1839:
1631:
correlate with the existence of more enzyme in the reduced state, lead to a greater inhibition of cyanide. At these basal concentrations, NO inhibition of
Complex IV is known to have beneficial effects, such as increasing oxygen levels in blood vessel tissues. The inability of the enzyme to reduce oxygen to water results in a buildup of oxygen, which can diffuse deeper into surrounding tissues. NO inhibition of Complex IV has a larger effect at lower oxygen concentrations, increasing its utility as a vasodilator in tissues of need.
1819:
correlation between COX enzyme amount and activity, which indicates the regulation of COX at the level of gene expression. COX distribution is inconsistent across different regions of the animal brain, but its pattern of its distribution is consistent across animals. This pattern has been observed in the monkey, mouse, and calf brain. One isozyme of COX has been consistently detected in histochemical analysis of the brain. Such brain mapping has been accomplished in spontaneous mutant mice with cerebellar disease such as
1673:
40:
1452:
1416:
with other components of translation machinery, but exact molecular mechanisms are unclear due to difficulties associated with synthesizing translation machinery in-vitro. Though the interactions between subunits I, II, and III encoded within the mitochondrial genome make a lesser contribution to enzyme stability than interactions between bigenomic subunits, these subunits are more conserved, indicating potential unexplored roles for enzyme activity.
1851:
1430:
1790:. Mutations in these proteins can result in altered functionality of sub-complex assembly, copper transport, or translational regulation. Each gene mutation is associated with the etiology of a specific disease, with some having implications in multiple disorders. Disorders involving dysfunctional COX assembly via gene mutations include
265:
1730:
secretory granules. The extramitochondrial function of these cytochrome c oxidase subunits has not yet been characterized. Besides cytochrome c oxidase subunits, extramitochondrial localization has also been observed for large numbers of other mitochondrial proteins. This raises the possibility about
1623:
Cyanide is a non-competitive inhibitor for COX, binding with high affinity to the partially-reduced state of the enzyme and hindering further reduction of the enzyme. In the pulsed state, cyanide binds slowly, but with high affinity. The ligand is posited to electrostatically stabilize both metals at
1754:
The vast majority of COX disorders are linked to mutations in nuclear-encoded proteins referred to as assembly factors, or assembly proteins. These assembly factors contribute to COX structure and functionality, and are involved in several essential processes, including transcription and translation
1630:
can reversibly bind to either metal ion in the binuclear center to be oxidized to nitrite. NO and CN will compete with oxygen to bind at the site, reducing the rate of cellular respiration. Endogenous NO, however, which is produced at lower levels, augments CN inhibition. Higher levels of NO, which
1408:
molecule, which has been found to play a key role in stabilization of the holoenzyme complex. The dissociation of subunits VIIa and III in conjunction with the removal of cardiolipin results in total loss of enzyme activity. Subunits encoded in the nuclear genome are known to play a role in enzyme
1403:
Cofactors, including hemes, are inserted into subunits I & II. The two heme molecules reside in subunit I, helping with transport to subunit II where two copper molecules aid with the continued transfer of electrons. Subunits I and IV initiate assembly. Different subunits may associate to form
1619:
of cells. Higher concentrations of molecular oxygen are needed to compensate for increasing inhibitor concentrations, leading to an overall decrease in metabolic activity in the cell in the presence of an inhibitor. Other ligands, such as nitric oxide and hydrogen sulfide, can also inhibit COX by
1415:
Synthesis and assembly of COX subunits I, II, and III are facilitated by translational activators, which interact with the 5’ untranslated regions of mitochondrial mRNA transcripts. Translational activators are encoded in the nucleus. They can operate through either direct or indirect interaction
1592:
nuclear centers are oxidized; this is the conformation of the enzyme that has the highest activity. A two-electron reduction initiates a conformational change that allows oxygen to bind at the active site to the partially-reduced enzyme. Four electrons bind to COX to fully reduce the enzyme. Its
1399:
are a complex process that is not entirely understood due to the rapid and irreversible aggregation of hydrophobic subunits that form the holoenzyme complex, as well as aggregation of mutant subunits with exposed hydrophobic patches. COX subunits are encoded in both the nuclear and mitochondrial
1818:
The increased reliance of neurons on oxidative phosphorylation for energy facilitates the use of COX histochemistry in mapping regional brain metabolism in animals, since it establishes a direct and positive correlation between enzyme activity and neuronal activity. This can be seen in the
1637:
will bind COX in a noncompetitive fashion at a regulatory site on the enzyme, similar to carbon monoxide. Sulfide has the highest affinity to either the pulsed or partially reduced states of the enzyme, and is capable of partially reducing the enzyme at the heme
1539:
of Tyr(244), which becomes a tyrosyl radical. The second oxygen is converted to a hydroxide ion by picking up two electrons and a proton. A third electron from another cytochrome c is passed through the first two electron carriers to the cytochrome
3011:
Sadacharan SK, Singh B, Bowes T, Gupta RS (November 2005). "Localization of mitochondrial DNA encoded cytochrome c oxidase subunits I and II in rat pancreatic zymogen granules and pituitary growth hormone granules".
407:, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton
1992:
Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S (August 1995). "Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A".
1404:
sub-complex intermediates that later bind to other subunits to form the COX complex. In post-assembly modifications, COX will form a homodimer. This is required for activity. Dimers are connected by a
2266:
Kozjak-Pavlovic V, Prell F, Thiede B, Götz M, Wosiek D, Ott C, Rudel T (February 2014). "C1orf163/RESA1 is a novel mitochondrial intermembrane space protein connected to respiratory chain assembly".
1584:
COX exists in three conformational states: fully oxidized (pulsed), partially reduced, and fully reduced. Each inhibitor has a high affinity to a different state. In the pulsed state, both the heme a
517:
production. However, the currently accepted mechanism involves a rapid four-electron reduction involving immediate oxygen–oxygen bond cleavage, avoiding any intermediate likely to form superoxide.
501:
of cytochrome c oxidase show an unusual post-translational modification, linking C6 of Tyr(244) and the ε-N of His(240) (bovine enzyme numbering). It plays a vital role in enabling the cytochrome a
4008:
3632:
3482:
Strazielle C, Sturchler-Pierrat C, Staufenbiel M, Lalonde R (2003). "Regional brain cytochrome oxidase activity in beta-amyloid precursor protein transgenic mice with the
Swedish mutation".
1564:
binuclear center, reducing the Fe=O to Fe, with the oxygen atom picking up a proton simultaneously, regenerating this oxygen as a hydroxide ion coordinated in the middle of the cytochrome a
1412:
Assembly is known to occur in at least three distinct rate-determining steps. The products of these steps have been found, though specific subunit compositions have not been determined.
1400:
genomes. The three subunits that form the COX catalytic core are encoded in the mitochondrial genome. Over 30 different nuclear-encoded chaperone proteins are required for COX assembly.
3525:
Conejo NM, González-Pardo H, Gonzalez-Lima F, Arias JL (March 2010). "Spatial learning of the water maze: progression of brain circuits mapped with cytochrome oxidase histochemistry".
2166:"Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase"
2813:
2851:
1523:
binuclear center, reducing the metals to the Fe form and Cu. The hydroxide ligand is protonated and lost as water, creating a void between the metals that is filled by O
3300:"Neuron-specific specificity protein 4 bigenomically regulates the transcription of all mitochondria- and nucleus-encoded cytochrome c oxidase subunit genes in neurons"
4109:
4001:
3971:
2164:
Szklarczyk R, Wanschers BF, Cuypers TD, Esseling JJ, Riemersma M, van den Brand MA, Gloerich J, Lasonder E, van den Heuvel LP, Nijtmans LG, Huynen MA (February 2012).
2491:
Sedlák E, Robinson NC (September 2015). "Destabilization of the
Quaternary Structure of Bovine Heart Cytochrome c Oxidase upon Removal of Tightly Bound Cardiolipin".
219:
3625:
2351:
Fontanesi F, Soto IC, Horn D, Barrientos A (December 2006). "Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process".
3590:
354:
310:
298:
1747:. Such disorders usually manifest in early childhood and affect predominantly tissues with high energy demands (brain, heart, muscle). Among the many classified
3905:
238:
2072:
Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH (April 1998). "Electron transfer by domain movement in cytochrome bc1".
509:
binuclear center to accept four electrons in reducing molecular oxygen and four protons to water. The mechanism of reduction was formerly thought to involve a
440:
and 14 protein subunits in mammals. In mammals, eleven subunits are nuclear in origin, and three are synthesized in the mitochondria. The complex contains two
3119:
Soltys BJ, Gupta RS (1999). "Mitochondrial proteins at unexpected cellular locations: export of proteins from mitochondria from an evolutionary perspective".
1624:
once by positioning itself between them. A high nitric oxide concentration, such as one added exogenously to the enzyme, reverses cyanide inhibition of COX.
1441:
about names of the six traditional intermediate states (APFOER); 2021 Cyro-EM result proposing an RPFOE mechanism with reversed assignment of red-ox phases (
3994:
3439:
Strazielle C, Hayzoun K, Derer M, Mariani J, Lalonde R (April 2006). "Regional brain variations of cytochrome oxidase activity in Relnrl-orl mutant mice".
4590:
3618:
2215:
Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P (December 2012).
4585:
3055:
Gupta RS, Ramachandra NB, Bowes T, Singh B (2008). "Unusual
Cellular Disposition of the Mitochondrial Molecular Chaperones Hsp60, Hsp70 and Hsp10".
4676:
4258:
4045:
3876:
1838:
3239:
4344:
2056:
4153:
2618:"Protein-protein interfaces from cytochrome c oxidase I evolve faster than nonbinding surfaces, yet negative selection is the driving force"
4102:
1646:
S levels are sufficient to inhibit the enzyme. There is no interaction between hydrogen sulfide and the fully reduced conformation of COX.
4917:
4025:
4444:
4379:
3758:
3677:
2432:
4389:
4214:
3140:
3072:
2845:
2807:
2797:
2303:"The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast"
2835:
1755:
of mitochondrion-encoded subunits, processing of preproteins and membrane insertion, and cofactor biosynthesis and incorporation.
4715:
4513:
4148:
1877:
1711:
231:
4882:
4710:
4374:
4095:
1872:
1707:
475:, which is reduced by the preceding component of the respiratory chain (cytochrome bc1 complex, Complex III), docks near the Cu
2753:
Nicholls P, Marshall DC, Cooper CE, Wilson MT (October 2013). "Sulfide inhibition of and metabolism by cytochrome c oxidase".
1731:
existence of yet unidentified specific mechanisms for protein translocation from mitochondria to other cellular destinations.
4705:
4491:
4449:
3915:
3745:
3733:
3682:
3580:
1867:
1703:
158:
3392:"Brain cytochrome oxidase: purification, antibody production, and immunohistochemical/histochemical correlations in the CNS"
182:
4902:
3754:
3584:
1548:
binuclear center, and this electron and two protons convert the tyrosyl radical back to Tyr, and the hydroxide bound to Cu
4295:
4237:
4127:
3741:
3571:
4349:
3986:
4523:
4518:
4322:
4188:
483:
binuclear center now passes an electron on to cytochrome a, which in turn passes an electron on to the cytochrome a
1714:). Of these 3 subunits encoded by mitochondrial DNA, two have been identified in extramitochondrial locations. In
4248:
4071:
3842:
2125:"A combined quantum chemical and crystallographic study on the oxidized binuclear center of cytochrome c oxidase"
1459:
433:
408:
479:
binuclear center and passes an electron to it, being oxidized back to cytochrome c containing Fe. The reduced Cu
4439:
4317:
3837:
3797:
3672:
3604:
384:
347:
176:
69:
39:
3164:
Soltys BJ, Gupta RS (May 1999). "Mitochondrial-matrix proteins at unexpected locations: are they exported?".
2217:"MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation"
4922:
4486:
4273:
3862:
3778:
3689:
2670:"Interaction of cyanide and nitric oxide with cytochrome c oxidase: implications for acute cyanide toxicity"
437:
163:
3263:
Zee JM, Glerum DM (December 2006). "Defects in cytochrome oxidase assembly in humans: lessons from yeast".
1672:
1451:
4481:
4138:
4066:
3821:
3770:
3737:
1824:
1807:
1748:
416:
1951:
Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA (September 2012).
4912:
4855:
4595:
4471:
4307:
4285:
3715:
3347:
Wong-Riley MT (March 1989). "Cytochrome oxidase: an endogenous metabolic marker for neuronal activity".
1661:
498:
358:
243:
151:
1615:
all bind to cytochrome c oxidase, inhibiting the protein from functioning and leading to the chemical
1572:
center as it was at the start of this cycle. Overall, four reduced cytochrome c's are oxidized while O
4602:
4459:
4408:
4354:
3816:
3704:
3200:
2711:"Carbon monoxide specifically inhibits cytochrome c oxidase of human mitochondrial respiratory chain"
2081:
2002:
381:
86:
3966:
1744:
322:
81:
179:
48:
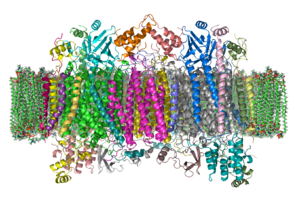
oxidase in a phospholipid bilayer. The intermembrane space lies to top of the image. Adapted from
4626:
4394:
4268:
4040:
3857:
3550:
3507:
3464:
3372:
3231:
3090:
3037:
2778:
2388:"A Human SCO2 Mutation Helps Define the Role of Sco1p in the Cytochrome Oxidase Assembly Pathway"
2105:
2026:
1723:
103:
3600:
2928:"The ligand binding battle at cytochrome c oxidase: how NO regulates oxygen gradients in tissue"
491:
binuclear center. The two metal ions in this binuclear center are 4.5 Å apart and coordinate a
4907:
4616:
4419:
4200:
3645:
3542:
3499:
3456:
3421:
3364:
3329:
3280:
3223:
3181:
3146:
3136:
3078:
3068:
3029:
2990:
2949:
2903:
2841:
2803:
2770:
2732:
2691:
2647:
2598:
2549:
2508:
2470:
2409:
2386:
Dickinson, Elizabeth K.; Adams, Denise L.; Schon, Eric A.; Glerum, D. Moira (September 2000).
2368:
2324:
2283:
2248:
2197:
2146:
2097:
2052:
2018:
1974:
1933:
1699:
1653:
908:
271:
170:
50:
2528:"Control of protein synthesis in yeast mitochondria: the concept of translational activators"
2123:
Kaila VR, Oksanen E, Goldman A, Bloch DA, Verkhovsky MI, Sundholm D, Wikström M (July 2011).
4429:
4222:
3940:
3935:
3808:
3662:
3534:
3491:
3448:
3411:
3403:
3356:
3319:
3311:
3272:
3215:
3173:
3128:
3060:
3021:
2980:
2939:
2893:
2885:
2762:
2722:
2681:
2637:
2629:
2588:
2580:
2539:
2500:
2462:
2399:
2360:
2314:
2275:
2238:
2228:
2187:
2177:
2136:
2089:
2010:
1964:
1923:
1915:
1634:
1442:
2985:
2968:
2428:
1347:
1323:
1275:
1203:
1028:
4334:
4227:
4017:
3575:
3102:
2243:
1850:
1612:
1107:
1059:
139:
3059:. Novartis Foundation Symposia. Vol. 291. pp. 59–68, discussion 69–73, 137–40.
3610:
2085:
2006:
1676:
Location of the 3 cytochrome c oxidase subunit genes in the human mitochondrial genome:
1458:
Please expand the section to include this information. Further details may exist on the
115:
3594:
3416:
3407:
3391:
3324:
3299:
3219:
2969:"Cell respiration s controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase"
2898:
2873:
2642:
2617:
2593:
2568:
2192:
2165:
1919:
1799:
1795:
1791:
1727:
388:
214:
74:
3495:
3177:
3132:
2727:
2710:
2404:
2387:
1928:
1903:
1620:
binding to regulatory sites on the enzyme, reducing the rate of cellular respiration.
194:
4896:
4118:
3360:
2319:
2302:
1396:
492:
303:
189:
3554:
3468:
3376:
3041:
2782:
2030:
1382:
1358:
1334:
1310:
1286:
1262:
1238:
1214:
1190:
1166:
1142:
1118:
1094:
1070:
1039:
1015:
991:
967:
943:
919:
895:
871:
847:
823:
799:
775:
751:
727:
703:
679:
655:
631:
607:
600:
576:
4751:
4726:
4384:
4263:
4176:
3881:
3867:
3789:
3511:
3235:
2109:
1627:
1616:
1552:
to a water molecule. The fourth electron from another cytochrome c flows through Cu
1527:. The oxygen is rapidly reduced, with two electrons coming from the Fe-cytochrome a
472:
449:
445:
412:
396:
370:
2944:
2927:
1827:. This technique has also been used to map learning activity in the animal brain.
327:
315:
17:
2874:"Cyanide inhibition of cytochrome c oxidase. A rapid-freeze e.p.r. investigation"
2584:
2544:
2527:
2141:
2124:
4021:
2504:
1751:, those involving dysfunctional COX assembly are thought to be the most severe.
1657:
1409:
dimerization and stability. Mutations to these subunits eliminate COX function.
1405:
198:
2466:
2453:
Khalimonchuk O, Rödel G (December 2005). "Biogenesis of cytochrome c oxidase".
2364:
2233:
2216:
1969:
1952:
1531:, which is converted to the ferryl oxo form (Fe=O). The oxygen atom close to Cu
1446:
1328:
Mitochondrial inner membrane protein (Cytochrome c oxidase assembly protein 18)
4503:
4165:
3725:
3641:
3538:
3064:
3025:
2279:
1715:
514:
374:
2182:
1953:"NDUFA4 is a subunit of complex IV of the mammalian electron transport chain"
1601:
binuclear center, is considered the inactive or resting state of the enzyme.
4434:
3667:
2686:
2669:
2014:
1664:
inhibit cytochrome c oxidase, binding from within the mitochondrial matrix.
3546:
3503:
3460:
3333:
3284:
3227:
3185:
3150:
3082:
3033:
2953:
2774:
2736:
2695:
2651:
2602:
2569:"Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core"
2553:
2512:
2474:
2413:
2372:
2328:
2287:
2252:
2201:
2150:
1978:
1743:
oxidase (COX) functionality or structure can result in severe, often fatal
1535:
picks up one electron from Cu, and a second electron and a proton from the
1429:
3425:
3368:
2994:
2907:
2616:
Aledo JC, Valverde H, Ruíz-Camacho M, Morilla I, López FD (October 2014).
2101:
2022:
1937:
1904:"Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen"
4580:
4454:
4087:
4076:
3699:
2709:
Alonso JR, Cardellach F, López S, Casademont J, Miró O (September 2003).
2633:
1758:
Currently, mutations have been identified in seven COX assembly factors:
1649:
1536:
510:
392:
362:
27:
Complex enzyme found in bacteria, archaea, and mitochondria of eukaryotes
2766:
2668:
Leavesley HB, Li L, Prabhakaran K, Borowitz JL, Isom GE (January 2008).
4740:
4735:
4655:
1719:
1604:
1593:
fully reduced state, which consists of a reduced Fe at the cytochrome a
980:
546:
366:
350:
146:
127:
3452:
3315:
2889:
1660:, which also inhibits the same oxidase system. High levels of ATP can
275:
54:
4845:
4840:
4810:
4805:
4685:
4665:
4660:
4650:
4645:
4640:
4635:
3648:
1882:
1820:
1803:
1787:
1690:
1684:
1678:
884:
860:
812:
788:
764:
740:
668:
644:
620:
589:
565:
404:
400:
336:
226:
122:
110:
98:
3276:
4865:
4860:
4850:
4835:
4830:
4825:
4820:
4815:
4800:
4795:
4790:
4785:
4780:
4775:
4770:
4765:
4760:
4556:
4543:
3950:
3945:
2093:
1779:
1775:
1771:
1759:
1608:
1371:
1299:
1251:
1227:
1004:
956:
932:
836:
716:
692:
539:
264:
1280:
Cytochrome c oxidase assembly protein COX16 homolog mitochondrial
3930:
3925:
3920:
3910:
3900:
3693:
3298:
Johar K, Priya A, Dhar S, Liu Q, Wong-Riley MT (November 2013).
1783:
1767:
1763:
1511:
Two electrons are passed from two cytochrome c's, through the Cu
1379:
1355:
1331:
1307:
1283:
1259:
1235:
1211:
1187:
1179:
1163:
1155:
1139:
1131:
1115:
1091:
1083:
1067:
1036:
1012:
988:
964:
940:
916:
892:
868:
844:
820:
796:
772:
748:
724:
700:
676:
652:
628:
604:
597:
573:
551:
441:
134:
4091:
3990:
3614:
3199:
Pecina P, Houstková H, Hansíková H, Zeman J, Houstek J (2004).
1423:
985:
Cytochrome c oxidase subunit 7A-related protein, mitochondrial
471:
form a binuclear center that is the site of oxygen reduction.
2532:
Biochimica et
Biophysica Acta (BBA) - Molecular Cell Research
1112:
Cytochrome c oxidase assembly factor 4 homolog, mitochondrial
1088:
Cytochrome c oxidase assembly factor 3 homolog, mitochondrial
269:
Subunit I and II of
Complex IV excluding all other subunits,
526:
Table of conserved subunits of cytochrome c oxidase complex
2872:
Jensen P, Wilson MT, Aasa R, Malmström BG (December 1984).
1902:
Castresana J, Lübben M, Saraste M, Higgins DG (June 1994).
1726:, relatively high amounts of these subunits were found in
1698:
Cytochrome c oxidase has 3 subunits which are encoded by
1208:
Cytochrome c oxidase assembly protein COX11 mitochondrial
1739:
Defects involving genetic mutations altering cytochrome
913:
Putative cytochrome c oxidase subunit 7A3, mitochondrial
2567:
Soto IC, Fontanesi F, Liu J, Barrientos A (June 2012).
673:
Cytochrome c oxidase subunit 4 isoform 2, mitochondrial
649:
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial
2526:
Herrmann JM, Woellhaf MW, Bonnefoy N (February 2013).
1576:
and four protons are reduced to two water molecules.
2051:(4th ed.). Hoboken, NJ: John Wiley & Sons.
4749:
4724:
4694:
4674:
4624:
4615:
4569:
4532:
4500:
4468:
4416:
4407:
4363:
4331:
4304:
4282:
4245:
4236:
4213:
4185:
4162:
4135:
4126:
4059:
4033:
3959:
3850:
3834:
3806:
3787:
3768:
3723:
3714:
3655:
3581:
Interactive
Molecular model of cytochrome c oxidase
2573:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
2129:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
1256:
Cytochrome c oxidase assembly protein COX15 homolog
524:
321:
309:
297:
289:
284:
257:
237:
225:
213:
208:
188:
169:
157:
145:
133:
121:
109:
97:
92:
80:
68:
63:
32:
3201:"Genetic defects of cytochrome c oxidase assembly"
3057:The Biology of Extracellular Molecular Chaperones
3972:Electron-transferring-flavoprotein dehydrogenase
1668:Extramitochondrial and subcellular localizations
1009:Cytochrome c oxidase subunit 8A, mitochondrial P
3877:Complex III/Coenzyme Q - cytochrome c reductase
2921:
2919:
2917:
2748:
2746:
2663:
2661:
2486:
2484:
2353:American Journal of Physiology. Cell Physiology
2346:
2344:
2342:
2340:
2338:
889:Cytochrome c oxidase subunit 7A2, mitochondrial
865:Cytochrome c oxidase subunit 7A1, mitochondrial
769:Cytochrome c oxidase subunit 6A2, mitochondrial
745:Cytochrome c oxidase subunit 6A1, mitochondrial
2431:. University of Illinois at Urbana-Champaign.
1160:Cytochrome c oxidase assembly factor 6 homolog
1064:Cytochrome c oxidase assembly factor 1 homolog
1033:Cytochrome c oxidase subunit 8C, mitochondrial
961:Cytochrome c oxidase subunit 7C, mitochondrial
937:Cytochrome c oxidase subunit 7B, mitochondrial
721:Cytochrome c oxidase subunit 5B, mitochondrial
697:Cytochrome c oxidase subunit 5A, mitochondrial
4103:
4002:
3626:
3006:
3004:
58: (It is a homodimer in this structure)
8:
2042:
2040:
1718:acinar tissue, these subunits were found in
513:intermediate, which was believed to lead to
395:. It receives an electron from each of four
44:The crystal structure of bovine cytochrome
4621:
4591:Mitochondrial permeability transition pore
4573:
4413:
4242:
4132:
4110:
4096:
4088:
4009:
3995:
3987:
3847:
3720:
3633:
3619:
3611:
3591:UMich Orientation of Proteins in Membranes
3390:Hevner RF, Wong-Riley MT (November 1989).
3114:
3112:
1642:center. It is unclear whether endogenous H
1515:and cytochrome a sites to the cytochrome a
205:
3603:at the U.S. National Library of Medicine
3415:
3323:
2984:
2943:
2897:
2726:
2685:
2641:
2592:
2543:
2403:
2318:
2242:
2232:
2191:
2181:
2140:
1968:
1927:
4586:Mitochondrial membrane transport protein
1671:
2301:Gaisne M, Bonnefoy N (September 2006).
1894:
1834:
1376:Cytochrome c oxidase protein 20 homolog
1184:Cytochrome c oxidase assembly factor 7,
4345:Cholesterol side-chain cleavage enzyme
4024:: Oxidoreduction-driven transporters (
3098:
3088:
2986:10.1111/j.1432-1033.1997.t01-1-00350.x
2967:Arnold S, Kadenbach B (October 1997).
2796:Roberts M, Reiss MJ, Monger G (2000).
1136:Cytochrome c oxidase assembly factor 5
254:
29:
1352:Cytochrome c oxidase assembly protein
1304:Cytochrome c oxidase copper chaperone
1232:Cytochrome c oxidase assembly protein
7:
1556:and cytochrome a to the cytochrome a
399:molecules and transfers them to one
353:, now reclassified as a translocase
4259:Coenzyme Q – cytochrome c reductase
3527:Neurobiology of Learning and Memory
4445:Oxoglutarate dehydrogenase complex
4380:Glycerol-3-phosphate dehydrogenase
3863:Complex II/Succinate dehydrogenase
3759:Pyruvate dehydrogenase phosphatase
3408:10.1523/jneurosci.09-11-03884.1989
3220:10.33549/physiolres.930000.53.S213
1920:10.1002/j.1460-2075.1994.tb06541.x
463:centers. In fact, the cytochrome a
25:
4390:Carnitine palmitoyltransferase II
2728:10.1034/j.1600-0773.2003.930306.x
4514:Carbamoyl phosphate synthetase I
4154:Long-chain-fatty-acid—CoA ligase
4149:Carnitine palmitoyltransferase I
3572:The Cytochrome Oxidase home page
3441:Journal of Neuroscience Research
3121:International Review of Cytology
2926:Gladwin MT, Shiva S (May 2009).
2755:Biochemical Society Transactions
2429:"Cytochrome oxidase: Complex IV"
2320:10.1111/j.1567-1364.2006.00083.x
1878:Cytochrome c oxidase subunit III
1849:
1837:
1450:
1428:
817:Cytochrome c oxidase subunit 6B2
793:Cytochrome c oxidase subunit 6B1
455:, and two copper centers, the Cu
263:
38:
4375:Glutamate aspartate transporter
3887:Complex IV/Cytochrome c oxidase
3245:from the original on 2011-07-18
3014:Histochemistry and Cell Biology
2854:from the original on 2022-02-24
2816:from the original on 2022-02-24
2435:from the original on 2018-01-23
2392:Journal of Biological Chemistry
1873:Cytochrome c oxidase subunit II
1722:granules. Additionally, in the
841:Cytochrome c oxidase subunit 6C
4492:Pyruvate dehydrogenase complex
4450:Succinyl coenzyme A synthetase
3734:Pyruvate dehydrogenase complex
3166:Trends in Biochemical Sciences
2837:Biology: A Functional Approach
2244:11858/00-001M-0000-000E-DDDF-4
1868:Cytochrome c oxidase subunit I
625:Cytochrome c oxidase subunit 3
594:Cytochrome c oxidase subunit 2
570:Cytochrome c oxidase subunit 1
1:
3755:Pyruvate dehydrogenase kinase
3496:10.1016/S0306-4522(03)00037-X
3265:Biochemistry and Cell Biology
3178:10.1016/s0968-0004(99)01390-0
3133:10.1016/S0074-7696(08)62396-7
2945:10.1161/CIRCRESAHA.109.198911
2715:Pharmacology & Toxicology
2405:10.1016/S0021-9258(19)61443-2
1735:Genetic defects and disorders
495:in the fully oxidized state.
380:It is the last enzyme in the
4296:Dihydroorotate dehydrogenase
3858:Complex I/NADH dehydrogenase
3361:10.1016/0166-2236(89)90165-3
2622:Genome Biology and Evolution
2585:10.1016/j.bbabio.2011.09.005
2545:10.1016/j.bbamcr.2012.03.007
2268:Journal of Molecular Biology
2142:10.1016/j.bbabio.2010.12.016
4350:Steroid 11-beta-hydroxylase
3396:The Journal of Neuroscience
2505:10.1021/acs.biochem.5b00540
1597:heme group and a reduced Cu
4939:
4918:Integral membrane proteins
4524:N-Acetylglutamate synthase
4519:Ornithine transcarbamylase
4323:Glycerol phosphate shuttle
4189:monoamine neurotransmitter
3678:Oxoglutarate dehydrogenase
2467:10.1016/j.mito.2005.08.002
2365:10.1152/ajpcell.00233.2006
2234:10.1016/j.cell.2012.11.053
1970:10.1016/j.cmet.2012.07.015
1823:and a transgenic model of
1447:10.1038/s41467-021-27174-y
554:family with Human protein
4878:
4576:
4552:
4249:oxidative phosphorylation
4072:Inorganic pyrophosphatase
3843:oxidative phosphorylation
3539:10.1016/j.nlm.2009.12.002
3304:Journal of Neurochemistry
3065:10.1002/9780470754030.ch5
3026:10.1007/s00418-005-0056-2
2280:10.1016/j.jmb.2013.12.001
545:Protein description from
434:integral membrane protein
409:electrochemical potential
262:
204:
37:
4440:Isocitrate dehydrogenase
4318:Malate-aspartate shuttle
3838:electron transport chain
3798:Methylmalonyl-CoA mutase
3673:Isocitrate dehydrogenase
3605:Medical Subject Headings
2183:10.1186/gb-2012-13-2-r12
2047:Voet D, Voet JG (2011).
1479:The overall reaction is
499:Crystallographic studies
415:then uses to synthesize
385:electron transport chain
4487:Glutamate dehydrogenase
4274:Succinate dehydrogenase
3779:Glutamate dehydrogenase
3690:Succinate dehydrogenase
3683:Succinyl CoA synthetase
3349:Trends in Neurosciences
2878:The Biochemical Journal
2015:10.1126/science.7652554
432:The complex is a large
4883:mitochondrial diseases
4482:Aspartate transaminase
4139:fatty acid degradation
4067:Cytochrome b6f complex
3822:Aspartate transaminase
3595:families/superfamily-4
3208:Physiological Research
2674:Toxicological Sciences
1808:sensorineural deafness
1749:mitochondrial diseases
1702:(cytochrome c oxidase
1695:
1439:is missing information
521:The conserved subunits
438:metal prosthetic sites
4596:Mitochondrial carrier
4472:anaplerotic reactions
4308:mitochondrial shuttle
4286:pyrimidine metabolism
2687:10.1093/toxsci/kfm254
1675:
359:transmembrane protein
4903:Cellular respiration
4603:Translocator protein
4460:Malate dehydrogenase
4355:Aldosterone synthase
3817:Pyruvate carboxylase
3705:Malate dehydrogenase
3601:Cytochrome-c+Oxidase
3214:(Suppl 1): S213-23.
2932:Circulation Research
1491:→ 4 Fe – cytochrome
436:composed of several
340:cytochrome c oxidase
293:Cytochrome c oxidase
258:Cytochrome c oxidase
33:Cytochrome c oxidase
4215:Intermembrane space
3967:Alternative oxidase
3771:α-ketoglutaric acid
2834:Roberts MB (1986).
2767:10.1042/BST20130070
2398:(35): 26780–26785.
2307:FEMS Yeast Research
2086:1998Natur.392..677Z
2007:1995Sci...269.1069T
1825:Alzheimer's disease
1745:metabolic disorders
1503:' = - 218 kJ/mol,
527:
4570:Other/to be sorted
4535:alcohol metabolism
4395:Uncoupling protein
4269:NADH dehydrogenase
2840:. Nelson Thornes.
2802:. Nelson Thornes.
2634:10.1093/gbe/evu240
1724:anterior pituitary
1696:
1656:is converted into
1654:methylated spirits
1499:O Δ
1483:4 Fe – cytochrome
525:
403:molecule and four
18:Cytochrome oxidase
4890:
4889:
4874:
4873:
4617:Mitochondrial DNA
4611:
4610:
4565:
4564:
4420:citric acid cycle
4403:
4402:
4209:
4208:
4201:Monoamine oxidase
4085:
4084:
3984:
3983:
3980:
3979:
3830:
3829:
3646:Citric acid cycle
3453:10.1002/jnr.20772
3316:10.1111/jnc.12433
2890:10.1042/bj2240829
2427:Crofts A (1996).
2058:978-0-470-57095-1
2001:(5227): 1069–74.
1914:(11): 2516–2525.
1831:Additional images
1700:mitochondrial DNA
1477:
1476:
1388:
1387:
1047:Assembly subunits
361:complex found in
333:
332:
253:
252:
249:
248:
152:metabolic pathway
16:(Redirected from
4930:
4754:
4729:
4699:
4679:
4629:
4622:
4574:
4537:
4507:
4475:
4430:Citrate synthase
4423:
4414:
4368:
4338:
4311:
4289:
4252:
4243:
4223:Adenylate kinase
4194:
4170:
4142:
4133:
4112:
4105:
4098:
4089:
4011:
4004:
3997:
3988:
3848:
3809:oxaloacetic acid
3721:
3663:Citrate synthase
3635:
3628:
3621:
3612:
3559:
3558:
3522:
3516:
3515:
3479:
3473:
3472:
3436:
3430:
3429:
3419:
3387:
3381:
3380:
3344:
3338:
3337:
3327:
3295:
3289:
3288:
3260:
3254:
3253:
3251:
3250:
3244:
3205:
3196:
3190:
3189:
3161:
3155:
3154:
3116:
3107:
3106:
3100:
3096:
3094:
3086:
3052:
3046:
3045:
3008:
2999:
2998:
2988:
2964:
2958:
2957:
2947:
2923:
2912:
2911:
2901:
2869:
2863:
2862:
2860:
2859:
2831:
2825:
2824:
2822:
2821:
2799:Advanced Biology
2793:
2787:
2786:
2750:
2741:
2740:
2730:
2706:
2700:
2699:
2689:
2665:
2656:
2655:
2645:
2613:
2607:
2606:
2596:
2564:
2558:
2557:
2547:
2523:
2517:
2516:
2488:
2479:
2478:
2450:
2444:
2443:
2441:
2440:
2424:
2418:
2417:
2407:
2383:
2377:
2376:
2348:
2333:
2332:
2322:
2298:
2292:
2291:
2263:
2257:
2256:
2246:
2236:
2212:
2206:
2205:
2195:
2185:
2161:
2155:
2154:
2144:
2120:
2114:
2113:
2080:(6677): 677–84.
2069:
2063:
2062:
2044:
2035:
2034:
1989:
1983:
1982:
1972:
1948:
1942:
1941:
1931:
1908:The EMBO Journal
1899:
1853:
1841:
1635:Hydrogen sulfide
1472:
1469:
1463:
1455:
1454:
1432:
1424:
1395:COX assembly in
528:
278:
267:
255:
206:
57:
42:
30:
21:
4938:
4937:
4933:
4932:
4931:
4929:
4928:
4927:
4893:
4892:
4891:
4886:
4870:
4750:
4745:
4725:
4720:
4695:
4690:
4675:
4670:
4625:
4607:
4561:
4548:
4533:
4528:
4501:
4496:
4469:
4464:
4417:
4399:
4364:
4359:
4335:steroidogenesis
4332:
4327:
4305:
4300:
4283:
4278:
4246:
4232:
4228:Creatine kinase
4205:
4191:
4186:
4181:
4163:
4158:
4136:
4122:
4116:
4086:
4081:
4055:
4046:ETC Complex III
4029:
4015:
3985:
3976:
3955:
3897:
3871:
3841:
3836:
3826:
3802:
3783:
3764:
3710:
3651:
3639:
3576:Rice University
3568:
3563:
3562:
3524:
3523:
3519:
3481:
3480:
3476:
3438:
3437:
3433:
3402:(11): 3884–98.
3389:
3388:
3384:
3346:
3345:
3341:
3297:
3296:
3292:
3277:10.1139/o06-201
3262:
3261:
3257:
3248:
3246:
3242:
3203:
3198:
3197:
3193:
3163:
3162:
3158:
3143:
3118:
3117:
3110:
3097:
3087:
3075:
3054:
3053:
3049:
3010:
3009:
3002:
2966:
2965:
2961:
2925:
2924:
2915:
2871:
2870:
2866:
2857:
2855:
2848:
2833:
2832:
2828:
2819:
2817:
2810:
2795:
2794:
2790:
2752:
2751:
2744:
2708:
2707:
2703:
2667:
2666:
2659:
2628:(11): 3064–76.
2615:
2614:
2610:
2566:
2565:
2561:
2525:
2524:
2520:
2499:(36): 5569–77.
2490:
2489:
2482:
2452:
2451:
2447:
2438:
2436:
2426:
2425:
2421:
2385:
2384:
2380:
2359:(6): C1129-47.
2350:
2349:
2336:
2300:
2299:
2295:
2265:
2264:
2260:
2214:
2213:
2209:
2163:
2162:
2158:
2122:
2121:
2117:
2071:
2070:
2066:
2059:
2046:
2045:
2038:
1991:
1990:
1986:
1957:Cell Metabolism
1950:
1949:
1945:
1901:
1900:
1896:
1891:
1864:
1857:
1854:
1845:
1842:
1833:
1816:
1737:
1694:(orange boxes).
1670:
1645:
1641:
1613:carbon monoxide
1600:
1596:
1591:
1587:
1582:
1575:
1571:
1567:
1563:
1559:
1555:
1551:
1547:
1543:
1534:
1530:
1526:
1522:
1518:
1514:
1498:
1490:
1473:
1467:
1464:
1457:
1449:
1433:
1422:
1393:
523:
508:
504:
490:
486:
482:
478:
470:
466:
462:
458:
453:
430:
425:
391:located in the
299:OPM superfamily
280:
270:
59:
49:
28:
23:
22:
15:
12:
11:
5:
4936:
4934:
4926:
4925:
4923:Copper enzymes
4920:
4915:
4910:
4905:
4895:
4894:
4888:
4887:
4879:
4876:
4875:
4872:
4871:
4869:
4868:
4863:
4858:
4853:
4848:
4843:
4838:
4833:
4828:
4823:
4818:
4813:
4808:
4803:
4798:
4793:
4788:
4783:
4778:
4773:
4768:
4763:
4757:
4755:
4747:
4746:
4744:
4743:
4738:
4732:
4730:
4722:
4721:
4719:
4718:
4713:
4708:
4702:
4700:
4692:
4691:
4689:
4688:
4682:
4680:
4672:
4671:
4669:
4668:
4663:
4658:
4653:
4648:
4643:
4638:
4632:
4630:
4619:
4613:
4612:
4609:
4608:
4606:
4605:
4600:
4599:
4598:
4593:
4583:
4577:
4571:
4567:
4566:
4563:
4562:
4560:
4559:
4553:
4550:
4549:
4547:
4546:
4540:
4538:
4530:
4529:
4527:
4526:
4521:
4516:
4510:
4508:
4498:
4497:
4495:
4494:
4489:
4484:
4478:
4476:
4466:
4465:
4463:
4462:
4457:
4452:
4447:
4442:
4437:
4432:
4426:
4424:
4411:
4405:
4404:
4401:
4400:
4398:
4397:
4392:
4387:
4382:
4377:
4371:
4369:
4361:
4360:
4358:
4357:
4352:
4347:
4341:
4339:
4329:
4328:
4326:
4325:
4320:
4314:
4312:
4302:
4301:
4299:
4298:
4292:
4290:
4280:
4279:
4277:
4276:
4271:
4266:
4261:
4255:
4253:
4240:
4238:Inner membrane
4234:
4233:
4231:
4230:
4225:
4219:
4217:
4211:
4210:
4207:
4206:
4204:
4203:
4197:
4195:
4183:
4182:
4180:
4179:
4173:
4171:
4160:
4159:
4157:
4156:
4151:
4145:
4143:
4130:
4128:Outer membrane
4124:
4123:
4117:
4115:
4114:
4107:
4100:
4092:
4083:
4082:
4080:
4079:
4074:
4069:
4063:
4061:
4057:
4056:
4054:
4053:
4051:ETC Complex IV
4048:
4043:
4037:
4035:
4031:
4030:
4016:
4014:
4013:
4006:
3999:
3991:
3982:
3981:
3978:
3977:
3975:
3974:
3969:
3963:
3961:
3957:
3956:
3954:
3953:
3948:
3943:
3938:
3933:
3928:
3923:
3918:
3913:
3908:
3903:
3895:
3890:
3889:
3884:
3879:
3874:
3869:
3865:
3860:
3854:
3852:
3845:
3832:
3831:
3828:
3827:
3825:
3824:
3819:
3813:
3811:
3804:
3803:
3801:
3800:
3794:
3792:
3785:
3784:
3782:
3781:
3775:
3773:
3766:
3765:
3763:
3762:
3753:(regulated by
3750:
3749:
3730:
3728:
3718:
3712:
3711:
3709:
3708:
3702:
3697:
3686:
3685:
3680:
3675:
3670:
3665:
3659:
3657:
3653:
3652:
3640:
3638:
3637:
3630:
3623:
3615:
3609:
3608:
3598:
3588:
3578:
3567:
3566:External links
3564:
3561:
3560:
3517:
3490:(4): 1151–63.
3474:
3431:
3382:
3339:
3310:(4): 496–508.
3290:
3255:
3191:
3156:
3141:
3108:
3099:|journal=
3073:
3047:
3000:
2979:(1): 350–354.
2959:
2938:(10): 1136–8.
2913:
2864:
2846:
2826:
2808:
2788:
2742:
2701:
2657:
2608:
2559:
2518:
2480:
2445:
2419:
2378:
2334:
2293:
2258:
2227:(7): 1528–41.
2207:
2170:Genome Biology
2156:
2115:
2064:
2057:
2036:
1984:
1943:
1893:
1892:
1890:
1887:
1886:
1885:
1880:
1875:
1870:
1863:
1860:
1859:
1858:
1855:
1848:
1846:
1843:
1836:
1832:
1829:
1815:
1814:Histochemistry
1812:
1800:leukodystrophy
1796:cardiomyopathy
1792:Leigh syndrome
1736:
1733:
1728:growth hormone
1669:
1666:
1662:allosterically
1643:
1639:
1598:
1594:
1589:
1585:
1581:
1578:
1573:
1569:
1565:
1561:
1557:
1553:
1549:
1545:
1541:
1532:
1528:
1524:
1520:
1516:
1512:
1509:
1508:
1496:
1488:
1475:
1474:
1436:
1434:
1427:
1421:
1418:
1392:
1389:
1386:
1385:
1377:
1374:
1369:
1366:
1362:
1361:
1353:
1350:
1345:
1342:
1338:
1337:
1329:
1326:
1321:
1318:
1314:
1313:
1305:
1302:
1297:
1294:
1290:
1289:
1281:
1278:
1273:
1270:
1266:
1265:
1257:
1254:
1249:
1246:
1242:
1241:
1233:
1230:
1225:
1222:
1218:
1217:
1209:
1206:
1201:
1198:
1194:
1193:
1185:
1182:
1177:
1174:
1170:
1169:
1161:
1158:
1153:
1150:
1146:
1145:
1137:
1134:
1129:
1126:
1122:
1121:
1113:
1110:
1105:
1102:
1098:
1097:
1089:
1086:
1081:
1078:
1074:
1073:
1065:
1062:
1057:
1054:
1050:
1049:
1043:
1042:
1034:
1031:
1026:
1023:
1019:
1018:
1010:
1007:
1002:
999:
995:
994:
986:
983:
978:
975:
971:
970:
962:
959:
954:
951:
947:
946:
938:
935:
930:
927:
923:
922:
914:
911:
906:
903:
899:
898:
890:
887:
882:
879:
875:
874:
866:
863:
858:
855:
851:
850:
842:
839:
834:
831:
827:
826:
818:
815:
810:
807:
803:
802:
794:
791:
786:
783:
779:
778:
770:
767:
762:
759:
755:
754:
746:
743:
738:
735:
731:
730:
722:
719:
714:
711:
707:
706:
698:
695:
690:
687:
683:
682:
674:
671:
666:
663:
659:
658:
650:
647:
642:
639:
635:
634:
626:
623:
618:
615:
611:
610:
595:
592:
587:
584:
580:
579:
571:
568:
563:
560:
556:
555:
549:
543:
537:
534:
522:
519:
506:
502:
488:
484:
480:
476:
468:
464:
460:
456:
451:
429:
426:
424:
421:
331:
330:
325:
319:
318:
313:
307:
306:
301:
295:
294:
291:
287:
286:
282:
281:
268:
260:
259:
251:
250:
247:
246:
241:
235:
234:
229:
223:
222:
217:
211:
210:
202:
201:
192:
186:
185:
174:
167:
166:
161:
155:
154:
149:
143:
142:
137:
131:
130:
125:
119:
118:
113:
107:
106:
101:
95:
94:
90:
89:
84:
78:
77:
72:
66:
65:
61:
60:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4935:
4924:
4921:
4919:
4916:
4914:
4911:
4909:
4906:
4904:
4901:
4900:
4898:
4885:
4884:
4877:
4867:
4864:
4862:
4859:
4857:
4854:
4852:
4849:
4847:
4844:
4842:
4839:
4837:
4834:
4832:
4829:
4827:
4824:
4822:
4819:
4817:
4814:
4812:
4809:
4807:
4804:
4802:
4799:
4797:
4794:
4792:
4789:
4787:
4784:
4782:
4779:
4777:
4774:
4772:
4769:
4767:
4764:
4762:
4759:
4758:
4756:
4753:
4748:
4742:
4739:
4737:
4734:
4733:
4731:
4728:
4723:
4717:
4714:
4712:
4709:
4707:
4704:
4703:
4701:
4698:
4693:
4687:
4684:
4683:
4681:
4678:
4673:
4667:
4664:
4662:
4659:
4657:
4654:
4652:
4649:
4647:
4644:
4642:
4639:
4637:
4634:
4633:
4631:
4628:
4623:
4620:
4618:
4614:
4604:
4601:
4597:
4594:
4592:
4589:
4588:
4587:
4584:
4582:
4579:
4578:
4575:
4572:
4568:
4558:
4555:
4554:
4551:
4545:
4542:
4541:
4539:
4536:
4531:
4525:
4522:
4520:
4517:
4515:
4512:
4511:
4509:
4506:
4505:
4499:
4493:
4490:
4488:
4485:
4483:
4480:
4479:
4477:
4474:
4473:
4467:
4461:
4458:
4456:
4453:
4451:
4448:
4446:
4443:
4441:
4438:
4436:
4433:
4431:
4428:
4427:
4425:
4422:
4421:
4415:
4412:
4410:
4406:
4396:
4393:
4391:
4388:
4386:
4383:
4381:
4378:
4376:
4373:
4372:
4370:
4367:
4362:
4356:
4353:
4351:
4348:
4346:
4343:
4342:
4340:
4337:
4336:
4330:
4324:
4321:
4319:
4316:
4315:
4313:
4310:
4309:
4303:
4297:
4294:
4293:
4291:
4288:
4287:
4281:
4275:
4272:
4270:
4267:
4265:
4262:
4260:
4257:
4256:
4254:
4251:
4250:
4244:
4241:
4239:
4235:
4229:
4226:
4224:
4221:
4220:
4218:
4216:
4212:
4202:
4199:
4198:
4196:
4193:
4190:
4184:
4178:
4175:
4174:
4172:
4169:
4167:
4161:
4155:
4152:
4150:
4147:
4146:
4144:
4141:
4140:
4134:
4131:
4129:
4125:
4120:
4119:Mitochondrial
4113:
4108:
4106:
4101:
4099:
4094:
4093:
4090:
4078:
4075:
4073:
4070:
4068:
4065:
4064:
4062:
4058:
4052:
4049:
4047:
4044:
4042:
4041:ETC Complex I
4039:
4038:
4036:
4032:
4027:
4023:
4019:
4012:
4007:
4005:
4000:
3998:
3993:
3992:
3989:
3973:
3970:
3968:
3965:
3964:
3962:
3958:
3952:
3949:
3947:
3944:
3942:
3939:
3937:
3934:
3932:
3929:
3927:
3924:
3922:
3919:
3917:
3914:
3912:
3909:
3907:
3904:
3902:
3898:
3892:
3891:
3888:
3885:
3883:
3880:
3878:
3875:
3873:
3866:
3864:
3861:
3859:
3856:
3855:
3853:
3849:
3846:
3844:
3839:
3835:Mitochondrial
3833:
3823:
3820:
3818:
3815:
3814:
3812:
3810:
3805:
3799:
3796:
3795:
3793:
3791:
3786:
3780:
3777:
3776:
3774:
3772:
3767:
3760:
3756:
3752:
3751:
3747:
3743:
3739:
3735:
3732:
3731:
3729:
3727:
3722:
3719:
3717:
3713:
3706:
3703:
3701:
3698:
3695:
3691:
3688:
3687:
3684:
3681:
3679:
3676:
3674:
3671:
3669:
3666:
3664:
3661:
3660:
3658:
3654:
3650:
3647:
3643:
3636:
3631:
3629:
3624:
3622:
3617:
3616:
3613:
3606:
3602:
3599:
3597:
3596:
3592:
3589:
3586:
3582:
3579:
3577:
3573:
3570:
3569:
3565:
3556:
3552:
3548:
3544:
3540:
3536:
3533:(3): 362–71.
3532:
3528:
3521:
3518:
3513:
3509:
3505:
3501:
3497:
3493:
3489:
3485:
3478:
3475:
3470:
3466:
3462:
3458:
3454:
3450:
3447:(5): 821–31.
3446:
3442:
3435:
3432:
3427:
3423:
3418:
3413:
3409:
3405:
3401:
3397:
3393:
3386:
3383:
3378:
3374:
3370:
3366:
3362:
3358:
3355:(3): 94–101.
3354:
3350:
3343:
3340:
3335:
3331:
3326:
3321:
3317:
3313:
3309:
3305:
3301:
3294:
3291:
3286:
3282:
3278:
3274:
3271:(6): 859–69.
3270:
3266:
3259:
3256:
3241:
3237:
3233:
3229:
3225:
3221:
3217:
3213:
3209:
3202:
3195:
3192:
3187:
3183:
3179:
3175:
3171:
3167:
3160:
3157:
3152:
3148:
3144:
3142:9780123645982
3138:
3134:
3130:
3126:
3122:
3115:
3113:
3109:
3104:
3092:
3084:
3080:
3076:
3074:9780470754030
3070:
3066:
3062:
3058:
3051:
3048:
3043:
3039:
3035:
3031:
3027:
3023:
3020:(5): 409–21.
3019:
3015:
3007:
3005:
3001:
2996:
2992:
2987:
2982:
2978:
2974:
2973:Eur J Biochem
2970:
2963:
2960:
2955:
2951:
2946:
2941:
2937:
2933:
2929:
2922:
2920:
2918:
2914:
2909:
2905:
2900:
2895:
2891:
2887:
2884:(3): 829–37.
2883:
2879:
2875:
2868:
2865:
2853:
2849:
2847:9780174480198
2843:
2839:
2838:
2830:
2827:
2815:
2811:
2809:9780174387329
2805:
2801:
2800:
2792:
2789:
2784:
2780:
2776:
2772:
2768:
2764:
2761:(5): 1312–6.
2760:
2756:
2749:
2747:
2743:
2738:
2734:
2729:
2724:
2720:
2716:
2712:
2705:
2702:
2697:
2693:
2688:
2683:
2680:(1): 101–11.
2679:
2675:
2671:
2664:
2662:
2658:
2653:
2649:
2644:
2639:
2635:
2631:
2627:
2623:
2619:
2612:
2609:
2604:
2600:
2595:
2590:
2586:
2582:
2579:(6): 883–97.
2578:
2574:
2570:
2563:
2560:
2555:
2551:
2546:
2541:
2538:(2): 286–94.
2537:
2533:
2529:
2522:
2519:
2514:
2510:
2506:
2502:
2498:
2494:
2487:
2485:
2481:
2476:
2472:
2468:
2464:
2461:(6): 363–88.
2460:
2456:
2455:Mitochondrion
2449:
2446:
2434:
2430:
2423:
2420:
2415:
2411:
2406:
2401:
2397:
2393:
2389:
2382:
2379:
2374:
2370:
2366:
2362:
2358:
2354:
2347:
2345:
2343:
2341:
2339:
2335:
2330:
2326:
2321:
2316:
2313:(6): 869–82.
2312:
2308:
2304:
2297:
2294:
2289:
2285:
2281:
2277:
2274:(4): 908–20.
2273:
2269:
2262:
2259:
2254:
2250:
2245:
2240:
2235:
2230:
2226:
2222:
2218:
2211:
2208:
2203:
2199:
2194:
2189:
2184:
2179:
2175:
2171:
2167:
2160:
2157:
2152:
2148:
2143:
2138:
2135:(7): 769–78.
2134:
2130:
2126:
2119:
2116:
2111:
2107:
2103:
2099:
2095:
2094:10.1038/33612
2091:
2087:
2083:
2079:
2075:
2068:
2065:
2060:
2054:
2050:
2043:
2041:
2037:
2032:
2028:
2024:
2020:
2016:
2012:
2008:
2004:
2000:
1996:
1988:
1985:
1980:
1976:
1971:
1966:
1963:(3): 378–86.
1962:
1958:
1954:
1947:
1944:
1939:
1935:
1930:
1925:
1921:
1917:
1913:
1909:
1905:
1898:
1895:
1888:
1884:
1881:
1879:
1876:
1874:
1871:
1869:
1866:
1865:
1861:
1852:
1847:
1840:
1835:
1830:
1828:
1826:
1822:
1813:
1811:
1809:
1805:
1801:
1797:
1793:
1789:
1785:
1781:
1777:
1773:
1769:
1765:
1761:
1756:
1752:
1750:
1746:
1742:
1734:
1732:
1729:
1725:
1721:
1717:
1713:
1709:
1705:
1701:
1693:
1692:
1687:
1686:
1681:
1680:
1674:
1667:
1665:
1663:
1659:
1655:
1651:
1647:
1636:
1632:
1629:
1625:
1621:
1618:
1614:
1610:
1606:
1602:
1579:
1577:
1538:
1506:
1502:
1494:
1486:
1482:
1481:
1480:
1471:
1468:December 2021
1461:
1453:
1448:
1444:
1440:
1437:This section
1435:
1431:
1426:
1425:
1419:
1417:
1413:
1410:
1407:
1401:
1398:
1390:
1384:
1381:
1378:
1375:
1373:
1370:
1367:
1364:
1363:
1360:
1357:
1354:
1351:
1349:
1346:
1343:
1340:
1339:
1336:
1333:
1330:
1327:
1325:
1322:
1319:
1316:
1315:
1312:
1309:
1306:
1303:
1301:
1298:
1295:
1292:
1291:
1288:
1285:
1282:
1279:
1277:
1274:
1271:
1268:
1267:
1264:
1261:
1258:
1255:
1253:
1250:
1247:
1244:
1243:
1240:
1237:
1234:
1231:
1229:
1226:
1223:
1220:
1219:
1216:
1213:
1210:
1207:
1205:
1202:
1199:
1196:
1195:
1192:
1189:
1186:
1183:
1181:
1178:
1175:
1172:
1171:
1168:
1165:
1162:
1159:
1157:
1154:
1151:
1148:
1147:
1144:
1141:
1138:
1135:
1133:
1130:
1127:
1124:
1123:
1120:
1117:
1114:
1111:
1109:
1106:
1103:
1100:
1099:
1096:
1093:
1090:
1087:
1085:
1082:
1079:
1076:
1075:
1072:
1069:
1066:
1063:
1061:
1058:
1055:
1052:
1051:
1048:
1045:
1044:
1041:
1038:
1035:
1032:
1030:
1027:
1024:
1021:
1020:
1017:
1014:
1011:
1008:
1006:
1003:
1000:
997:
996:
993:
990:
987:
984:
982:
979:
976:
973:
972:
969:
966:
963:
960:
958:
955:
952:
949:
948:
945:
942:
939:
936:
934:
931:
928:
925:
924:
921:
918:
915:
912:
910:
907:
904:
901:
900:
897:
894:
891:
888:
886:
883:
880:
877:
876:
873:
870:
867:
864:
862:
859:
856:
853:
852:
849:
846:
843:
840:
838:
835:
832:
829:
828:
825:
822:
819:
816:
814:
811:
808:
805:
804:
801:
798:
795:
792:
790:
787:
784:
781:
780:
777:
774:
771:
768:
766:
763:
760:
757:
756:
753:
750:
747:
744:
742:
739:
736:
733:
732:
729:
726:
723:
720:
718:
715:
712:
709:
708:
705:
702:
699:
696:
694:
691:
688:
685:
684:
681:
678:
675:
672:
670:
667:
664:
661:
660:
657:
654:
651:
648:
646:
643:
640:
637:
636:
633:
630:
627:
624:
622:
619:
616:
613:
612:
609:
606:
602:
599:
596:
593:
591:
588:
585:
582:
581:
578:
575:
572:
569:
567:
564:
561:
558:
557:
553:
550:
548:
544:
541:
538:
536:Subunit name
535:
533:
530:
529:
520:
518:
516:
512:
500:
496:
494:
493:hydroxide ion
474:
454:
447:
443:
439:
435:
427:
422:
420:
418:
414:
410:
406:
402:
398:
394:
390:
386:
383:
378:
376:
372:
368:
364:
360:
357:) is a large
356:
352:
349:
345:
341:
338:
329:
326:
324:
320:
317:
314:
312:
308:
305:
302:
300:
296:
292:
288:
283:
277:
273:
266:
261:
256:
245:
242:
240:
236:
233:
230:
228:
224:
221:
218:
216:
212:
207:
203:
200:
196:
193:
191:
190:Gene Ontology
187:
184:
181:
178:
175:
172:
168:
165:
162:
160:
156:
153:
150:
148:
144:
141:
138:
136:
132:
129:
128:NiceZyme view
126:
124:
120:
117:
114:
112:
108:
105:
102:
100:
96:
91:
88:
85:
83:
79:
76:
73:
71:
67:
62:
56:
52:
47:
41:
36:
31:
19:
4913:Hemoproteins
4880:
4727:ATP synthase
4696:
4534:
4502:
4470:
4418:
4385:ATP synthase
4365:
4333:
4306:
4284:
4264:Cytochrome c
4247:
4187:
4177:Kynureninase
4164:
4137:
4050:
4022:proton pumps
3893:
3886:
3882:Cytochrome c
3790:succinyl-CoA
3593:
3530:
3526:
3520:
3487:
3484:Neuroscience
3483:
3477:
3444:
3440:
3434:
3399:
3395:
3385:
3352:
3348:
3342:
3307:
3303:
3293:
3268:
3264:
3258:
3247:. Retrieved
3211:
3207:
3194:
3172:(5): 174–7.
3169:
3165:
3159:
3124:
3120:
3056:
3050:
3017:
3013:
2976:
2972:
2962:
2935:
2931:
2881:
2877:
2867:
2856:. Retrieved
2836:
2829:
2818:. Retrieved
2798:
2791:
2758:
2754:
2721:(3): 142–6.
2718:
2714:
2704:
2677:
2673:
2625:
2621:
2611:
2576:
2572:
2562:
2535:
2531:
2521:
2496:
2493:Biochemistry
2492:
2458:
2454:
2448:
2437:. Retrieved
2422:
2395:
2391:
2381:
2356:
2352:
2310:
2306:
2296:
2271:
2267:
2261:
2224:
2220:
2210:
2173:
2169:
2159:
2132:
2128:
2118:
2077:
2073:
2067:
2049:Biochemistry
2048:
1998:
1994:
1987:
1960:
1956:
1946:
1911:
1907:
1897:
1817:
1757:
1753:
1740:
1738:
1697:
1689:
1683:
1677:
1648:
1633:
1628:Nitric oxide
1626:
1622:
1617:asphyxiation
1603:
1583:
1510:
1504:
1500:
1492:
1484:
1478:
1465:
1438:
1420:Biochemistry
1414:
1411:
1402:
1394:
1372:COX20_HUMAN
1348:COX19_HUMAN
1324:COX18_HUMAN
1300:COX17_HUMAN
1276:COX16_HUMAN
1252:COX15_HUMAN
1228:COX14_HUMAN
1204:COX11_HUMAN
1046:
717:COX5B_HUMAN
693:COX5A_HUMAN
531:
497:
473:Cytochrome c
450:cytochrome a
446:cytochrome a
431:
413:ATP synthase
411:, which the
397:cytochrome c
379:
371:mitochondria
343:
339:
334:
116:BRENDA entry
45:
4677:Complex III
3899:synthesis:
3716:Anaplerotic
1712:subunit III
1658:formic acid
1507:' = +565 mV
1406:cardiolipin
1180:COA7_HUMAN
1156:COA6_HUMAN
1132:COA5_HUMAN
1108:COA4_HUMAN
1084:COA3_HUMAN
1060:COA1_HUMAN
1029:COX8C_HUMAN
1005:COX8A_HUMAN
981:COX7R_HUMAN
957:COX7C_HUMAN
933:COX7B_HUMAN
909:COX7S_HUMAN
885:CX7A2_HUMAN
861:CX7A1_HUMAN
837:COX6C_HUMAN
813:CX6B2_HUMAN
789:CX6B1_HUMAN
765:CX6A2_HUMAN
741:CX6A1_HUMAN
669:COX42_HUMAN
645:COX41_HUMAN
428:The complex
382:respiratory
311:OPM protein
285:Identifiers
104:IntEnz view
64:Identifiers
4897:Categories
4697:Complex IV
4504:urea cycle
4192:metabolism
4168:metabolism
4166:tryptophan
3894:Coenzyme Q
3868:Coenzyme Q
3726:acetyl-CoA
3642:Metabolism
3583:(Requires
3249:2010-11-17
3127:: 133–96.
2858:2020-10-25
2820:2020-10-25
2439:2018-01-28
2176:(2): R12.
1889:References
1856:Complex IV
1716:pancreatic
1708:subunit II
1588:and the Cu
1580:Inhibition
621:COX3_HUMAN
590:COX2_HUMAN
566:COX1_HUMAN
515:superoxide
375:eukaryotes
369:, and the
355:EC 7.1.1.9
344:Complex IV
323:Membranome
173:structures
140:KEGG entry
87:9001-16-5
4881:see also
4627:Complex I
4435:Aconitase
3668:Aconitase
3585:MDL Chime
3101:ignored (
3091:cite book
1704:subunit I
1487:+ 4 H + O
1460:talk page
423:Structure
93:Databases
4908:EC 1.9.3
4581:Frataxin
4455:Fumarase
4121:proteins
4077:V-ATPase
4018:Ion pump
3700:Fumarase
3555:24271956
3547:19969098
3504:12732258
3469:45787322
3461:16511878
3377:42996304
3334:24032355
3285:17215873
3240:Archived
3228:15119951
3186:10322429
3151:10494626
3083:18575266
3042:24440427
3034:16133117
2954:19461104
2852:Archived
2814:Archived
2783:11554252
2775:24059525
2737:12969439
2696:17906319
2652:25359921
2603:21958598
2554:22450032
2513:26284624
2475:16199211
2433:Archived
2414:10854440
2373:16760263
2329:16911509
2288:24333015
2253:23260140
2202:22356826
2151:21211513
2031:27210776
1979:22902835
1862:See also
1650:Methanol
1537:hydroxyl
1391:Assembly
542:protein
511:peroxide
393:membrane
363:bacteria
244:proteins
232:articles
220:articles
177:RCSB PDB
4741:MT-ATP8
4736:MT-ATP6
4656:MT-ND4L
3872:(CoQ10)
3851:Primary
3707:and ETC
3649:enzymes
3512:9366458
3426:2555458
3417:6569932
3369:2469224
3325:3820366
3236:8119738
2995:9363790
2908:6098268
2899:1144519
2643:4255772
2594:3262112
2193:3334569
2110:4380033
2102:9565029
2082:Bibcode
2023:7652554
2003:Bibcode
1995:Science
1938:8013452
1720:zymogen
1605:Cyanide
1383:PF12597
1359:PF06747
1335:PF02096
1311:PF05051
1287:PF14138
1263:PF02628
1239:PF14880
1215:PF04442
1191:PF08238
1167:PF02297
1143:PF10203
1119:PF06747
1095:PF09813
1071:PF08695
1040:PF02285
1016:PF02285
992:PF02238
968:PF02935
944:PF05392
920:PF02238
896:PF02238
872:PF02238
848:PF02937
824:PF02297
800:PF02297
776:PF02046
752:PF02046
728:PF01215
704:PF02284
680:PF02936
656:PF02936
632:PF00510
608:PF00116
601:PF02790
577:PF00115
547:UniProt
487:>-Cu
405:protons
367:archaea
351:1.9.3.1
279:
199:QuickGO
164:profile
147:MetaCyc
82:CAS no.
75:1.9.3.1
4846:MT-TS2
4841:MT-TS1
4811:MT-TL2
4806:MT-TL1
4716:MT-CO3
4711:MT-CO2
4706:MT-CO1
4686:MT-CYB
4666:MT-ND6
4661:MT-ND5
4651:MT-ND4
4646:MT-ND3
4641:MT-ND2
4636:MT-ND1
4409:Matrix
3941:COQ10B
3936:COQ10A
3607:(MeSH)
3553:
3545:
3510:
3502:
3467:
3459:
3424:
3414:
3375:
3367:
3332:
3322:
3283:
3234:
3226:
3184:
3149:
3139:
3081:
3071:
3040:
3032:
2993:
2952:
2906:
2896:
2844:
2806:
2781:
2773:
2735:
2694:
2650:
2640:
2601:
2591:
2552:
2511:
2473:
2412:
2371:
2327:
2286:
2251:
2200:
2190:
2149:
2108:
2100:
2074:Nature
2055:
2029:
2021:
1977:
1936:
1929:395125
1926:
1883:Heme a
1821:reeler
1806:, and
1804:anemia
1788:LRPPRC
1710:, and
1691:COXIII
1688:, and
1611:, and
905:Cox7a3
881:Cox7a2
857:Cox7a1
809:Cox6b2
785:Cox6b1
761:Cox6a2
737:Cox6a1
665:Cox4a2
641:Cox4i1
467:and Cu
459:and Cu
401:oxygen
337:enzyme
290:Symbol
227:PubMed
209:Search
195:AmiGO
183:PDBsum
123:ExPASy
111:BRENDA
99:IntEnz
70:EC no.
4866:MT-TY
4861:MT-TW
4856:MT-TV
4851:MT-TT
4836:MT-TR
4831:MT-TQ
4826:MT-TP
4821:MT-TN
4816:MT-TM
4801:MT-TK
4796:MT-TI
4791:MT-TH
4786:MT-TG
4781:MT-TF
4776:MT-TE
4771:MT-TD
4766:MT-TC
4761:MT-TA
4557:PMPCB
4544:ALDH2
4366:other
4060:Other
4026:TC 3D
3960:Other
3951:PDSS2
3946:PDSS1
3656:Cycle
3551:S2CID
3508:S2CID
3465:S2CID
3373:S2CID
3243:(PDF)
3232:S2CID
3204:(PDF)
3038:S2CID
2779:S2CID
2106:S2CID
2027:S2CID
1780:COX20
1776:COX15
1772:COX10
1760:SURF1
1685:COXII
1609:azide
1495:+ 2 H
1397:yeast
1368:Cox20
1344:Cox19
1320:Cox18
1296:Cox17
1272:Cox16
1248:Cox15
1224:Cox14
1200:Cox11
1025:Cox8c
1001:Cox8a
977:Cox7r
953:Cox7c
929:Cox7b
833:Cox6c
713:Cox5b
689:Cox5a
540:Human
442:hemes
389:cells
346:(was
159:PRIAM
4752:tRNA
3931:COQ9
3926:COQ7
3921:COQ6
3916:COQ5
3911:COQ4
3906:COQ3
3901:COQ2
3757:and
3694:SDHA
3543:PMID
3500:PMID
3457:PMID
3422:PMID
3365:PMID
3330:PMID
3281:PMID
3224:PMID
3182:PMID
3147:PMID
3137:ISBN
3103:help
3079:PMID
3069:ISBN
3030:PMID
2991:PMID
2950:PMID
2904:PMID
2842:ISBN
2804:ISBN
2771:PMID
2733:PMID
2692:PMID
2648:PMID
2599:PMID
2577:1817
2550:PMID
2536:1833
2509:PMID
2471:PMID
2410:PMID
2369:PMID
2325:PMID
2284:PMID
2249:PMID
2221:Cell
2198:PMID
2147:PMID
2133:1807
2098:PMID
2053:ISBN
2019:PMID
1975:PMID
1934:PMID
1786:and
1784:COA5
1768:SCO2
1764:SCO1
1679:COXI
1380:Pfam
1356:Pfam
1332:Pfam
1308:Pfam
1284:Pfam
1260:Pfam
1236:Pfam
1212:Pfam
1188:Pfam
1176:Coa7
1164:Pfam
1152:Coa6
1140:Pfam
1128:Coa5
1116:Pfam
1104:Coa4
1092:Pfam
1080:Coa3
1068:Pfam
1056:Coa1
1037:Pfam
1013:Pfam
989:Pfam
965:Pfam
941:Pfam
917:Pfam
893:Pfam
869:Pfam
845:Pfam
821:Pfam
797:Pfam
773:Pfam
749:Pfam
725:Pfam
701:Pfam
677:Pfam
653:Pfam
629:Pfam
617:Cox3
605:Pfam
598:Pfam
586:Cox2
574:Pfam
562:Cox1
552:Pfam
505:- Cu
448:and
444:, a
335:The
316:2dyr
276:2EIK
239:NCBI
180:PDBe
135:KEGG
55:1OCC
4034:ETC
3807:to
3788:to
3769:to
3724:to
3574:at
3535:doi
3492:doi
3488:118
3449:doi
3412:PMC
3404:doi
3357:doi
3320:PMC
3312:doi
3308:127
3273:doi
3216:doi
3174:doi
3129:doi
3125:194
3061:doi
3022:doi
3018:124
2981:doi
2977:249
2940:doi
2936:104
2894:PMC
2886:doi
2882:224
2763:doi
2723:doi
2682:doi
2678:101
2638:PMC
2630:doi
2589:PMC
2581:doi
2540:doi
2501:doi
2463:doi
2400:doi
2396:275
2361:doi
2357:291
2315:doi
2276:doi
2272:426
2239:hdl
2229:doi
2225:151
2188:PMC
2178:doi
2137:doi
2090:doi
2078:392
2011:doi
1999:269
1965:doi
1924:PMC
1916:doi
1844:ETC
1652:in
1568:–Cu
1560:–Cu
1544:–Cu
1519:–Cu
1443:doi
532:No.
417:ATP
387:of
373:of
342:or
328:257
272:PDB
215:PMC
171:PDB
51:PDB
4899::
4020::
3896:10
3870:10
3746:E3
3744:,
3742:E2
3740:,
3738:E1
3644::
3549:.
3541:.
3531:93
3529:.
3506:.
3498:.
3486:.
3463:.
3455:.
3445:83
3443:.
3420:.
3410:.
3398:.
3394:.
3371:.
3363:.
3353:12
3351:.
3328:.
3318:.
3306:.
3302:.
3279:.
3269:84
3267:.
3238:.
3230:.
3222:.
3212:53
3210:.
3206:.
3180:.
3170:24
3168:.
3145:.
3135:.
3123:.
3111:^
3095::
3093:}}
3089:{{
3077:.
3067:.
3036:.
3028:.
3016:.
3003:^
2989:.
2975:.
2971:.
2948:.
2934:.
2930:.
2916:^
2902:.
2892:.
2880:.
2876:.
2850:.
2812:.
2777:.
2769:.
2759:41
2757:.
2745:^
2731:.
2719:93
2717:.
2713:.
2690:.
2676:.
2672:.
2660:^
2646:.
2636:.
2624:.
2620:.
2597:.
2587:.
2575:.
2571:.
2548:.
2534:.
2530:.
2507:.
2497:54
2495:.
2483:^
2469:.
2457:.
2408:.
2394:.
2390:.
2367:.
2355:.
2337:^
2323:.
2309:.
2305:.
2282:.
2270:.
2247:.
2237:.
2223:.
2219:.
2196:.
2186:.
2174:13
2172:.
2168:.
2145:.
2131:.
2127:.
2104:.
2096:.
2088:.
2076:.
2039:^
2025:.
2017:.
2009:.
1997:.
1973:.
1961:16
1959:.
1955:.
1932:.
1922:.
1912:13
1910:.
1906:.
1810:.
1802:,
1798:,
1794:,
1782:,
1778:,
1774:,
1770:,
1766:,
1762:,
1706:,
1682:,
1607:,
1456:).
1365:14
1341:13
1317:12
1293:11
1269:10
1022:20
998:19
974:18
950:17
926:16
902:15
878:14
854:13
830:12
806:11
782:10
603:,
419:.
377:.
365:,
348:EC
274::
197:/
53::
4111:e
4104:t
4097:v
4028:)
4010:e
4003:t
3996:v
3840:/
3761:)
3748:)
3736:(
3696:)
3692:(
3634:e
3627:t
3620:v
3587:)
3557:.
3537::
3514:.
3494::
3471:.
3451::
3428:.
3406::
3400:9
3379:.
3359::
3336:.
3314::
3287:.
3275::
3252:.
3218::
3188:.
3176::
3153:.
3131::
3105:)
3085:.
3063::
3044:.
3024::
2997:.
2983::
2956:.
2942::
2910:.
2888::
2861:.
2823:.
2785:.
2765::
2739:.
2725::
2698:.
2684::
2654:.
2632::
2626:6
2605:.
2583::
2556:.
2542::
2515:.
2503::
2477:.
2465::
2459:5
2442:.
2416:.
2402::
2375:.
2363::
2331:.
2317::
2311:6
2290:.
2278::
2255:.
2241::
2231::
2204:.
2180::
2153:.
2139::
2112:.
2092::
2084::
2061:.
2033:.
2013::
2005::
1981:.
1967::
1940:.
1918::
1741:c
1644:2
1640:3
1638:a
1599:B
1595:3
1590:B
1586:3
1574:2
1570:B
1566:3
1562:B
1558:3
1554:A
1550:B
1546:B
1542:3
1540:a
1533:B
1529:3
1525:2
1521:B
1517:3
1513:A
1505:E
1501:G
1497:2
1493:c
1489:2
1485:c
1470:)
1466:(
1462:.
1445::
1245:9
1221:8
1197:7
1173:6
1149:5
1125:4
1101:3
1077:2
1053:1
758:9
734:8
710:7
686:6
662:5
638:4
614:3
583:2
559:1
507:B
503:3
489:B
485:3
481:A
477:A
469:B
465:3
461:B
457:A
452:3
304:4
46:c
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.