121:
signal the end of the loop, thus defining this hairpin as a three-residue loop. This single hydrogen bond is then removed to create the tertiary hairpin; a five-residue loop with doubly bound residues. This pattern continues indefinitely and defines all beta hairpins within the class. Class 2 follows the same pattern beginning with a two-residue loop with terminating residues that share two hydrogen bonds. Class 3 begins with a three-residue, and class 4 with a four-residue. Class 5 does not exist as that primary hairpin is already defined in class 1. Pi This classification scheme not only accounts for various degrees of hydrogen bonding, but also says something about the biological behavior of the hairpin. Single amino acid replacements may destroy a particular hydrogen bond, but will not unfold the hairpin or change its class. On the other hand, amino acid insertions and deletions will have to unfold and reform the entire
270:, β-hairpins are not stabilized by a regular hydrogen bonding pattern. As a result, early attempts required at least 20–30 amino acid residues to attain stable tertiary folds of β-hairpins. However, this lower limit was reduced to 12 amino acids by the stability gains conferred by the incorporation of tryptophan-tryptophan cross-strand pairs. Two nonhydrogen-bonding tryptophan pairs have been shown to interlock in a zipper-like motif, stabilizing the β-hairpin structure while still allowing it to remain
297:, which can be induced to switch from the trans to the cis conformation by light at 360 nm. When the azobenzene moiety is in the cis conformation, the amino acid residues align correctly to adopt a β-hairpin formation. However, the trans conformation does not have proper turn geometry for the β-hairpin. This phenomenon can be used to investigate peptide conformational dynamics with femtosecond absorption spectroscopy.
147:
249:
170:) have uncovered a stepwise folding process that drives beta-hairpin folding. This hairpin has sequence features similar to over 13,000 known hairpins, and thus may serve as a more general model for beta hairpin formation. The formation of a native turn region signals the folding cascade to start, where a
193:
Researchers believe that turns do not originate in the N-strand, due to increased rigidity (often caused by a proline leading up to the native turn region) and less conformational options. The initial turn formation takes place in about 1 μs. Once the initial turn has been established, two mechanisms
150:
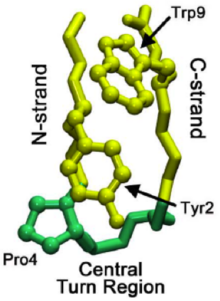
The Pin1 Domain. Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) – a 34-residue protein – is depicted above in two different ways. On the left, the reverse turns are easily seen in green, while the β-strands are seen in yellow. These come together to create a β-hairpin motif. The figure
115:
residues in their loop sequences, such that they were named one-residue, two-residue, etc. This system, however, is somewhat ambiguous as it does not take into account whether the residues that signal the end of the hairpin are singly or doubly hydrogen bonded to one another. An improved means of
120:
in the beta sheet. The primary hairpin of class 1 is a one-residue loop where the bound residues share two hydrogen bonds. One hydrogen bond is then removed to create a three-residue loop, which is the secondary hairpin of class 1. Singly bound residues are counted in the loop sequence but also
239:
116:
classification has since been proposed by Milner-White and Poet. Beta hairpins are broken into four distinct classes as depicted in the publication's Figure 1. Each class begins with the smallest possible number of loop residues and progressively increases the loop size by removing
177:
In the folding of overall proteins, the turn may originate not in the native turn region but in the C-strand of the beta-hairpin. This turn then propagates through the C-strand (the beta strand leading to C-terminus) until it reaches the native turn region. Sometimes the
129:
in the secondary structure. This will change the class of the hairpin in the process. As substitutions are the most common amino acid mutations, a protein could potentially undergo a conversion without affecting the functionality of the beta hairpin.
19:
452:
Jager, Marcus; Deechongkit, Songpon; Koepf, Edward K.; Nguyen, Houbi; Gao, Jianmin; Powers, Evan T.; Gruebele, Martin; Kelly, Jeffery W. (2008). "Understanding the mechanism of β-sheet folding from a chemical and biological perspective".
139:
230:
residues within the actual loop portion of the β-hairpin, since this amino acid is rigid and contributes to the "turn" formation. These proline residues can be seen as red side chains in the image of the Pin1 WW domain below (left).
265:
The design of peptides that adopt β-hairpin structure (without relying on metal binding, unusual amino acids, or disulfide crosslinks) has made significant progress and yielded insights into protein dynamics. Unlike
286:
487:
Kay, B.K.; Williamson, M.P.; Sudol, M. The
Importance of Being Proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. The FASEB Journal. 2000, 14, 231–241.
567:
Dong, Shou-Liang; Löweneck, Markus; Schrader, Tobias E.; Schreier, Wolfgang J.; Zinth, Wolfgang; Moroder, Luis; Renner, Christian (23 January 2006). "A Photocontrolled β-Hairpin
Peptide".
223:
of the conserved tryptophans and the proline-rich areas of the ligand. Other amino acids can then associate with the hydrophobic core of the β-hairpin structure to enforce secure binding.
216:(W) residues that are conserved within the sequence and aid in the folding of the β-sheets to produce a small hydrophobic core. These tryptophan residues can be seen below (right) in red.
628:
194:
have been proposed as to how the rest of the beta-hairpin folds: a hydrophobic collapse with side-chain level rearrangements, or the more accepted zipper-like mechanism.
293:
The synthesis of trpzip β-hairpin peptides has incorporated photoswitches that facilitate precise control over folding. Several amino acids in the turn are replaced by
197:
The β-hairpin loop motif can be found in many macromolecular proteins. However, small and simple β-hairpins can exist on their own as well. To see this clearly, the
182:
interactions leading up to the native turn region are too strong, causing reverse propagation. However, once the native turn does form, interactions between
621:
614:
314:
Blanco, F. J.; Rivas, G.; Serrano, L. (1994). "A short linear peptide that folds into a native stable beta-hairpin in aqueous solution".
776:
209:
278:
structure of a tryptophan zipper (trpzip) β-peptide shows the stabilizing effect of favorable interactions between adjacent
155:
Understanding the mechanism through which micro-domains fold can help to shed light onto the folding patterns of whole
643:
58:
190:
residues (seen in image at right) in the region help to stabilize the turn, preventing "roll back" or dissolution.
356:
Sibanda, B.L.; Blundell, T.L.; Thorton, J.M. (1985). "Conformations of Beta-Hairpins in
Protein Structures".
510:
408:
220:
708:
85:
339:
397:"β-hairpin forms by rolling up from C-terminal: Topological guidance of early folding dynamics"
682:
592:
584:
546:
528:
470:
434:
331:
54:
50:
751:
576:
536:
518:
462:
424:
416:
323:
42:
100:
514:
412:
687:
429:
396:
164:
770:
541:
498:
271:
117:
74:
343:
267:
208:, function by adhering to proline-rich and/or phosphorylated peptides to mediate
746:
730:
658:
497:
Cochran, Andrea G.; Skelton, Nicholas J.; Starovasnik, Melissa A. (8 May 2001).
375:
Milner-White, J.; Poet, R. (1986). "Four
Classes of Beta-Hairpins in Proteins".
122:
92:
46:
725:
703:
677:
606:
395:
Enemark, Søren; Kurniawan, Nicholas A.; Rajagopalan, Raj (11 September 2012).
294:
213:
187:
179:
126:
112:
99:
in aqueous solution, suggesting that hairpins could form nucleation sites for
78:
70:
66:
62:
588:
532:
713:
663:
205:
596:
580:
550:
523:
474:
438:
335:
18:
672:
156:
327:
151:
on the right depicts the same enzyme in a more three-dimensional aspect.
138:
227:
183:
96:
39:
466:
420:
279:
284:
247:
237:
145:
137:
111:
Beta hairpins were originally categorized solely by the number of
17:
73:. Beta hairpins can occur in isolation or as part of a series of
198:
610:
285:
275:
248:
238:
146:
95:
to show that beta-hairpins can be formed from isolated short
174:
turn is one that is present in the final folded structure.
53:. The motif consists of two strands that are adjacent in
69:
of the next), and linked by a short loop of two to five
390:
388:
739:
696:
651:
642:
201:Domain protein is shown to the left as an example.
499:"Tryptophan zippers: Stable, monomeric β-hairpins"
503:Proceedings of the National Academy of Sciences
371:
369:
622:
8:
204:Proteins that are β-sheet rich, also called
648:
629:
615:
607:
540:
522:
428:
306:
219:This enzyme binds its ligand through
77:strands that collectively comprise a
7:
562:
560:
142:Native turn region of a beta-hairpin
252:Pin1 wwdomain-Conserved Tryptophans
159:. Studies of a beta hairpin called
261:Artificially designed beta-hairpin
14:
22:CGI representation of a β-hairpin
242:Pin1 wwdomain-Proline-rich loops
65:of one sheet is adjacent to the
569:Chemistry – A European Journal
1:
210:protein–protein interactions
134:Folding and binding dynamics
644:Protein secondary structure
637:Protein secondary structure
793:
226:It is also common to find
777:Protein structural motifs
212:. The "WW" refers to two
30:(sometimes also called
581:10.1002/chem.200500986
524:10.1073/pnas.091100898
290:
253:
243:
152:
143:
23:
288:
251:
241:
149:
141:
21:
377:Biochemical Journal
221:van der Waals forces
125:in order to avoid a
84:Researchers such as
515:2001PNAS...98.5578C
413:2012NatSR...2E.649E
328:10.1038/nsb0994-584
401:Scientific Reports
291:
289:azobenzene hairpin
254:
244:
153:
144:
24:
764:
763:
760:
759:
683:Polyproline helix
509:(10): 5578–5583.
467:10.1002/bip.21101
421:10.1038/srep00649
258:
257:
57:, oriented in an
55:primary structure
49:that look like a
784:
752:Helix-turn-helix
649:
631:
624:
617:
608:
601:
600:
575:(4): 1114–1120.
564:
555:
554:
544:
526:
494:
488:
485:
479:
478:
449:
443:
442:
432:
392:
383:
373:
364:
354:
348:
347:
311:
234:
233:
86:Francisco Blanco
43:structural motif
792:
791:
787:
786:
785:
783:
782:
781:
767:
766:
765:
756:
740:Supersecondary:
735:
692:
667:
638:
635:
605:
604:
566:
565:
558:
496:
495:
491:
486:
482:
451:
450:
446:
394:
393:
386:
374:
367:
358:Nature(London)
355:
351:
316:Nat Struct Biol
313:
312:
308:
303:
263:
136:
109:
101:protein folding
75:hydrogen bonded
61:direction (the
12:
11:
5:
790:
788:
780:
779:
769:
768:
762:
761:
758:
757:
755:
754:
749:
743:
741:
737:
736:
734:
733:
728:
723:
718:
717:
716:
706:
700:
698:
694:
693:
691:
690:
688:Collagen helix
685:
680:
675:
670:
665:
661:
655:
653:
646:
640:
639:
636:
634:
633:
626:
619:
611:
603:
602:
556:
489:
480:
461:(6): 751–758.
444:
384:
365:
349:
322:(9): 584–590.
305:
304:
302:
299:
262:
259:
256:
255:
245:
168:on Proteopedia
135:
132:
118:hydrogen bonds
108:
107:Classification
105:
45:involving two
38:) is a simple
36:beta-beta unit
13:
10:
9:
6:
4:
3:
2:
789:
778:
775:
774:
772:
753:
750:
748:
745:
744:
742:
738:
732:
729:
727:
724:
722:
719:
715:
712:
711:
710:
707:
705:
702:
701:
699:
695:
689:
686:
684:
681:
679:
676:
674:
671:
669:
662:
660:
657:
656:
654:
650:
647:
645:
641:
632:
627:
625:
620:
618:
613:
612:
609:
598:
594:
590:
586:
582:
578:
574:
570:
563:
561:
557:
552:
548:
543:
538:
534:
530:
525:
520:
516:
512:
508:
504:
500:
493:
490:
484:
481:
476:
472:
468:
464:
460:
456:
448:
445:
440:
436:
431:
426:
422:
418:
414:
410:
406:
402:
398:
391:
389:
385:
381:
378:
372:
370:
366:
362:
359:
353:
350:
345:
341:
337:
333:
329:
325:
321:
317:
310:
307:
300:
298:
296:
287:
283:
281:
277:
273:
272:water-soluble
269:
260:
250:
246:
240:
236:
235:
232:
229:
224:
222:
217:
215:
211:
207:
202:
200:
195:
191:
189:
185:
181:
175:
173:
169:
167:
162:
158:
148:
140:
133:
131:
128:
124:
119:
114:
106:
104:
102:
98:
94:
90:
87:
82:
80:
76:
72:
68:
64:
60:
56:
52:
48:
44:
41:
37:
33:
29:
20:
16:
721:Beta hairpin
720:
572:
568:
506:
502:
492:
483:
458:
454:
447:
404:
400:
379:
376:
360:
357:
352:
319:
315:
309:
292:
264:
225:
218:
203:
196:
192:
176:
171:
165:
160:
154:
110:
88:
83:
59:antiparallel
47:beta strands
35:
31:
28:beta hairpin
27:
25:
15:
747:Coiled coil
455:Biopolymers
123:beta strand
93:protein NMR
71:amino acids
32:beta-ribbon
726:Beta bulge
301:References
295:azobenzene
214:tryptophan
206:WW domains
188:tryptophan
127:beta bulge
113:amino acid
91:have used
79:beta sheet
67:C-terminus
63:N-terminus
714:Beta turn
697:Extended:
589:1521-3765
533:0027-8424
268:α-helices
166:Chignolin
161:chignolin
771:Category
731:α-strand
704:β-strand
652:Helices:
597:16294349
551:11331745
475:18844292
439:22970341
382:289–292.
363:170–174.
344:35065527
184:prolines
157:proteins
97:peptides
678:β-helix
673:π-helix
659:α-helix
511:Bibcode
430:3438464
409:Bibcode
407:: 649.
336:7634098
282:rings.
228:proline
180:residue
51:hairpin
40:protein
595:
587:
549:
539:
531:
473:
437:
427:
342:
334:
280:indole
274:. The
172:native
89:et al.
668:helix
542:33255
340:S2CID
163:(see
709:Turn
593:PMID
585:ISSN
547:PMID
529:ISSN
471:PMID
435:PMID
380:240
361:316
332:PMID
199:Pin1
186:and
26:The
577:doi
537:PMC
519:doi
463:doi
425:PMC
417:doi
324:doi
276:NMR
34:or
773::
666:10
591:.
583:.
573:12
571:.
559:^
545:.
535:.
527:.
517:.
507:98
505:.
501:.
469:.
459:90
457:.
433:.
423:.
415:.
403:.
399:.
387:^
368:^
338:.
330:.
318:.
103:.
81:.
664:3
630:e
623:t
616:v
599:.
579::
553:.
521::
513::
477:.
465::
441:.
419::
411::
405:2
346:.
326::
320:1
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.