284:
280:
276:
272:
268:
260:
256:
245:
244:
106:BRENDA entry
34:
3147:Active site
3045:Iduronidase
2979:Pullulanase
2646:mannosidase
2396:sphingosine
2292:ganglioside
1397:Swiss-model
1281:Identifiers
1250:Swiss-model
1128:Identifiers
1076:ytoplasmic
787:Swiss-model
665:Identifiers
634:Swiss-model
512:Identifiers
477:; whereas,
374:Functional
293:β-N-acetyl-
261:N-acetyl-β-
94:IntEnz view
54:Identifiers
3419:Categories
3350:Isomerases
3324:Hydrolases
3191:Regulation
3055:Heparanase
3023:Fucosidase
2841:Neutral AB
2669:Fucosidase
2623:Catabolism
2579:Metabolism
2404:Ceramidase
2237:glycolipid
2229:Metabolism
2128:Biochem. J
1979:Biochem. J
1677:MSD Manual
1657:2021-07-11
1511:Biochem. J
1454:Biochem. J
1423:References
1393:Structures
1388:Search for
1350:Other data
1246:Structures
1241:Search for
1230:q24.1-24.3
1203:Other data
837:-282, and
783:Structures
778:Search for
740:Other data
630:Structures
625:Search for
587:Other data
487:hydrolysis
386:isoenzyme
343:-acetyl-β-
337:hexosamine
325:hydrolysis
163:structures
130:KEGG entry
77:9012-33-3
3229:EC number
2989:lysosomal
2893:Chitinase
2858:cytosolic
2846:Neutral C
2826:Cellulase
2803:Trehalase
2793:Invertase
2751:Hydrolase
2683:Transport
2597:Anabolism
2374:sulfatide
2322:globoside
1356:EC number
1332:NM_173620
1303:NCBI gene
1209:EC number
1185:NM_012215
1144:NCBI gene
992:pneumonia
903:aspartate
895:glutamate
746:EC number
722:NM_000521
681:NCBI gene
593:EC number
569:NM_000520
528:NCBI gene
479:mutations
392:function
376:lysosomal
83:Databases
3445:EC 3.2.1
3253:Kinetics
3177:Cofactor
3140:Activity
2898:Lysozyme
2282:ceramide
2193:3.2.1.52
2158:19040401
2109:16882729
2050:11148210
1960:16356930
1850:16698036
1788:12662933
1407:InterPro
1361:3.2.1.52
1297:FLJ23825
1260:InterPro
1214:3.2.1.52
1004:lysosome
813:Function
797:InterPro
751:3.2.1.52
644:InterPro
598:3.2.1.52
331:-acetyl-
319:) is an
253:3.2.1.52
234:proteins
222:articles
210:articles
167:RCSB PDB
65:3.2.1.52
3409:Biology
3363:Ligases
3133:Enzymes
2881:Amylase
2808:Lactase
2798:Maltase
2785:Sucrase
2702:tagging
2690:SLC17A5
2590:enzymes
2585:·
2240:enzymes
2149:2850170
2100:1567679
2077:Bibcode
2000:1172820
1919:7887427
1910:1801160
1841:2910082
1779:2910754
1712:8672428
1633:5506280
1588:6055190
1532:1186125
1507:Müller"
1484:2529847
1475:1138940
1403:Domains
1373:Chr. 17
1339:UniProt
1256:Domains
1226:Chr. 10
1192:UniProt
1072:uclear
793:Domains
729:UniProt
640:Domains
610:Chr. 15
576:UniProt
409:in vivo
189:QuickGO
154:profile
137:MetaCyc
72:CAS no.
3395:Portal
3337:Lyases
2967:Klotho
2526:SPTLC1
2439:ASAH2C
2434:ASAH2B
2186:(MeSH)
2156:
2146:
2107:
2097:
2048:
2009:945735
2007:
1997:
1958:
1917:
1907:
1874:
1848:
1838:
1786:
1776:
1710:
1631:
1586:
1539:
1529:
1482:
1472:
1344:Q8IYN4
1327:RefSeq
1308:284004
1286:Symbol
1197:O60502
1180:RefSeq
1173:604039
1133:Symbol
867:lipids
833:-281,
829:-280,
763:Chr. 5
734:P07686
717:RefSeq
710:606873
670:Symbol
581:P06865
564:RefSeq
557:606869
517:Symbol
321:enzyme
217:PubMed
199:Search
185:AmiGO
173:PDBsum
113:ExPASy
101:BRENDA
89:IntEnz
60:EC no.
3289:Types
3076:3.2.2
3060:HPSE2
2945:Alpha
2874:Other
2764:3.2.1
2450:Other
2429:ASAH2
2424:ASAH1
2419:ACER3
2414:ACER2
2409:ACER1
2372:From
2350:From
2320:From
2290:From
1541:33660
1377:q25.3
1368:Locus
1320:26307
1289:HEXDC
1221:Locus
1149:10724
1137:MGEA5
1111:HEXDC
1091:MGEA5
1084:cetyl
985:codon
767:q13.3
758:Locus
614:q24.1
605:Locus
149:PRIAM
3381:list
3374:EC7
3368:list
3361:EC6
3355:list
3348:EC5
3342:list
3335:EC4
3329:list
3322:EC3
3316:list
3309:EC2
3303:list
3296:EC1
3038:HEXB
3033:HEXA
3011:NAGA
2950:Beta
2923:NEU4
2918:NEU3
2913:NEU2
2908:NEU1
2836:Acid
2757:3.2)
2674:NAGA
2609:GCS1
2536:UGCG
2504:CLN8
2499:CLN6
2494:CLN5
2489:CLN3
2154:PMID
2105:PMID
2046:PMID
2005:PMID
1956:PMID
1915:PMID
1872:ISBN
1846:PMID
1784:PMID
1708:PMID
1629:PMID
1584:PMID
1537:PMID
1480:PMID
1315:HGNC
1168:OMIM
1161:7056
1156:HGNC
1053:2GK1
1031:2GK1
705:OMIM
698:4879
693:HGNC
686:3074
674:HEXB
552:OMIM
545:4878
540:HGNC
533:3073
521:HEXA
489:of G
483:HEXA
471:HEXB
447:HEXB
444:and
441:HEXA
357:and
229:NCBI
170:PDBe
125:KEGG
2700:M6P
2471:NCL
2394:To
2280:To
2256:To
2144:PMC
2136:doi
2132:419
2095:PMC
2085:doi
2073:103
2036:doi
2032:276
1995:PMC
1987:doi
1983:155
1946:doi
1942:281
1905:PMC
1836:PMC
1828:doi
1824:359
1774:PMC
1766:doi
1762:327
1735:doi
1731:118
1700:doi
1619:doi
1615:245
1576:doi
1527:PMC
1519:doi
1515:175
1470:PMC
1462:doi
1458:261
1049:PDB
1027:PDB
839:Pro
835:Glu
831:Ser
827:Gly
823:Arg
481:in
205:PMC
161:PDB
3421::
3089::
2766::
2755:EC
2581::
2235:,
2231:,
2190:EC
2152:.
2142:.
2130:.
2126:.
2103:.
2093:.
2083:.
2071:.
2067:.
2044:.
2030:.
2026:.
2003:.
1993:.
1981:.
1977:.
1954:.
1940:.
1936:.
1913:.
1901:56
1899:.
1895:.
1858:^
1844:.
1834:.
1822:.
1818:.
1796:^
1782:.
1772:.
1760:.
1756:.
1729:.
1706:.
1696:35
1694:.
1675:.
1650:.
1627:.
1613:.
1609:.
1582:.
1570:.
1535:.
1525:.
1513:.
1509:.
1478:.
1468:.
1456:.
1452:.
1106:.
1051::
1029::
1008:M2
978:M2
974:M2
954:M3
950:M2
934:M3
930:M2
912:M3
899:M2
883:M2
875:M3
871:M2
863:M2
859:M2
855:M2
847:M2
843:M2
819:M2
497:.
491:M2
460:M2
454:M2
426:S
415:B
405:M2
397:A
361:.
313:β-
311:,
307:,
303:,
299:,
291:,
285:β-
283:,
279:,
275:,
271:,
267:,
259:,
255:,
250:EC
187:/
33:β-
3397::
3383:)
3379:(
3370:)
3366:(
3357:)
3353:(
3344:)
3340:(
3331:)
3327:(
3318:)
3314:(
3305:)
3301:(
3125:e
3118:t
3111:v
2791:/
2787:/
2743:e
2736:t
2729:v
2571:e
2564:t
2557:v
2538:)
2534:(
2528:)
2524:(
2221:e
2214:t
2207:v
2160:.
2138::
2111:.
2087::
2079::
2052:.
2038::
2011:.
1989::
1962:.
1948::
1921:.
1880:.
1852:.
1830::
1790:.
1768::
1741:.
1737::
1714:.
1702::
1679:.
1660:.
1635:.
1621::
1605:N
1590:.
1578::
1572:6
1564:N
1560:N
1556:N
1543:.
1521::
1501:N
1486:.
1464::
1448:N
1444:N
1440:N
1086:t
1082:a
1078:O
1074:c
1070:n
908:N
879:N
851:N
467:N
452:G
345:D
341:N
335:-
333:D
329:N
315:D
295:D
287:D
263:D
248:(
35:N
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.