287:, a theory which posits that codon bias exists because of nonrandomness in the mutational patterns. In other words, some codons can undergo more changes and therefore result in lower equilibrium frequencies, also known as “rare” codons. Different organisms also exhibit different mutational biases, and there is growing evidence that the level of genome-wide GC content is the most significant parameter in explaining codon bias differences between organisms. Additional studies have demonstrated that codon biases can be statistically predicted in
20:
272:. Although it has been shown that the rate of amino acid incorporation at more frequent codons occurs at a much higher rate than that of rare codons, the speed of translation has not been shown to be directly affected and therefore the bias towards more frequent codons may not be directly advantageous. However, the increase in translation elongation speed may still be indirectly advantageous by increasing the cellular concentration of free
219:, codons already present in high frequencies drive up the expression of their corresponding tRNAs, and tRNAs normally expressed at high levels drive up the frequency of their corresponding codons). However, this model does not seem to yet have experimental confirmation. Another problem is that the evolution of tRNA genes has been a very inactive area of research.
126:, as is indeed the case for the above-mentioned organisms. In other organisms that do not show high growing rates or that present small genomes, codon usage optimization is normally absent, and codon preferences are determined by the characteristic mutational biases seen in that particular genome. Examples of this are
439:
regions. As a result, co-translational protein folding introduces several spatial and temporal constraints on the nascent polypeptide chain in its folding trajectory. Because mRNA translation rates are coupled to protein folding, and codon adaptation is linked to translation elongation, it has been
403:
enzymes preferentially use codons that are poorly adapted to normal tRNA abundances, but have codons that are adapted to tRNA pools under starvation conditions. Thus, codon usage can introduce an additional level of transcriptional regulation for appropriate gene expression under specific cellular
440:
hypothesized that manipulation at the sequence level may be an effective strategy to regulate or improve protein folding. Several studies have shown that pausing of translation as a result of local mRNA structure occurs for certain proteins, which may be necessary for proper folding. Furthermore,
412:
Generally speaking for highly expressed genes, translation elongation rates are faster along transcripts with higher codon adaptation to tRNA pools, and slower along transcripts with rare codons. This correlation between codon translation rates and cognate tRNA concentrations provides additional
379:
can lead to inefficient use and depletion of ribosomes and ultimately reduce levels of heterologous protein production. In addition, the composition of the gene (e.g. the total number of rare codons and the presence of consecutive rare codons) may also affect translation accuracy. However, using
74:
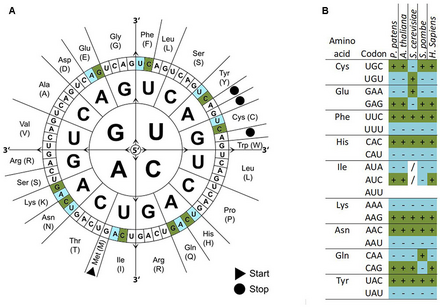
is degenerate. The genetic codes of different organisms are often biased towards using one of the several codons that encode the same amino acid over the others—that is, a greater frequency of one will be found than expected by chance. How such biases arise is a much debated area of
69:
There are 64 different codons (61 codons encoding for amino acids and 3 stop codons) but only 20 different translated amino acids. The overabundance in the number of codons allows many amino acids to be encoded by more than one codon. Because of such redundancy it is said that the
91:, which contains two distinct databases, CoCoPUTs and TissueCoCoPUTs. Together, these two databases provide comprehensive, up-to-date codon, codon pair and dinucleotide usage statistics for all organisms with available sequence information and 52 human tissues, respectively.
489:, are widely used to analyze variations in codon usage among genes. There are many computer programs to implement the statistical analyses enumerated above, including CodonW, GCUA, INCA, etc. Codon optimization has applications in designing synthetic genes and
1822:
Ikemura T (September 1981). "Correlation between the abundance of
Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system".
413:
modulation of translation elongation rates, which can provide several advantages to the organism. Specifically, codon usage can allow for global regulation of these rates, and rare codons may contribute to the accuracy of translation at the expense of speed.
210:
The nature of the codon usage-tRNA optimization has been fiercely debated. It is not clear whether codon usage drives tRNA evolution or vice versa. At least one mathematical model has been developed where both codon usage and tRNA expression co-evolve in
380:
codons that are optimized for tRNA pools in a particular host to overexpress a heterologous gene may also cause amino acid starvation and alter the equilibrium of tRNA pools. This method of adjusting codons to match host tRNA abundances, called
560:
Alexaki, Aikaterini; Kames, Jacob; Holcomb, David D.; Athey, John; Santana-Quintero, Luis V.; Lam, Phuc Vihn Nguyen; Hamasaki-Katagiri, Nobuko; Osipova, Ekaterina; Simonyan, Vahan; Bar, Haim; Komar, Anton A.; Kimchi-Sarfaty, Chava (June 2019).
384:, has traditionally been used for expression of a heterologous gene. However, new strategies for optimization of heterologous expression consider global nucleotide content such as local mRNA folding, codon pair bias, a codon ramp,
610:
Kames, Jacob; Alexaki, Aikaterini; Holcomb, David D.; Santana-Quintero, Luis V.; Athey, John C.; Hamasaki-Katagiri, Nobuko; Katneni, Upendra; Golikov, Anton; Ibla, Juan C.; Bar, Haim; Kimchi-Sarfaty, Chava (January 2020).
1568:
Correddu, D.; Montaño López, J. d. J.; Angermayr, S. A.; Middleditch, M. J.; Payne, L. S.; Leung, I. K. H. (2019). "Effect of
Consecutive Rare Codons on the Recombinant Production of Human Proteins in Escherichia coli".
444:
have been shown to have significant consequences in the folding process of the nascent protein and can even change substrate specificity of enzymes. These studies suggest that codon usage influences the speed at which
344:
regions can therefore play a major role in RNA secondary structure and downstream protein expression, which can undergo further selective pressures. In particular, strong secondary structure at the
122:(tRNA) pool. It is thought that optimal codons help to achieve faster translation rates and high accuracy. As a result of these factors, translational selection is expected to be stronger in highly
227:
Different factors have been proposed to be related to codon usage bias, including gene expression level (reflecting selection for optimizing the translation process by tRNA abundance),
465:, many statistical methods have been proposed and used to analyze codon usage bias. Methods such as the 'frequency of optimal codons' (Fop), the relative codon adaptation (RCA) or the
268:. The selectionist model also explains why more frequent codons are recognized by more abundant tRNA molecules, as well as the correlation between preferred codons, tRNA levels, and
260:
Although the mechanism of codon bias selection remains controversial, possible explanations for this bias fall into two general categories. One explanation revolves around the
509:
Athey, John; Alexaki, Aikaterini; Osipova, Ekaterina; Rostovtsev, Alexandre; Santana-Quintero, Luis V.; Katneni, Upendra; Simonyan, Vahan; Kimchi-Sarfaty, Chava (2017-09-02).
881:
Kanaya, Shigehiko; Yamada, Yuko; Kudo, Yoshihiro; Ikemura, Toshimichi (1999). "Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of
773:
319:. It also suggests that selection is generally weak, but that selection intensity scales to higher expression and more functional constraints of coding sequences.
340:
of mRNA influences translational efficiency, synonymous changes at this region on the mRNA can result in profound effects on gene expression. Codon usage in
299:
and further supporting the mutation bias model. However, this model alone cannot fully explain why preferred codons are recognized by more abundant tRNAs.
315:. This hypothesis states that selection favors major codons over minor codons, but minor codons are able to persist due to mutation pressure and
838:
Sharp, Paul M.; Stenico, Michele; Peden, John F.; Lloyd, Andrew T. (1993). "Codon usage: mutational bias, translational selection, or both?".
99:
1624:
Mignon, C.; Mariano, N.; Stadthagen, G.; Lugari, A.; Lagoutte, P.; Donnat, S.; Chenavas, S.; Perot, C.; Sodoyer, R.; Werle, B. (2018).
732:"Codon Usage Bias in Animals: Disentangling the Effects of Natural Selection, Effective Population Size, and GC-Biased Gene Conversion"
449:
emerge vectorially from the ribosome, which may further impact protein folding pathways throughout the available structural space.
333:
371:
and translation of a particular coding sequence can be less efficient when placed in a non-native context. For an overexpressed
1271:"Genome-Wide Patterns of Codon Bias Are Shaped by Natural Selection in the Purple Sea Urchin, Strongylocentrotus purpuratus"
613:"TissueCoCoPUTs: Novel Human Tissue-Specific Codon and Codon-Pair Usage Tables Based on Differential Tissue Gene Expression"
157:
1220:"Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes"
486:
654:"Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift"
1521:"Inhibition of translation by consecutive rare leucine codons in E. coli: absence of effect of varying mRNA stability"
247:, transcriptional selection, RNA stability, optimal growth temperature, hypersaline adaptation, and dietary nitrogen.
207:. The suggestion has been made that these codon biases play a role in the temporal regulation of their late proteins.
2143:
563:"Codon and Codon-Pair Usage Tables (CoCoPUTs): Facilitating Genetic Variation Analyses and Recombinant Gene Design"
470:
2108:
264:, in which codon bias contributes to the efficiency and/or accuracy of protein expression and therefore undergoes
1912:"The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications"
389:
352:
can inhibit translation, and mRNA folding at the 5’ end generates a large amount of variation in protein levels.
375:, the corresponding mRNA makes a large percent of total cellular RNA, and the presence of rare codons along the
2148:
2128:
2098:
2041:
368:
232:
228:
114:
106:
for codons that are favorable in regard to translation. Optimal codons in fast-growing microorganisms, like
2089:
2113:
2103:
885:
tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis".
703:
L. Duret and N. Galtier (2009). "Biased gene conversion and the evolution of mammalian genomic landscapes".
482:
360:
141:
36:
1916:
466:
345:
147:
385:
767:
462:
364:
236:
59:
25:
1484:
Novoa, E. M.; Ribas De
Pouplana, L (2012). "Speeding with control: Codon usage, tRNAs, and ribosomes".
730:
N. Galtier, C. Roux, M. Rousselle, J. Romiguier, E. Figuet, S. Glemin, N. Bierne and L. Duret (2018).
1780:
1331:
1096:
441:
400:
167:
1771:
Comeron JM, Aguadé M (September 1998). "An evaluation of measures of synonymous codon usage bias".
244:
200:
134:
76:
1804:
1606:
1466:
863:
592:
478:
381:
376:
265:
240:
977:
Bornelöv, Susanne; Selmi, Tommaso; Flad, Sophia; Dietmann, Sabine; Frye, Michaela (2019-06-07).
19:
2018:
1943:
1889:
1840:
1796:
1753:
1701:
1649:
1598:
1550:
1501:
1458:
1418:
1367:
1349:
1300:
1251:
1200:
1151:
1124:
1065:
1057:
1018:
1000:
959:
951:
910:
902:
855:
820:
812:
685:
634:
584:
542:
308:
292:
103:
493:. Several software packages are available online for this purpose (refer to external links).
2008:
2000:
1933:
1925:
1879:
1871:
1832:
1788:
1743:
1735:
1691:
1683:
1639:
1588:
1580:
1540:
1532:
1493:
1450:
1408:
1398:
1357:
1339:
1290:
1282:
1241:
1231:
1190:
1182:
1114:
1104:
1049:
1008:
990:
941:
894:
847:
804:
753:
743:
712:
675:
665:
624:
574:
532:
522:
269:
108:
94:
It is generally acknowledged that codon biases reflect the contributions of 3 main factors:
1454:
2093:
2045:
1083:
Shin, Young C.; Bischof, Georg F.; Lauer, William A.; Desrosiers, Ronald C. (2015-09-10).
788:
474:
428:
421:
123:
95:
1860:"Relative codon adaptation: a generic codon bias index for prediction of gene expression"
1784:
1335:
1100:
930:"Absence of translationally selected synonymous codon usage bias in Helicobacter pylori"
716:
435:
of a protein exits the translating ribosome and becomes solvent-exposed before its more
2013:
1988:
1884:
1859:
1748:
1723:
1696:
1671:
1545:
1520:
1413:
1387:"Dietary nitrogen alters codon bias and genome composition in parasitic microorganisms"
1386:
1295:
1270:
1246:
1219:
1119:
1084:
1013:
978:
680:
653:
537:
510:
458:
296:
184:
1989:"Comparison of correspondence analysis methods for synonymous codon usage in bacteria"
1938:
1911:
1195:
1170:
1053:
898:
2137:
1907:
1836:
1610:
1362:
1319:
596:
341:
316:
277:
1808:
1320:"Replicational and transcriptional selection on codon usage in Borrelia burgdorferi"
481:
are used to measure codon usage evenness. Multivariate statistical methods, such as
1630:
1470:
867:
490:
188:
128:
71:
2051:
1085:"Importance of codon usage for the temporal regulation of viral gene expression"
363:
is used in many biotechnological applications, including protein production and
349:
311:
and selection, the prevailing hypothesis for codon bias can be explained by the
288:
176:
172:
55:
2123:
1536:
946:
929:
658:
Proceedings of the
National Academy of Sciences of the United States of America
88:
1571:
1497:
1403:
995:
758:
629:
612:
579:
562:
527:
436:
432:
396:
196:
192:
180:
162:
139:
Organisms that show an intermediate level of codon usage optimization include
63:
51:
47:
43:
1962:
1353:
1344:
1061:
1004:
955:
906:
816:
469:(CAI) are used to predict gene expression levels, while methods such as the '
102:
reflecting mutational preferences (typically favoring AT-ending codons), and
2004:
1929:
1875:
1644:
1625:
1236:
1109:
748:
731:
670:
372:
273:
204:
2022:
1893:
1757:
1739:
1705:
1653:
1626:"Codon harmonization - going beyond the speed limit for protein expression"
1602:
1554:
1505:
1462:
1422:
1304:
1255:
1204:
1155:
1128:
1069:
1022:
963:
914:
808:
689:
638:
588:
546:
337:
1947:
1844:
1800:
1371:
1286:
1040:
genome are co-adapted for optimal translation of highly expressed genes".
859:
824:
1186:
1171:"Synonymous codon usage is subject to selection in thermophilic bacteria"
388:
or codon correlations. With the number of nucleotide changes introduced,
212:
79:. Codon usage tables detailing genomic codon usage bias for organisms in
2124:
Online
Synonymous Codon Usage Analyses with the ade4 and seqinR packages
2066:
1672:"Synonymous but not the same: The causes and consequences of codon bias"
1593:
1792:
1724:"Genetic Code Redundancy and Its Influence on the Encoded Polypeptides"
851:
446:
80:
1584:
118:(baker's yeast), reflect the composition of their respective genomic
84:
2038:
1687:
399:
genes such as those involved in amino acid starvation. For example,
2081:
1142:
Ermolaeva MD (October 2001). "Synonymous codon usage in bacteria".
367:. Because tRNA pools vary between different organisms, the rate of
2086:
928:
Atherton, John C.; Sharp, Paul M.; Lafay, Bénédicte (2000-04-01).
39:
18:
2119:
scRCA - Automatic determination of translational codon usage bias
2061:
2056:
2118:
1036:
Duret, Laurent (2000). "tRNA gene number and codon usage in the
152:
119:
1441:
Hershberg, R; Petrov, D. A. (2008). "Selection on codon bias".
392:
is often necessary for the creation of such an optimized gene.
2048:: estimating codon usage bias and its statistical significance
979:"Codon usage optimization in pluripotent embryonic stem cells"
791:(1996). "Co-variation of tRNA abundance and codon usage in
283:
The second explanation for codon usage can be explained by
2071:
203:
that display heavily skewed codon usage compared to the
35:
refers to differences in the frequency of occurrence of
1987:
Suzuki H, Brown CJ, Forney LJ, Top EM (December 2008).
239:(GC skew, reflecting strand-specific mutational bias),
2076:
2114:
CAIcal -Set of tools to assess codon usage adaptation
2129:
Genetic
Algorithm Simulation for Codon Optimization
1218:Paul S, Bag SK, Das S, Harvill ET, Dutta C (2008).
511:"A new and updated resource for codon usage tables"
98:that favors GC-ending codons in diploid organisms,
1728:Computational and Structural Biotechnology Journal
295:, arguing against the idea of selective forces on
2109:E-CAI - Expected value of Codon Adaptation Index
1665:
1663:
1519:Shu, P.; Dai, H.; Gao, W.; Goldman, E. (2006).
1436:
1434:
1432:
1324:Proceedings of the National Academy of Sciences
1089:Proceedings of the National Academy of Sciences
395:Specialized codon bias is further seen in some
1169:Lynn DJ, Singer GA, Hickey DA (October 2002).
8:
772:: CS1 maint: multiple names: authors list (
276:and potentially the rate of initiation for
89:HIVE-Codon Usage Tables (HIVE-CUTs) project
2087:ACUA - Automated Codon Usage Analysis Tool
2082:INCA - Interactive Codon Analysis software
356:Effect on transcription or gene expression
2012:
1937:
1883:
1747:
1695:
1643:
1592:
1544:
1412:
1402:
1361:
1343:
1294:
1245:
1235:
1194:
1118:
1108:
1012:
994:
945:
757:
747:
679:
669:
628:
578:
536:
526:
408:Effect on speed of translation elongation
2104:HEG-DB - Highly Expressed Genes Database
2077:JCat - Java Codon Usage Adaptation Tool
1717:
1715:
501:
1967:Correspondence Analysis of Codon Usage
1722:Spencer, P. S.; Barral, J. M. (2012).
1455:10.1146/annurev.genet.42.110807.091442
765:
313:mutation-selection-drift balance model
303:Mutation-selection-drift balance model
7:
2099:OPTIMIZER - Codon usage optimization
1385:Seward, Emily; Kelly, Steve (2016).
1269:Kober, K. M.; Pogson, G. H. (2013).
652:P. Shah and M. A. Gilchrist (2011).
307:To reconcile the evidence from both
50:(a triplet) that encodes a specific
2067:GCUA - General Codon Usage Analysis
717:10.1146/annurev-genom-082908-150001
1318:McInerney, James O. (1998-09-01).
14:
1670:Plotkin, J. B.; Kudla, G (2011).
328:Effect on RNA secondary structure
323:Consequences of codon composition
787:Dong, Hengjiang; Nilsson, Lars;
256:Mutational bias versus selection
58:chain or for the termination of
2052:HIVE-Codon Usage Table database
46:. A codon is a series of three
2072:Graphical Codon Usage Analyser
1:
1858:Fox JM, Erill I (June 2010).
1054:10.1016/s0168-9525(00)02041-2
899:10.1016/s0378-1119(99)00225-5
158:Strongylocentrotus purpuratus
2039:Composition Analysis Toolkit
1837:10.1016/0022-2836(81)90003-6
797:Journal of Molecular Biology
795:at different growth rates".
617:Journal of Molecular Biology
567:Journal of Molecular Biology
487:principal component analysis
361:Heterologous gene expression
705:Annu Rev Genomics Hum Genet
175:). Several viral families (
2165:
1537:10.3727/000000006783991881
947:10.1099/00221287-146-4-851
471:effective number of codons
16:Genetic bias in coding DNA
1910:; Li, Wen-Hsiung (1987).
1498:10.1016/j.tig.2012.07.006
1443:Annual Review of Genetics
1404:10.1186/s13059-016-1087-9
996:10.1186/s13059-019-1726-z
630:10.1016/j.jmb.2020.01.011
580:10.1016/j.jmb.2019.04.021
528:10.1186/s12859-017-1793-7
417:Effect on protein folding
390:artificial gene synthesis
96:GC-biased gene conversion
1345:10.1073/pnas.95.18.10698
233:horizontal gene transfer
231:(GC content, reflecting
229:guanine-cytosine content
115:Saccharomyces cerevisiae
1676:Nature Reviews Genetics
1645:10.1002/1873-3468.13046
1237:10.1186/gb-2008-9-4-r70
1110:10.1073/pnas.1515387112
671:10.1073/pnas.1016719108
483:correspondence analysis
401:amino acid biosynthetic
241:amino acid conservation
142:Drosophila melanogaster
1961:Peden J (2005-04-15).
1917:Nucleic Acids Research
1740:10.5936/csbj.201204006
809:10.1006/jmbi.1996.0428
467:codon adaptation index
199:) are known to encode
148:Caenorhabditis elegans
29:
2005:10.1093/dnares/dsn028
1963:"Codon usage indices"
1930:10.1093/nar/15.3.1281
1876:10.1093/dnares/dsq012
1287:10.1534/g3.113.005769
749:10.1093/molbev/msy015
463:computational biology
365:metabolic engineering
346:ribosome-binding site
251:Evolutionary theories
237:guanine-cytosine skew
235:or mutational bias),
26:Physcomitrella patens
22:
2057:Codon Usage Database
1144:Curr Issues Mol Biol
442:synonymous mutations
309:mutational pressures
293:intergenic sequences
223:Contributing factors
168:Arabidopsis thaliana
87:can be found in the
23:Codon usage bias in
1785:1998JMolE..47..268C
1336:1998PNAS...9510698M
1330:(18): 10698–10703.
1101:2015PNAS..11214030S
1095:(45): 14030–14035.
840:Biochem. Soc. Trans
789:Kurland, Charles G.
453:Methods of analysis
386:codon harmonization
334:secondary structure
262:selectionist theory
201:structural proteins
135:Helicobacter pylori
77:molecular evolution
2092:2020-07-26 at the
2044:2020-07-26 at the
1793:10.1007/PL00006384
1486:Trends in Genetics
1187:10.1093/nar/gkf546
1042:Trends in Genetics
852:10.1042/bst0210835
759:20.500.12210/34500
515:BMC Bioinformatics
479:information theory
382:codon optimization
266:positive selection
245:protein hydropathy
30:
2144:Molecular biology
1175:Nucleic Acids Res
883:Bacillus subtilis
623:(11): 3369–3378.
573:(13): 2434–2441.
270:gene copy numbers
104:natural selection
2156:
2027:
2026:
2016:
1984:
1978:
1977:
1975:
1974:
1958:
1952:
1951:
1941:
1924:(3): 1281–1295.
1904:
1898:
1897:
1887:
1855:
1849:
1848:
1819:
1813:
1812:
1768:
1762:
1761:
1751:
1719:
1710:
1709:
1699:
1667:
1658:
1657:
1647:
1638:(9): 1554–1564.
1621:
1615:
1614:
1596:
1585:10.1002/iub.2162
1565:
1559:
1558:
1548:
1516:
1510:
1509:
1481:
1475:
1474:
1438:
1427:
1426:
1416:
1406:
1382:
1376:
1375:
1365:
1347:
1315:
1309:
1308:
1298:
1281:(7): 1069–1083.
1266:
1260:
1259:
1249:
1239:
1215:
1209:
1208:
1198:
1166:
1160:
1159:
1139:
1133:
1132:
1122:
1112:
1080:
1074:
1073:
1033:
1027:
1026:
1016:
998:
974:
968:
967:
949:
925:
919:
918:
878:
872:
871:
835:
829:
828:
793:Escherichia coli
784:
778:
777:
771:
763:
761:
751:
742:(5): 1092–1103.
727:
721:
720:
700:
694:
693:
683:
673:
649:
643:
642:
632:
607:
601:
600:
582:
557:
551:
550:
540:
530:
506:
457:In the field of
431:, such that the
350:initiation codon
109:Escherichia coli
33:Codon usage bias
2164:
2163:
2159:
2158:
2157:
2155:
2154:
2153:
2149:Gene expression
2134:
2133:
2094:Wayback Machine
2046:Wayback Machine
2035:
2030:
1986:
1985:
1981:
1972:
1970:
1960:
1959:
1955:
1906:
1905:
1901:
1857:
1856:
1852:
1821:
1820:
1816:
1770:
1769:
1765:
1721:
1720:
1713:
1688:10.1038/nrg2899
1669:
1668:
1661:
1623:
1622:
1618:
1567:
1566:
1562:
1518:
1517:
1513:
1483:
1482:
1478:
1440:
1439:
1430:
1384:
1383:
1379:
1317:
1316:
1312:
1268:
1267:
1263:
1217:
1216:
1212:
1168:
1167:
1163:
1141:
1140:
1136:
1082:
1081:
1077:
1035:
1034:
1030:
976:
975:
971:
927:
926:
922:
880:
879:
875:
837:
836:
832:
786:
785:
781:
764:
729:
728:
724:
702:
701:
697:
664:(25): 10231–6.
651:
650:
646:
609:
608:
604:
559:
558:
554:
508:
507:
503:
499:
475:Shannon entropy
455:
422:Protein folding
419:
410:
358:
330:
325:
305:
285:mutational bias
258:
253:
225:
124:expressed genes
17:
12:
11:
5:
2162:
2160:
2152:
2151:
2146:
2136:
2135:
2132:
2131:
2126:
2121:
2116:
2111:
2106:
2101:
2096:
2084:
2079:
2074:
2069:
2064:
2059:
2054:
2049:
2034:
2033:External links
2031:
2029:
2028:
1979:
1953:
1908:Sharp, Paul M.
1899:
1850:
1831:(3): 389–409.
1814:
1763:
1711:
1659:
1616:
1579:(2): 266–274.
1560:
1511:
1492:(11): 574–81.
1476:
1428:
1391:Genome Biology
1377:
1310:
1261:
1210:
1181:(19): 4272–7.
1161:
1134:
1075:
1048:(7): 287–289.
1028:
983:Genome Biology
969:
940:(4): 851–860.
920:
893:(1): 143–155.
873:
846:(4): 835–841.
830:
803:(5): 649–663.
779:
722:
695:
644:
602:
552:
500:
498:
495:
459:bioinformatics
454:
451:
418:
415:
409:
406:
357:
354:
329:
326:
324:
321:
304:
301:
297:coding regions
278:messenger RNAs
257:
254:
252:
249:
224:
221:
185:papillomavirus
100:arrival biases
15:
13:
10:
9:
6:
4:
3:
2:
2161:
2150:
2147:
2145:
2142:
2141:
2139:
2130:
2127:
2125:
2122:
2120:
2117:
2115:
2112:
2110:
2107:
2105:
2102:
2100:
2097:
2095:
2091:
2088:
2085:
2083:
2080:
2078:
2075:
2073:
2070:
2068:
2065:
2063:
2060:
2058:
2055:
2053:
2050:
2047:
2043:
2040:
2037:
2036:
2032:
2024:
2020:
2015:
2010:
2006:
2002:
1999:(6): 357–65.
1998:
1994:
1990:
1983:
1980:
1969:. SourceForge
1968:
1964:
1957:
1954:
1949:
1945:
1940:
1935:
1931:
1927:
1923:
1919:
1918:
1913:
1909:
1903:
1900:
1895:
1891:
1886:
1881:
1877:
1873:
1870:(3): 185–96.
1869:
1865:
1861:
1854:
1851:
1846:
1842:
1838:
1834:
1830:
1826:
1818:
1815:
1810:
1806:
1802:
1798:
1794:
1790:
1786:
1782:
1779:(3): 268–74.
1778:
1774:
1767:
1764:
1759:
1755:
1750:
1745:
1741:
1737:
1733:
1729:
1725:
1718:
1716:
1712:
1707:
1703:
1698:
1693:
1689:
1685:
1681:
1677:
1673:
1666:
1664:
1660:
1655:
1651:
1646:
1641:
1637:
1633:
1632:
1627:
1620:
1617:
1612:
1608:
1604:
1600:
1595:
1590:
1586:
1582:
1578:
1574:
1573:
1564:
1561:
1556:
1552:
1547:
1542:
1538:
1534:
1531:(2): 97–106.
1530:
1526:
1522:
1515:
1512:
1507:
1503:
1499:
1495:
1491:
1487:
1480:
1477:
1472:
1468:
1464:
1460:
1456:
1452:
1448:
1444:
1437:
1435:
1433:
1429:
1424:
1420:
1415:
1410:
1405:
1400:
1397:(226): 3–15.
1396:
1392:
1388:
1381:
1378:
1373:
1369:
1364:
1359:
1355:
1351:
1346:
1341:
1337:
1333:
1329:
1325:
1321:
1314:
1311:
1306:
1302:
1297:
1292:
1288:
1284:
1280:
1276:
1272:
1265:
1262:
1257:
1253:
1248:
1243:
1238:
1233:
1229:
1225:
1221:
1214:
1211:
1206:
1202:
1197:
1192:
1188:
1184:
1180:
1176:
1172:
1165:
1162:
1157:
1153:
1149:
1145:
1138:
1135:
1130:
1126:
1121:
1116:
1111:
1106:
1102:
1098:
1094:
1090:
1086:
1079:
1076:
1071:
1067:
1063:
1059:
1055:
1051:
1047:
1043:
1039:
1032:
1029:
1024:
1020:
1015:
1010:
1006:
1002:
997:
992:
988:
984:
980:
973:
970:
965:
961:
957:
953:
948:
943:
939:
935:
931:
924:
921:
916:
912:
908:
904:
900:
896:
892:
888:
884:
877:
874:
869:
865:
861:
857:
853:
849:
845:
841:
834:
831:
826:
822:
818:
814:
810:
806:
802:
798:
794:
790:
783:
780:
775:
769:
760:
755:
750:
745:
741:
737:
736:Mol Biol Evol
733:
726:
723:
718:
714:
710:
706:
699:
696:
691:
687:
682:
677:
672:
667:
663:
659:
655:
648:
645:
640:
636:
631:
626:
622:
618:
614:
606:
603:
598:
594:
590:
586:
581:
576:
572:
568:
564:
556:
553:
548:
544:
539:
534:
529:
524:
520:
516:
512:
505:
502:
496:
494:
492:
488:
484:
480:
476:
472:
468:
464:
460:
452:
450:
448:
443:
438:
434:
430:
426:
423:
416:
414:
407:
405:
402:
398:
393:
391:
387:
383:
378:
374:
370:
369:transcription
366:
362:
355:
353:
351:
347:
343:
342:noncoding DNA
339:
335:
327:
322:
320:
318:
317:genetic drift
314:
310:
302:
300:
298:
294:
290:
286:
281:
279:
275:
271:
267:
263:
255:
250:
248:
246:
242:
238:
234:
230:
222:
220:
218:
214:
208:
206:
202:
198:
194:
190:
186:
182:
178:
174:
170:
169:
164:
160:
159:
154:
150:
149:
145:(fruit fly),
144:
143:
138:
136:
131:
130:
125:
121:
117:
116:
111:
110:
105:
101:
97:
92:
90:
86:
82:
78:
73:
67:
65:
61:
57:
54:residue in a
53:
49:
45:
41:
38:
34:
28:
27:
21:
1996:
1992:
1982:
1971:. Retrieved
1966:
1956:
1921:
1915:
1902:
1867:
1863:
1853:
1828:
1825:J. Mol. Biol
1824:
1817:
1776:
1773:J. Mol. Evol
1772:
1766:
1731:
1727:
1682:(1): 32–42.
1679:
1675:
1635:
1631:FEBS Letters
1629:
1619:
1594:11343/286411
1576:
1570:
1563:
1528:
1524:
1514:
1489:
1485:
1479:
1446:
1442:
1394:
1390:
1380:
1327:
1323:
1313:
1278:
1274:
1264:
1227:
1223:
1213:
1178:
1174:
1164:
1147:
1143:
1137:
1092:
1088:
1078:
1045:
1041:
1037:
1031:
986:
982:
972:
937:
934:Microbiology
933:
923:
890:
886:
882:
876:
843:
839:
833:
800:
796:
792:
782:
768:cite journal
739:
735:
725:
708:
704:
698:
661:
657:
647:
620:
616:
605:
570:
566:
555:
521:(391): 391.
518:
514:
504:
491:DNA vaccines
456:
447:polypeptides
424:
420:
411:
404:conditions.
394:
359:
331:
312:
306:
284:
282:
261:
259:
226:
216:
209:
189:polyomavirus
166:
156:
146:
140:
133:
132:(human) and
129:Homo sapiens
127:
120:transfer RNA
113:
107:
93:
72:genetic code
68:
32:
31:
24:
1224:Genome Biol
1150:(4): 91–7.
711:: 285–311.
473:' (Nc) and
291:using only
289:prokaryotes
177:herpesvirus
173:thale cress
64:stop codons
60:translation
56:polypeptide
48:nucleotides
2138:Categories
1973:2010-10-20
1572:IUBMB Life
1449:: 287–99.
1230:(4): R70.
1038:C. elegans
989:(1): 119.
497:References
437:C-terminal
433:N-terminus
397:endogenous
377:transcript
197:parvovirus
193:adenovirus
181:lentivirus
163:sea urchin
151:(nematode
52:amino acid
44:coding DNA
37:synonymous
1611:202555575
1525:Gene Expr
1354:0027-8424
1062:0168-9525
1005:1474-760X
956:1350-0872
907:0378-1119
817:0022-2836
597:139104807
429:vectorial
373:transgene
280:(mRNAs).
274:ribosomes
215:fashion (
205:host cell
2090:Archived
2042:Archived
2023:18940873
1894:20453079
1809:21862217
1758:24688635
1706:21102527
1654:29624661
1603:31509345
1555:17017124
1506:22921354
1463:18983258
1423:27842572
1305:23637123
1256:18397532
1205:12364606
1156:11719972
1129:26504241
1070:10858656
1023:31174582
964:10784043
915:10570992
690:21646514
639:31982380
589:31029701
547:28865429
332:Because
213:feedback
2014:2608848
1993:DNA Res
1948:3547335
1885:2885275
1864:DNA Res
1845:6175758
1801:9732453
1781:Bibcode
1749:3962081
1734:: 1–8.
1697:3074964
1546:6032470
1471:7085012
1414:5109750
1372:9724767
1332:Bibcode
1296:3704236
1247:2643941
1120:4653223
1097:Bibcode
1014:6555954
868:8582630
860:8132077
825:8709146
681:3121864
538:5581930
425:in vivo
336:of the
165:), and
81:GenBank
2062:CodonW
2021:
2011:
1946:
1939:340524
1936:
1892:
1882:
1843:
1807:
1799:
1756:
1746:
1704:
1694:
1652:
1609:
1601:
1553:
1543:
1504:
1469:
1461:
1421:
1411:
1370:
1360:
1352:
1303:
1293:
1254:
1244:
1203:
1196:140546
1193:
1154:
1127:
1117:
1068:
1060:
1021:
1011:
1003:
962:
954:
913:
905:
866:
858:
823:
815:
688:
678:
637:
595:
587:
545:
535:
338:5’ end
195:, and
85:RefSeq
40:codons
1805:S2CID
1607:S2CID
1467:S2CID
1363:27958
864:S2CID
593:S2CID
477:from
2019:PMID
1944:PMID
1890:PMID
1841:PMID
1797:PMID
1754:PMID
1702:PMID
1650:PMID
1599:PMID
1551:PMID
1502:PMID
1459:PMID
1419:PMID
1368:PMID
1350:ISSN
1301:PMID
1252:PMID
1201:PMID
1152:PMID
1125:PMID
1066:PMID
1058:ISSN
1019:PMID
1001:ISSN
960:PMID
952:ISSN
911:PMID
903:ISSN
887:Gene
856:PMID
821:PMID
813:ISSN
774:link
686:PMID
635:PMID
585:PMID
543:PMID
485:and
461:and
217:i.e.
153:worm
83:and
2009:PMC
2001:doi
1934:PMC
1926:doi
1880:PMC
1872:doi
1833:doi
1829:151
1789:doi
1744:PMC
1736:doi
1692:PMC
1684:doi
1640:doi
1636:592
1589:hdl
1581:doi
1541:PMC
1533:doi
1494:doi
1451:doi
1409:PMC
1399:doi
1358:PMC
1340:doi
1291:PMC
1283:doi
1242:PMC
1232:doi
1191:PMC
1183:doi
1115:PMC
1105:doi
1093:112
1050:doi
1009:PMC
991:doi
942:doi
938:146
895:doi
891:238
848:doi
805:doi
801:260
754:hdl
744:doi
713:doi
676:PMC
666:doi
662:108
625:doi
621:432
575:doi
571:431
533:PMC
523:doi
427:is
348:or
155:),
112:or
66:).
42:in
2140::
2017:.
2007:.
1997:15
1995:.
1991:.
1965:.
1942:.
1932:.
1922:15
1920:.
1914:.
1888:.
1878:.
1868:17
1866:.
1862:.
1839:.
1827:.
1803:.
1795:.
1787:.
1777:47
1775:.
1752:.
1742:.
1730:.
1726:.
1714:^
1700:.
1690:.
1680:12
1678:.
1674:.
1662:^
1648:.
1634:.
1628:.
1605:.
1597:.
1587:.
1577:72
1575:.
1549:.
1539:.
1529:13
1527:.
1523:.
1500:.
1490:28
1488:.
1465:.
1457:.
1447:42
1445:.
1431:^
1417:.
1407:.
1395:17
1393:.
1389:.
1366:.
1356:.
1348:.
1338:.
1328:95
1326:.
1322:.
1299:.
1289:.
1277:.
1275:G3
1273:.
1250:.
1240:.
1226:.
1222:.
1199:.
1189:.
1179:30
1177:.
1173:.
1146:.
1123:.
1113:.
1103:.
1091:.
1087:.
1064:.
1056:.
1046:16
1044:.
1017:.
1007:.
999:.
987:20
985:.
981:.
958:.
950:.
936:.
932:.
909:.
901:.
889:.
862:.
854:.
844:21
842:.
819:.
811:.
799:.
770:}}
766:{{
752:.
740:35
738:.
734:.
709:10
707:.
684:.
674:.
660:.
656:.
633:.
619:.
615:.
591:.
583:.
569:.
565:.
541:.
531:.
519:18
517:.
513:.
243:,
191:,
187:,
183:,
179:,
2025:.
2003::
1976:.
1950:.
1928::
1896:.
1874::
1847:.
1835::
1811:.
1791::
1783::
1760:.
1738::
1732:1
1708:.
1686::
1656:.
1642::
1613:.
1591::
1583::
1557:.
1535::
1508:.
1496::
1473:.
1453::
1425:.
1401::
1374:.
1342::
1334::
1307:.
1285::
1279:3
1258:.
1234::
1228:9
1207:.
1185::
1158:.
1148:3
1131:.
1107::
1099::
1072:.
1052::
1025:.
993::
966:.
944::
917:.
897::
870:.
850::
827:.
807::
776:)
762:.
756::
746::
719:.
715::
692:.
668::
641:.
627::
599:.
577::
549:.
525::
171:(
161:(
137:.
62:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.