221:. It is an interesting organism to study because it inhabits nutrient-poor aquatic environments. Their ability to thrive in low levels of nutrients is facilitated by its dimorphic developmental cycle. The swarmer cell has a flagellum that protrudes from a single pole and is unable to initiate DNA replication unless differentiated into a stalked cell. The differentiation process includes a morphological transition characterized by ejection of its flagellum and growth of a stalk at the same pole. Stalked cells can elongate and replicate their DNA while growing a flagellum at the opposite pole, giving rise to a pre-divisional cell. Although the precise function of stalks is still being investigated, it is likely that the stalks are involved in the uptake of nutrients in nutrient-limited conditions. Its use as a model originated with developmental biologist
564:
of intracellular asymmetry to establish and maintain the orientation of the polarity axis, which is crucial for polar morphogenesis and division. Recruitment of TipN to the nascent poles at the end of the division cycle redefines the identity of the poles and resets the correct polarity in both future daughter cells (with a polarity reversal in the swarmer cell). The cell cycle–regulated synthesis and removal of these polarly localized structures have provided a rich playground for the identification of landmark proteins important for their proper localization. TipN has two transmembrane regions in the
412:. Underlying all these operations are the mechanisms for production of protein and structural components and energy production. The “housekeeping” metabolic and catabolic subsystems provide the energy and the molecular raw materials for protein synthesis, cell wall construction and other operations of the cell. The housekeeping functions are coupled bidirectionally to the cell cycle control system. However, they can adapt, somewhat independently of the cell cycle control logic, to changing composition and levels of the available nutrient sources.
357:
chromosome, the actual reaction time for each reaction varies widely around the average rate. This leads to a significant and inevitable cell-to-cell variation time to complete replication of the chromosome. There is similar random variation in the rates of progression of all the other subsystem reaction cascades. The net effect is that the time to complete the cell cycle varies widely over the cells in a population even when they all are growing in identical environmental conditions. Cell cycle regulation includes
325:. These five proteins directly control the timing of expression of over 200 genes. The five master regulatory proteins are synthesized and then eliminated from the cell one after the other over the course of the cell cycle. Several additional cell signaling pathways are also essential to the proper functioning of this cell cycle engine. The principal role of these signaling pathways is to ensure reliable production and elimination of the CtrA protein from the cell at just the right times in the cell cycle.
581:
46:
264:
404:, this is accomplished by the genetic regulatory circuit composed of five master regulators and an associated phospho-signaling network. The phosphosignaling network monitors the state of progression of the cell cycle and plays an essential role in accomplishing asymmetric cell division. The cell cycle control system manages the time and place of the initiation of chromosome replication and
33:
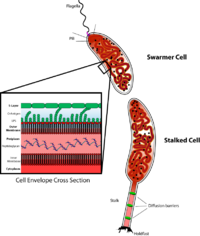
207:. The other daughter, called the "stalked" cell, has a tubular stalk structure protruding from one pole that has an adhesive holdfast material on its end, with which the stalked cell can adhere to surfaces. Swarmer cells differentiate into stalked cells after a short period of motility. Chromosome replication and cell division only occurs in the stalked cell stage.
548:, cell polarity is readily apparent by the assembly of polar organelles and by the polarization of the division plane, which results in the generation of stalked progeny that are longer than swarmer progeny. The formation of new cell poles at division implies that cell polarity must be re-established in the stalked progeny and reversed in the swarmer progeny.
419:
cell cycle control system are widely co-conserved across the alphaproteobacteria, but the ultimate function of this regulatory system varies widely in different species. These evolutionary changes reflect enormous differences between the individual species in fitness strategies and ecological niches.
246:
strain throughout the world. Additional phenotypic differences between the two strains have subsequently accumulated due to selective pressures on the NA1000 strain in the laboratory environment. The genetic basis of the phenotypic differences between the two strains results from coding, regulatory,
237:
strain CB15 (the strain originally isolated from a freshwater lake) and NA1000 (the primary experimental strain). In strain NA1000, which was derived from CB15 in the 1970s, the stalked and predivisional cells can be physically separated in the laboratory from new swarmer cells, while cell types from
563:
data strongly suggest a model in which TipN regulates the orientation of the polarity axis by providing a positional cue from the preceding cell cycle. In this model TipN specifies the site of the most recent division by identifying the new pole. The cell uses this positional information as a source
296:
has the swarmer cell stage that results in slower population growth. The swarmer cell is thought to provide cell dispersal, so that the organism constantly seeks out new environments. This may be particularly useful in severely nutrient-limited environments when the scant resources available can be
858:
Nierman, WC; Feldblyum, TV; Laub, MT; Paulsen, IT; Nelson, KE; Eisen, JA; Heidelberg, JF; Alley, MR; Ohta, N; Maddock, JR; Potocka, I; Nelson, WC; Newton, A; Stephens, C; Phadke, ND; Ely, B; DeBoy, RT; Dodson, RJ; Durkin, AS; Gwinn, ML; Haft, DH; Kolonay, JF; Smit, J; Craven, MB; Khouri, H; Shetty,
399:
The bacterial cell's control system has a hierarchical organization. The signaling and the control subsystem interfaces with the environment by means of sensory modules largely located on the cell surface. The genetic network logic responds to signals received from the environment and from internal
361:
signals that pace progression of the cell cycle engine to match progress of events at the regulatory subsystem level in each particular cell. This control system organization, with a controller (the cell cycle engine) driving a complex system, with modulation by feedback signals from the controlled
446:
cell cycle control are also found in these species. The specific coupling between the protein components of the cell cycle control network and the downstream readout of the circuit differ from species to species. The pattern is that the internal functionality of the network circuitry is conserved,
316:
constructed using biochemical and genetic logic circuitry organizes the timing of initiation of each of these subsystems. The central feature of the cell cycle regulation is a cyclical genetic circuit—a cell cycle engine—that is centered around the successive interactions of five master regulatory
274:
CB15 genome has 4,016,942 base pairs in a single circular chromosome encoding 3,767 genes. The genome contains multiple clusters of genes encoding proteins essential for survival in a nutrient-poor habitat. Included are those involved in chemotaxis, outer membrane channel function, degradation of
348:
to initiate the replication of the chromosome. The CtrA protein, in contrast, acts to block initiation of replication, so it must be removed from the cell before chromosome replication can begin. Multiple additional regulatory pathways integral to cell cycle regulation and involving both phospho
572:
coiled-coil domain. TipN homologues are present in other alpha-proteobacteria. TipN localizes to the new pole in both daughter cells after division and relocalizes to the cell division site in the late predivisional cell. Therefore, both daughter cells have TipN at the new pole after division.
482:
is a member of a group of bacteria that possess the stalk structure, a tubular extension from the cell body. However, the positioning of the stalk is not necessarily conserved at the pole of the cell body in different closely related species. Specifically, research has shown that not only the
356:
cells, replication of the chromosome involves about 2 million DNA synthesis reactions for each arm of the chromosome over 40 to 80 min depending on conditions. While the average time for each individual synthesis reaction can be estimated from the observed average total time to replicate the
291:
stalked cell stage provides a fitness advantage by anchoring the cell to surfaces to form biofilms and or to exploit nutrient sources. Generally, the bacterial species that divides fastest will be most effective at exploiting resources and effectively occupying ecological niches. Yet,
576:
The landmark protein TipN is essential for the proper placement of the flagellum. Mutants lacking TipN make serious mistakes in development. Instead of making a single flagellum at the correct cell pole , the cell makes multiple flagella at various locations, even on the stalk.
387:
cell cycle control system and its internal organization are co-conserved across many alphaproteobacteria species, but there are great differences in the regulatory apparatus' functionality and peripheral connectivity to other cellular subsystems from species to species. The
365:
The rate of progression of the cell cycle is further adjusted by additional signals arising from cellular sensors that monitor environmental conditions (for example, nutrient levels and the oxygen level) or the internal cell status (for example, presence of DNA damage).
238:
strain CB15 cannot be physically separated. The isolated swarmer cells can then be grown as a synchronized cell culture. Detailed study of the molecular development of these cells as they progress through the cell cycle has enabled researchers to understand
378:
cell cycle progression involves the entire cell operating as an integrated system. The control circuitry monitors the environment and the internal state of the cell, including the cell topology, as it orchestrates activation of cell cycle subsystems and
275:
aromatic ring compounds, and the breakdown of plant-derived carbon sources, in addition to many extracytoplasmic function sigma factors, providing the organism with the ability to respond to a wide range of environmental fluctuations. In 2010, the
460:
613:"Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundirnonas and Caulobacter"
297:
depleted very quickly. Many, perhaps most, of the swarmer daughter cells will not find a productive environment, but the obligate dispersal stage must increase the reproductive fitness of the species as a whole.
400:
cell status sensors to adapt the cell to current conditions. A major function of the top level control is to ensure that the operations involved in the cell cycle occur in the proper temporal order. In
611:
Abraham, Wolf-Rainer; Carsten Strömpl; Holger Meyer; Sabine
Lindholst; Edward R. B. Moore; Ruprecht Christ; Marc Vancanneyt; B. J. Tindali; Antonio Bennasar; John Smit; Michael Tesar (1999).
1998:
532:, Ackermann et al. suggested that aging is probably a fundamental property of all cellular organisms. A similar phenomenon has since been described in the bacterium
1972:
1445:
447:
but the coupling at the “edges” of the regulatory apparatus to the proteins controlling specific cellular functions differs widely among the different species.
352:
Each process activated by the proteins of the cell cycle engine involve a cascade of many reactions. The longest subsystem cascade is DNA replication. In
859:
J; Berry, K; Utterback, T; Tran, K; Wolf, A; Vamathevan, J; Ermolaeva, M; White, O; Salzberg, SL; Venter, JC; Shapiro, L; Fraser, CM (Mar 27, 2001).
1959:
349:
signaling pathways and regulated control of protein proteolysis act to assure that DnaA and CtrA are present in the cell just exactly when needed.
1985:
392:
cell cycle control system has been exquisitely optimized by evolutionary selection as a total system for robust operation in the face of internal
242:
cell cycle regulation in great detail. Due to this capacity to be physically synchronized, strain NA1000 has become the predominant experimental
702:
2031:
727:
1277:
Brilli, Matteo; Fondi, Marco; Fani, Renato; Mengoni, Alessio; Ferri, Lorenzo; Bazzicalupo, Marco; Biondi, Emanuele G. (2010).
336:
cell cycle where there can be overlapping rounds of chromosome replication simultaneously underway. The opposing roles of the
1990:
528:
was measured as the decline in the number of progeny produced over time. On the basis of experimental evolution studies in
592:
shows how TipN interact with two other polar proteins : the flagellar marker PodJ , and the stalk marker DivJ.
45:
312:
regulatory system controls many modular subsystems that organize the progression of cell growth and reproduction. A
1950:
505:
1788:
Huitema, Edgar; Pritchard, Sean; Matteson, David; Radhakrishnan, Sunish Kumar; Viollier, Patrick H. (2006-03-10).
422:
2026:
192:
188:
1279:"The diversity and evolution of cell cycle regulation in alpha-proteobacteria: A comparative genomic analysis"
332:
cell cycle is that the chromosome is replicated once and only once per cell cycle. This is in contrast to the
525:
499:
species. Presumably, It does so by a gain of function after protein expansion from around 400 amino acids in
703:"Top Canadian Prize Goes to Stanford Scientist Lucy Shapiro for Bringing Cell Biology into Three Dimensions"
1912:
434:
199:
daughter cells have two very different forms. One daughter is a mobile "swarmer" cell that has a single
1439:
345:
130:
1393:
1182:
1029:
872:
521:
1018:"An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation"
77:
1461:"Mechanisms of bacterial morphogenesis: evolutionary cell biology approaches provide new insights"
279:
NA1000 strain was sequenced and all differences with the CB15 "wild type" strain were identified.
1896:
1827:
1708:
1679:"A Landmark Protein Essential for Establishing and Perpetuating the Polarity of a Bacterial Cell"
1567:
1548:
1520:
683:
40:
1977:
483:
position of the stalk can change, but the number can vary as well in the closely related genus
1876:
1868:
1819:
1811:
1770:
1752:
1700:
1654:
1603:
1540:
1498:
1480:
1427:
1409:
1359:
1310:
1259:
1210:
1151:
1106:
1057:
998:
949:
900:
840:
788:
675:
634:
317:
proteins: DnaA, GcrA, CtrA, SciP, and CcrM whose roles were worked out by the laboratories of
1901:
1858:
1801:
1760:
1742:
1690:
1644:
1634:
1593:
1583:
1532:
1488:
1472:
1417:
1401:
1349:
1341:
1300:
1290:
1249:
1241:
1200:
1190:
1141:
1096:
1088:
1047:
1037:
988:
980:
939:
931:
890:
880:
830:
822:
778:
770:
665:
624:
428:
97:
580:
560:
409:
87:
169:
widely distributed in fresh water lakes and streams. The taxon is more properly known as
1397:
1382:"Sequential evolution of bacterial morphology by co-option of a developmental regulator"
1186:
1033:
876:
1765:
1730:
1598:
1571:
1493:
1460:
1422:
1381:
1354:
1329:
1305:
1278:
1254:
1229:
1205:
1170:
1101:
1076:
1052:
1017:
993:
968:
944:
919:
835:
810:
322:
313:
180:
67:
32:
2003:
1649:
1623:"Aging and Death in an Organism That Reproduces by Morphologically Symmetric Division"
1622:
783:
758:
670:
653:
2020:
1077:"DnaA couples DNA replication and the expression of two cell cycle master regulators"
895:
860:
495:
485:
159:
1831:
1712:
1552:
687:
1169:
Shen, X; Collier, J; Dill, D; Shapiro, L; Horowitz, M; McAdams, HH (Aug 12, 2008).
774:
735:
318:
263:
222:
935:
654:"The bacterial cytoskeleton: an intermediate filament-like function in cell shape"
1639:
1146:
1125:
984:
1944:
405:
107:
1935:
1863:
1846:
1806:
1789:
1695:
1678:
1175:
Proceedings of the
National Academy of Sciences of the United States of America
1171:"Architecture and inherent robustness of a bacterial cell-cycle control system"
865:
Proceedings of the
National Academy of Sciences of the United States of America
629:
612:
1380:
Jiang, Chao; Brown, Pamela J.B.; Ducret, Adrien; Brun1, Yves V. (2014-02-27).
1345:
1245:
569:
565:
393:
309:
218:
204:
184:
163:
1872:
1815:
1756:
1484:
1413:
1092:
1588:
1536:
1195:
1042:
200:
166:
1880:
1823:
1774:
1704:
1658:
1607:
1544:
1502:
1476:
1431:
1363:
1330:"The architecture and conservation pattern of whole-cell control circuitry"
1314:
1295:
1263:
1230:"The Architecture and Conservation Pattern of Whole-Cell Control Circuitry"
1214:
1155:
1110:
1061:
1002:
953:
904:
885:
844:
792:
679:
652:
Ausmees, Nora; Kuhn, Jeffrey R.; Jacobs-Wagner, Christine (December 2003).
638:
1747:
1523:; Urs Jenal (2003). "Senescence in a bacterium with asymmetric division".
1929:
1016:
Tan, M. H.; Kozdon, J. B.; Shen, X.; Shapiro, L.; McAdams, H. H. (2010).
442:
in, plant root nodules that fix nitrogen yet most of the proteins of the
439:
358:
214:
57:
1621:
Stewart, Eric J.; Richard Madden; Gregory Paul; Francois Taddei (2005).
1405:
826:
459:
1964:
811:"The Genetic Basis of Laboratory Adaptation in Caulobacter crescentus"
1906:
1790:"Bacterial Birth Scar Proteins Mark Future Flagellum Assembly Site"
759:"Biological Properties and Classification of the Caulobacter Group"
579:
458:
262:
1910:
809:
Marks ME; Castro-Rojas CM; Teiling C; et al. (July 2010).
588:
Cell development involves many such proteins working together.
247:
and insertion/deletion polymorphisms at five chromosomal loci.
536:, which gives rise to morphologically similar daughter cells.
340:
DnaA and CtrA proteins are essential to the tight control of
1729:
Treuner-Lange, Anke; Søgaard-Andersen, Lotte (2014-07-07).
1459:
Jiang, Chao; Caccamo, Paul D.; Brun, Yves V. (April 2015).
969:"Spatial complexity and control of a bacterial cell cycle"
370:
Evolutionary conservation of the cell cycle control system
493:
has been shown to manipulate stalk positioning in these
1677:
H, Lam; Wb, Schofield; C, Jacobs-Wagner (2006-03-10).
344:
chromosome replication. The DnaA protein acts at the
1847:"A Molecular Beacon Defines Bacterial Cell Asymmetry"
920:"System-level design of bacterial cell cycle control"
203:
at one cell pole that provides swimming motility for
861:"Complete genome sequence of Caulobacter crescentus"
1919:
1075:Collier, J; Murray, SR; Shapiro, L (Jan 25, 2006).
233:In the laboratory, researchers distinguish between
1845:Lawler, Melanie L.; Brun, Yves V. (2006-03-10).
1572:"Experimental evolution of aging in a bacterium"
617:International Journal of Systematic Bacteriology
438:is a soil bacterium that invades, and becomes a
1022:Proceedings of the National Academy of Sciences
383:asymmetric cell division. The proteins of the
555:life cycle is governed by regulators such as
374:The control circuitry that directs and paces
362:system creates a closed loop control system.
8:
606:
604:
1514:
1512:
1444:: CS1 maint: numeric names: authors list (
1907:
1228:McAdams, Harley H.; Shapiro, Lucy (2011).
451:The evolution of stalk positioning in the
31:
20:
1862:
1805:
1764:
1746:
1731:"Regulation of cell polarity in bacteria"
1694:
1648:
1638:
1597:
1587:
1492:
1421:
1353:
1304:
1294:
1253:
1204:
1194:
1145:
1100:
1051:
1041:
992:
943:
894:
884:
834:
804:
802:
782:
669:
628:
1566:Ackermann, Martin; Alexandra Schauerte;
918:McAdams, HH; Shapiro, L (Dec 17, 2009).
1902:Bacterium makes nature's strongest glue
600:
489:. SpmX, a polarly localized protein in
1437:
217:shape, which is caused by the protein
1724:
1722:
1672:
1670:
1668:
1375:
1373:
1328:McAdams, HH; Shapiro, L. (May 2011).
396:noise and environmental uncertainty.
7:
967:Collier, J; Shapiro, L (Aug 2007).
463:Diverse positioning of the stalks.
183:for studying the regulation of the
14:
701:Conger, Krista (March 31, 2009).
1126:"The role of proteolysis in the
973:Current Opinion in Biotechnology
503:to more than 800 amino acids in
44:
775:10.1128/mmbr.28.3.231-295.1964
408:as well as the development of
283:Role of the swarmer cell stage
1:
936:10.1016/j.febslet.2009.09.030
671:10.1016/S0092-8674(03)00935-8
1640:10.1371/journal.pbio.0030045
1234:Journal of Molecular Biology
1147:10.1016/j.resmic.2009.09.006
985:10.1016/j.copbio.2007.07.007
328:An essential feature of the
173:(Henrici and Johnson 1935).
1130:cell cycle and development"
757:Poindexter, JS (Sep 1964).
432:is an animal pathogen, and
2048:
2032:Bacteria described in 1964
1864:10.1016/j.cell.2006.02.027
1807:10.1016/j.cell.2006.01.019
1696:10.1016/j.cell.2005.12.040
630:10.1099/00207713-49-3-1053
473:Asticcacaulis biprosthecum
213:derives its name from its
1346:10.1016/j.jmb.2011.02.041
1246:10.1016/j.jmb.2011.02.041
763:Microbiol. Mol. Biol. Rev
471:(middle, sub-polar), and
469:Asticcacaulis excentricus
423:Agrobacterium tumefaciens
136:
129:
41:Scientific classification
39:
30:
23:
1576:BMC Evolutionary Biology
1134:Research in Microbiology
1093:10.1038/sj.emboj.7600927
559:, a cell cycle protein.
540:Cell polarity regulation
193:cellular differentiation
189:asymmetric cell division
1735:Journal of Cell Biology
1589:10.1186/1471-2148-7-126
1537:10.1126/science.1083532
1196:10.1073/pnas.0805258105
1043:10.1073/pnas.1014395107
526:Reproductive senescence
491:Caulobacter crescentus,
2004:caulobacter-crescentus
1951:Caulobacter crescentus
1921:Caulobacter crescentus
1897:Caulobacter crescentus
1477:10.1002/bies.201400098
1296:10.1186/1752-0509-4-52
1128:Caulobacter crescentus
886:10.1073/pnas.061029298
734:. 2014. Archived from
585:
501:Caulobacter crescentus
480:Caulobacter crescentus
476:
465:Caulobacter crescentus
435:Sinorhizobium meliloti
381:Caulobacter crescentus
267:
253:Caulobacter vibrioides
171:Caulobacter vibrioides
155:Caulobacter crescentus
140:Caulobacter crescentus
25:Caulobacter crescentus
1748:10.1083/jcb.201403136
1124:Jenal, U (Nov 2009).
583:
462:
426:is a plant pathogen,
346:origin of replication
266:
1570:; Urs Jenal (2007).
522:asymmetric bacterium
475:(right, bi-lateral).
415:The proteins of the
16:Species of bacterium
1519:Ackermann, Martin;
1406:10.1038/nature12900
1398:2014Natur.506..489J
1283:BMC Systems Biology
1187:2008PNAS..10511340S
1034:2010PNAS..10718985T
877:2001PNAS...98.4136N
827:10.1128/JB.00255-10
728:"2014 Lucy Shapiro"
251:is synonymous with
78:Alphaproteobacteria
1568:Stephen C. Stearns
1521:Stephen C. Stearns
586:
477:
268:
122:C. crescentus
2014:
2013:
1913:Taxon identifiers
1028:(44): 18985–990.
738:on 12 August 2017
566:N-terminal region
561:Yale University's
513:Caulobacter aging
151:
150:
2039:
2007:
2006:
1994:
1993:
1981:
1980:
1968:
1967:
1955:
1954:
1953:
1940:
1939:
1938:
1908:
1885:
1884:
1866:
1842:
1836:
1835:
1809:
1785:
1779:
1778:
1768:
1750:
1726:
1717:
1716:
1698:
1674:
1663:
1662:
1652:
1642:
1618:
1612:
1611:
1601:
1591:
1563:
1557:
1556:
1516:
1507:
1506:
1496:
1456:
1450:
1449:
1443:
1435:
1425:
1392:(7489): 489–93.
1377:
1368:
1367:
1357:
1325:
1319:
1318:
1308:
1298:
1274:
1268:
1267:
1257:
1225:
1219:
1218:
1208:
1198:
1181:(32): 11340–45.
1166:
1160:
1159:
1149:
1121:
1115:
1114:
1104:
1081:The EMBO Journal
1072:
1066:
1065:
1055:
1045:
1013:
1007:
1006:
996:
964:
958:
957:
947:
915:
909:
908:
898:
888:
855:
849:
848:
838:
806:
797:
796:
786:
754:
748:
747:
745:
743:
724:
718:
717:
715:
713:
698:
692:
691:
673:
649:
643:
642:
632:
608:
534:Escherichia coli
429:Brucella abortus
410:polar organelles
179:is an important
142:
98:Caulobacteraceae
49:
48:
35:
21:
2047:
2046:
2042:
2041:
2040:
2038:
2037:
2036:
2027:Caulobacterales
2017:
2016:
2015:
2010:
2002:
1997:
1989:
1984:
1976:
1971:
1963:
1958:
1949:
1948:
1943:
1934:
1933:
1928:
1915:
1893:
1888:
1844:
1843:
1839:
1787:
1786:
1782:
1728:
1727:
1720:
1676:
1675:
1666:
1620:
1619:
1615:
1565:
1564:
1560:
1518:
1517:
1510:
1458:
1457:
1453:
1436:
1379:
1378:
1371:
1327:
1326:
1322:
1276:
1275:
1271:
1227:
1226:
1222:
1168:
1167:
1163:
1123:
1122:
1118:
1074:
1073:
1069:
1015:
1014:
1010:
966:
965:
961:
930:(24): 3984–91.
917:
916:
912:
857:
856:
852:
821:(14): 3678–88.
808:
807:
800:
756:
755:
751:
741:
739:
732:Greengard Prize
726:
725:
721:
711:
709:
700:
699:
695:
651:
650:
646:
610:
609:
602:
598:
542:
524:shown to age.
515:
467:(left, polar),
457:
372:
303:
285:
261:
231:
147:
146:Poindexter 1964
144:
138:
125:
88:Caulobacterales
43:
17:
12:
11:
5:
2045:
2043:
2035:
2034:
2029:
2019:
2018:
2012:
2011:
2009:
2008:
1995:
1982:
1969:
1956:
1941:
1925:
1923:
1917:
1916:
1911:
1905:
1904:
1899:
1892:
1891:External links
1889:
1887:
1886:
1837:
1800:(5): 1025–37.
1780:
1718:
1689:(5): 1011–23.
1664:
1613:
1558:
1531:(5627): 1920.
1508:
1451:
1369:
1320:
1269:
1220:
1161:
1116:
1067:
1008:
959:
910:
871:(7): 4136–41.
850:
798:
749:
719:
693:
644:
623:(3): 1053–73.
599:
597:
594:
541:
538:
520:was the first
514:
511:
456:
449:
371:
368:
323:Harley McAdams
314:control system
302:
299:
284:
281:
260:
257:
230:
227:
181:model organism
149:
148:
145:
134:
133:
127:
126:
119:
117:
113:
112:
105:
101:
100:
95:
91:
90:
85:
81:
80:
75:
71:
70:
68:Pseudomonadota
65:
61:
60:
55:
51:
50:
37:
36:
28:
27:
15:
13:
10:
9:
6:
4:
3:
2:
2044:
2033:
2030:
2028:
2025:
2024:
2022:
2005:
2000:
1996:
1992:
1987:
1983:
1979:
1974:
1970:
1966:
1961:
1957:
1952:
1946:
1942:
1937:
1931:
1927:
1926:
1924:
1922:
1918:
1914:
1909:
1903:
1900:
1898:
1895:
1894:
1890:
1882:
1878:
1874:
1870:
1865:
1860:
1857:(5): 891–93.
1856:
1852:
1848:
1841:
1838:
1833:
1829:
1825:
1821:
1817:
1813:
1808:
1803:
1799:
1795:
1791:
1784:
1781:
1776:
1772:
1767:
1762:
1758:
1754:
1749:
1744:
1740:
1736:
1732:
1725:
1723:
1719:
1714:
1710:
1706:
1702:
1697:
1692:
1688:
1684:
1680:
1673:
1671:
1669:
1665:
1660:
1656:
1651:
1646:
1641:
1636:
1632:
1628:
1624:
1617:
1614:
1609:
1605:
1600:
1595:
1590:
1585:
1581:
1577:
1573:
1569:
1562:
1559:
1554:
1550:
1546:
1542:
1538:
1534:
1530:
1526:
1522:
1515:
1513:
1509:
1504:
1500:
1495:
1490:
1486:
1482:
1478:
1474:
1471:(4): 413–25.
1470:
1466:
1462:
1455:
1452:
1447:
1441:
1433:
1429:
1424:
1419:
1415:
1411:
1407:
1403:
1399:
1395:
1391:
1387:
1383:
1376:
1374:
1370:
1365:
1361:
1356:
1351:
1347:
1343:
1339:
1335:
1331:
1324:
1321:
1316:
1312:
1307:
1302:
1297:
1292:
1288:
1284:
1280:
1273:
1270:
1265:
1261:
1256:
1251:
1247:
1243:
1239:
1235:
1231:
1224:
1221:
1216:
1212:
1207:
1202:
1197:
1192:
1188:
1184:
1180:
1176:
1172:
1165:
1162:
1157:
1153:
1148:
1143:
1140:(9): 687–95.
1139:
1135:
1131:
1129:
1120:
1117:
1112:
1108:
1103:
1098:
1094:
1090:
1087:(2): 346–56.
1086:
1082:
1078:
1071:
1068:
1063:
1059:
1054:
1049:
1044:
1039:
1035:
1031:
1027:
1023:
1019:
1012:
1009:
1004:
1000:
995:
990:
986:
982:
979:(4): 333–40.
978:
974:
970:
963:
960:
955:
951:
946:
941:
937:
933:
929:
925:
921:
914:
911:
906:
902:
897:
892:
887:
882:
878:
874:
870:
866:
862:
854:
851:
846:
842:
837:
832:
828:
824:
820:
816:
812:
805:
803:
799:
794:
790:
785:
780:
776:
772:
769:(3): 231–95.
768:
764:
760:
753:
750:
737:
733:
729:
723:
720:
708:
707:Business Wire
704:
697:
694:
689:
685:
681:
677:
672:
667:
664:(6): 705–13.
663:
659:
655:
648:
645:
640:
636:
631:
626:
622:
618:
614:
607:
605:
601:
595:
593:
591:
582:
578:
574:
571:
567:
562:
558:
554:
553:C. crescentus
549:
547:
546:C. crescentus
539:
537:
535:
531:
530:C. crescentus
527:
523:
519:
512:
510:
508:
507:
506:Asticcacaulis
502:
498:
497:
496:Asticcacaulis
492:
488:
487:
486:Asticcacaulis
481:
474:
470:
466:
461:
454:
450:
448:
445:
441:
437:
436:
431:
430:
425:
424:
420:For example,
418:
413:
411:
407:
403:
397:
395:
391:
386:
382:
377:
369:
367:
363:
360:
355:
350:
347:
343:
339:
335:
331:
326:
324:
320:
315:
311:
308:
300:
298:
295:
290:
282:
280:
278:
273:
265:
258:
256:
254:
250:
249:C. crescentus
245:
241:
236:
235:C. crescentus
228:
226:
224:
220:
216:
212:
211:C. crescentus
208:
206:
202:
198:
194:
190:
186:
182:
178:
177:C. crescentus
174:
172:
168:
165:
161:
160:Gram-negative
157:
156:
143:
141:
135:
132:
131:Binomial name
128:
124:
123:
118:
115:
114:
111:
110:
106:
103:
102:
99:
96:
93:
92:
89:
86:
83:
82:
79:
76:
73:
72:
69:
66:
63:
62:
59:
56:
53:
52:
47:
42:
38:
34:
29:
26:
22:
19:
1920:
1854:
1850:
1840:
1797:
1793:
1783:
1738:
1734:
1686:
1682:
1630:
1627:PLOS Biology
1626:
1616:
1579:
1575:
1561:
1528:
1524:
1468:
1464:
1454:
1440:cite journal
1389:
1385:
1340:(1): 28–35.
1337:
1333:
1323:
1286:
1282:
1272:
1240:(1): 28–35.
1237:
1233:
1223:
1178:
1174:
1164:
1137:
1133:
1127:
1119:
1084:
1080:
1070:
1025:
1021:
1011:
976:
972:
962:
927:
924:FEBS Letters
923:
913:
868:
864:
853:
818:
815:J. Bacteriol
814:
766:
762:
752:
740:. Retrieved
736:the original
731:
722:
710:. Retrieved
706:
696:
661:
657:
647:
620:
616:
589:
587:
575:
568:and a large
556:
552:
550:
545:
543:
533:
529:
517:
516:
504:
500:
494:
490:
484:
479:
478:
472:
468:
464:
452:
443:
433:
427:
421:
416:
414:
401:
398:
389:
384:
380:
375:
373:
364:
353:
351:
341:
337:
333:
329:
327:
319:Lucy Shapiro
306:
304:
293:
288:
286:
276:
271:
269:
252:
248:
243:
239:
234:
232:
223:Lucy Shapiro
210:
209:
196:
176:
175:
170:
164:oligotrophic
154:
153:
152:
139:
137:
121:
120:
108:
24:
18:
1945:Wikispecies
1741:(1): 7–17.
518:Caulobacter
453:Caulobacter
444:Caulobacter
417:Caulobacter
406:cytokinesis
402:Caulobacter
390:Caulobacter
385:Caulobacter
376:Caulobacter
354:Caulobacter
342:Caulobacter
338:Caulobacter
330:Caulobacter
307:Caulobacter
294:Caulobacter
289:Caulobacter
277:Caulobacter
272:Caulobacter
244:Caulobacter
240:Caulobacter
197:Caulobacter
109:Caulobacter
2021:Categories
1633:(2): e45.
1334:J Mol Biol
596:References
570:C-terminal
394:stochastic
310:cell cycle
301:Cell cycle
219:crescentin
205:chemotaxis
185:cell cycle
1873:0092-8674
1816:0092-8674
1757:0021-9525
1485:1521-1878
1465:BioEssays
1414:0028-0836
509:species.
201:flagellum
167:bacterium
116:Species:
1978:10530223
1936:Q2943106
1930:Wikidata
1881:16530036
1832:15574493
1824:16530048
1775:25002676
1713:14200442
1705:16530047
1659:15685293
1608:17662151
1553:34770745
1545:12817142
1503:25664446
1432:24463524
1364:21371478
1315:20426835
1264:21371478
1215:18685108
1156:19781638
1111:16395331
1062:20956288
1003:17709236
954:19766635
905:11259647
845:20472802
793:14220656
688:14459851
680:14675535
639:10425763
440:symbiont
359:feedback
259:Genomics
215:crescent
94:Family:
64:Phylum:
58:Bacteria
54:Domain:
1965:3221693
1766:4085708
1599:2174458
1582:: 126.
1525:Science
1494:4368449
1423:4035126
1394:Bibcode
1355:3108490
1306:2877005
1255:3108490
1206:2516238
1183:Bibcode
1102:1383511
1053:2973855
1030:Bibcode
994:2716793
945:2795017
873:Bibcode
836:2897358
334:E. coli
229:Strains
104:Genus:
84:Order:
74:Class:
1991:968235
1879:
1871:
1830:
1822:
1814:
1773:
1763:
1755:
1711:
1703:
1657:
1650:546039
1647:
1606:
1596:
1551:
1543:
1501:
1491:
1483:
1430:
1420:
1412:
1386:Nature
1362:
1352:
1313:
1303:
1289:: 52.
1262:
1252:
1213:
1203:
1154:
1109:
1099:
1060:
1050:
1001:
991:
952:
942:
903:
893:
843:
833:
791:
784:441226
781:
742:14 May
712:14 May
686:
678:
637:
191:, and
1973:IRMNG
1828:S2CID
1709:S2CID
1549:S2CID
896:31192
684:S2CID
590:Fig#1
584:Fig#1
455:clade
158:is a
1999:LPSN
1986:ITIS
1960:GBIF
1877:PMID
1869:ISSN
1851:Cell
1820:PMID
1812:ISSN
1794:Cell
1771:PMID
1753:ISSN
1701:PMID
1683:Cell
1655:PMID
1604:PMID
1541:PMID
1499:PMID
1481:ISSN
1446:link
1428:PMID
1410:ISSN
1360:PMID
1311:PMID
1260:PMID
1211:PMID
1152:PMID
1107:PMID
1058:PMID
999:PMID
950:PMID
901:PMID
841:PMID
789:PMID
744:2015
714:2015
676:PMID
658:Cell
635:PMID
557:TipN
551:The
321:and
305:The
287:The
270:The
1859:doi
1855:124
1802:doi
1798:124
1761:PMC
1743:doi
1739:206
1691:doi
1687:124
1645:PMC
1635:doi
1594:PMC
1584:doi
1533:doi
1529:300
1489:PMC
1473:doi
1418:PMC
1402:doi
1390:506
1350:PMC
1342:doi
1338:409
1301:PMC
1291:doi
1250:PMC
1242:doi
1238:409
1201:PMC
1191:doi
1179:105
1142:doi
1138:160
1097:PMC
1089:doi
1048:PMC
1038:doi
1026:107
989:PMC
981:doi
940:PMC
932:doi
928:583
891:PMC
881:doi
831:PMC
823:doi
819:192
779:PMC
771:doi
666:doi
662:115
625:doi
544:In
195:.
2023::
2001::
1988::
1975::
1962::
1947::
1932::
1875:.
1867:.
1853:.
1849:.
1826:.
1818:.
1810:.
1796:.
1792:.
1769:.
1759:.
1751:.
1737:.
1733:.
1721:^
1707:.
1699:.
1685:.
1681:.
1667:^
1653:.
1643:.
1629:.
1625:.
1602:.
1592:.
1578:.
1574:.
1547:.
1539:.
1527:.
1511:^
1497:.
1487:.
1479:.
1469:37
1467:.
1463:.
1442:}}
1438:{{
1426:.
1416:.
1408:.
1400:.
1388:.
1384:.
1372:^
1358:.
1348:.
1336:.
1332:.
1309:.
1299:.
1285:.
1281:.
1258:.
1248:.
1236:.
1232:.
1209:.
1199:.
1189:.
1177:.
1173:.
1150:.
1136:.
1132:.
1105:.
1095:.
1085:25
1083:.
1079:.
1056:.
1046:.
1036:.
1024:.
1020:.
997:.
987:.
977:18
975:.
971:.
948:.
938:.
926:.
922:.
899:.
889:.
879:.
869:98
867:.
863:.
839:.
829:.
817:.
813:.
801:^
787:.
777:.
767:28
765:.
761:.
730:.
705:.
682:.
674:.
660:.
656:.
633:.
621:49
619:.
615:.
603:^
255:.
225:.
187:,
162:,
1883:.
1861::
1834:.
1804::
1777:.
1745::
1715:.
1693::
1661:.
1637::
1631:3
1610:.
1586::
1580:7
1555:.
1535::
1505:.
1475::
1448:)
1434:.
1404::
1396::
1366:.
1344::
1317:.
1293::
1287:4
1266:.
1244::
1217:.
1193::
1185::
1158:.
1144::
1113:.
1091::
1064:.
1040::
1032::
1005:.
983::
956:.
934::
907:.
883::
875::
847:.
825::
795:.
773::
746:.
716:.
690:.
668::
641:.
627::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.