279:
420:
428:
287:
560:
69:
24:
192:. The software is designed to be easy-to-learn for novice users, achieved by ensuring that tools for common tasks are 'discoverable' through familiar user interface elements (menus and toolbars), or by intuitive behaviour (mouse controls). Recent developments have enhanced the usability of the software for expert users, with customisable key bindings, extensions, and an extensive scripting interface.
254:, and Shelx files. The model may then be rotated in 3D and viewed from any viewpoint. The atomic model is represented by default using a stick-model, with vectors representing chemical bonds. The two halves of each bond are coloured according to the element of the atom at that end of the bond, allowing chemical structure and identity to be visualised in a manner familiar to most chemists.
270:
tools is the real space refinement engine, which will optimize the fit of a section of atomic model to the electron density in real time, with graphical feedback. The user may also intervene in this process, dragging the atoms into the right places if the initial model is too far away from the corresponding electron density.
435:
In macromolecular crystallography, the observed data is often weak and the observation-to-parameter ratio near 1. As a result, it is possible to build an incorrect atomic model into the electron density in some cases. To avoid this, careful validation is required. Coot provides a range of validation
257:
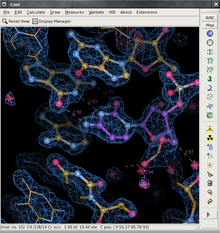
Coot can also display electron density, which is the result of structure determination experiments such as X-ray crystallography and EM reconstruction. The density is contoured using a 3D-mesh. The contour level controlled using the mouse wheel for easy manipulation - this provides a simple way for
269:
Coot provides extensive features for model building and refinement (i.e. adjusting the model to better fit the electron density), and for validation (i.e. checking that the atomic model agrees with the experimentally derived electron density and makes chemical sense). The most important of these
567:
Coot is built upon a number of libraries. Crystallographic tools include the
Clipper library for manipulating electron density and providing crystallographic algorithms, and the MMDB for the manipulation of atomic models. Other dependencies include
187:
Coot displays electron density maps and atomic models and allows model manipulations such as idealization, real space refinement, manual rotation/translation, rigid-body fitting, ligand search, solvation, mutations, rotamers, and
436:
tools, listed below. Having built an initial model, it is usual to check all of these and reconsider any parts of the model which are highlighted as problematic before deposition of the atomic coordinates with a public database.
591:
is a related project with which Coot shares some code. The projects are focused on slightly different problems, with CCP4mg dealing with presentation graphics and movies, whereas Coot deals with model building and validation.
600:
The software has gained considerable popularity, overtaking widely used packages such as 'O', XtalView, and Turbo Frodo. The primary publication has been cited in over 25,000 independent scientific papers since 2004.
889:
258:
the user to get an idea of the 3D electron density profile without the visual clutter of multiple contour levels. Electron density may be read into the program from
1021:
266:
map formats, though it is more common to calculate an electron density map directly from the X-ray diffraction data, read from an mtz, hkl, fcf or mmcif file.
588:
208:
579:
Much of the program's functionality is available through a scripting interface, which provides access from both the Python and Guile scripting languages.
933:
1083:
231:
1068:
1088:
862:
L. Potterton, S. McNicholas, E. Krissinel, J. Gruber, K. Cowtan, P. Emsley, G. N. Murshudov, S. Cohen, A. Perrakis and M. Noble (2004).
246:
Coot can be used to read files containing 3D atomic coordinate models of macromolecular structures in a number of formats, including
1063:
1078:
247:
223:
204:
1073:
263:
219:
39:
168:
278:
60:
163:) is used to display and manipulate atomic models of macromolecules, typically of proteins or nucleic acids, using
133:
419:
903:
427:
286:
832:
559:
996:
573:
474:
883:
172:
618:
235:
164:
762:
176:
121:
116:
68:
641:
259:
227:
215:
and CCP4. Additional support is available through the Coot wiki and an active COOT mailing list.
200:
23:
927:
720:
675:
441:
189:
97:
167:. It is primarily focused on building and validation of atomic models into three-dimensional
710:
665:
657:
128:
92:
971:
251:
807:
694:
453:
350:- optimize the fit of the model to the electron density, while preserving stereochemistry.
212:
199:, distributed under the GNU GPL. It is available from the Coot web site originally at the
1048:
738:
670:
207:. Pre-compiled binaries are also available for Linux and Windows from the web page and
786:
143:
1057:
446:
339:- find and fit a model to any small molecule which may be bound to the macromolecule.
196:
226:). Other contributors include Kevin Cowtan, Bernhard Lohkamp and Stuart McNicholas (
500:
372:
Rotamer tools (auto fit rotamer, manual rotamer, mutate and autofit, simple mutate)
301:- trace the main chain of a protein by placing correctly spaced alpha-carbon atoms.
324:
314:
307:- convert an initial trace of the alpha-carbon atoms to a full main-chain trace.
863:
715:
661:
538:- check for Hydrogen atoms with inappropriate environments (using Molprobity).
34:
377:
Torsion editing (edit chi angles, edit main chain torsions, general torsions)
724:
679:
521:
382:
Other protein tools (flip peptide, flip sidechain, cis <-> trans)
911:
479:- check for large differences between observed and calculated density.
1022:"Coot model building tools for molecular graphics - Google Scholar"
840:
558:
426:
418:
285:
277:
101:
470:- check for electron density not accounted for by existing atoms.
458:- examine differences between the torsions of NCS-related chains.
147:
947:
569:
109:
105:
544:- check for general differences between NCS related chains.
362:- optimize the fit of a rigid body to the electron density.
532:- identify parts of the model which don't fit the density.
485:- check for water molecules which do not fit the density.
175:
methods, although it has also been applied to data from
808:"Dr Kevin Cowtan - About staff, The University of York"
526:- check for unusual protein side-chain conformations.
464:- check for chiral centres with the wrong handedness.
866:
Developments in the CCP4 molecular-graphics project
144:
http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot
139:
127:
115:
91:
59:
47:
33:
640:P. Emsley; B. Lohkamp; W.G. Scott; Cowtan (2010).
596:Impact in the crystallographic computing community
697:Coot: model-building tools for molecular graphics
496:- check for improbable bond lengths, angles, etc.
8:
888:: CS1 maint: multiple names: authors list (
589:Collaborative Computational Project Number 4
587:The CCP4mg molecular graphics software from
394:- add ordered solvent molecules to the model
16:
550:- check for unusual DNA/RNA conformations.
67:
22:
15:
714:
669:
161:Crystallographic Object-Oriented Toolkit
610:
489:Check waters by difference map variance
932:: CS1 maint: archived copy as title (
925:
881:
423:Coot Ramachandran plot validation tool
400:- extend a protein or nucleotide chain
232:University of California at Santa Cruz
505:- check for non-planar peptide bonds.
387:Tools for adding atoms to the model:
333:- build an ideal DNA or RNA fragment.
28:the Coot main window (version 0.5pre)
7:
509:Temperature factor variance analysis
323:- fit a sequence of amino acids in
313:- fit a sequence of amino acids in
205:MRC Laboratory of Molecular Biology
294:Tools for general model building:
14:
833:"CCP4 Coordinate Library Project"
787:"Coot List At Www.Jiscmail.Ac.Uk"
767:Strucbio.biologie.uni-konstanz.de
368:- manually position a rigid body.
343:Tools for moving existing atoms:
644:Features and Development of Coot
431:Coot density fit validation tool
1084:Science software that uses GTK
148:http://www.biop.ox.ac.uk/coot/
1:
693:P. Emsley; K. Cowtan (2004).
515:GLN and ASN B-factor outliers
1069:Molecular modelling software
356:- optimize stereochemistry.
211:, and for Mac OS X through
1105:
1089:Science software for Linux
972:"CCMS Software - XtalView"
948:"Home Page of Alwyn Jones"
404:Add alternate conformation
327:conformation into density.
317:conformation into density.
282:Coot Real Space Refinement
134:GNU General Public License
997:"Turbo Frodo Description"
716:10.1107/s0907444904019158
662:10.1107/s0907444910007493
290:Coot Add Terminal Residue
234:), and Eugene Krissinel (
190:Ramachandran idealization
87:
55:
21:
1064:Crystallography software
462:Incorrect chiral volumes
1079:Free chemistry software
305:Ca Zone -> Mainchain
703:Acta Crystallographica
650:Acta Crystallographica
574:GNU Scientific Library
564:
432:
424:
348:Real space refine zone
291:
283:
218:The primary author is
1074:Free science software
562:
430:
422:
409:Place atom at pointer
366:Rotate/translate zone
289:
281:
173:X-ray crystallography
74:; 3 years ago
1026:Scholar.google.co.uk
555:Program architecture
530:Density fit analysis
398:Add terminal residue
274:Model building tools
236:Daresbury Laboratory
224:MRC-LMB at Cambridge
165:3D computer graphics
908:www.ysbl.york.ac.uk
483:Check/Delete waters
449:of a protein chain.
360:Rigid body fit zone
177:electron microscopy
122:Molecular modelling
18:
763:"Coot - CCP4 wiki"
583:Relation to CCP4mg
565:
433:
425:
299:C-alpha baton mode
292:
284:
230:), William Scott (
228:University of York
201:University of York
743:Mrc-lmb.cam.ac.uk
709:(12): 2126–2132.
621:. 2 February 2021
619:"Release 0.9.4.1"
494:Geometry analysis
442:Ramachandran plot
321:Place strand here
203:, and now at the
171:maps obtained by
153:
152:
72:/ 2 February 2021
1096:
1036:
1035:
1033:
1032:
1018:
1012:
1011:
1009:
1008:
993:
987:
986:
984:
983:
968:
962:
961:
959:
958:
944:
938:
937:
931:
923:
921:
919:
910:. Archived from
900:
894:
893:
887:
879:
872:Acta Crystallogr
859:
853:
852:
850:
848:
839:. Archived from
829:
823:
822:
820:
819:
804:
798:
797:
795:
794:
783:
777:
776:
774:
773:
759:
753:
752:
750:
749:
735:
729:
728:
718:
690:
684:
683:
673:
637:
631:
630:
628:
626:
615:
468:Unmodelled blobs
415:Validation tools
311:Place helix here
169:electron density
93:Operating system
82:
80:
75:
71:
26:
19:
1104:
1103:
1099:
1098:
1097:
1095:
1094:
1093:
1054:
1053:
1045:
1040:
1039:
1030:
1028:
1020:
1019:
1015:
1006:
1004:
995:
994:
990:
981:
979:
970:
969:
965:
956:
954:
946:
945:
941:
924:
917:
915:
914:on 10 June 2005
904:"Archived copy"
902:
901:
897:
880:
861:
860:
856:
846:
844:
843:on 10 June 2002
831:
830:
826:
817:
815:
812:Ysbl.york.ac.uk
806:
805:
801:
792:
790:
785:
784:
780:
771:
769:
761:
760:
756:
747:
745:
737:
736:
732:
692:
691:
687:
639:
638:
634:
624:
622:
617:
616:
612:
607:
598:
585:
557:
542:NCS differences
445:- validate the
417:
354:Regularize zone
276:
244:
185:
146:
83:
79:2 February 2021
78:
76:
73:
48:Initial release
43:Kevin D. Cowtan
42:
29:
12:
11:
5:
1102:
1100:
1092:
1091:
1086:
1081:
1076:
1071:
1066:
1056:
1055:
1052:
1051:
1044:
1043:External links
1041:
1038:
1037:
1013:
988:
963:
952:Xray.bmc.uu.se
939:
895:
854:
824:
799:
778:
754:
730:
685:
656:(4): 486–501.
632:
609:
608:
606:
603:
597:
594:
584:
581:
563:Coot structure
556:
553:
552:
551:
545:
539:
533:
527:
518:
512:
506:
503:omega analysis
497:
491:
486:
480:
475:Difference map
471:
465:
459:
450:
447:torsion angles
416:
413:
412:
411:
406:
401:
395:
385:
384:
379:
374:
369:
363:
357:
351:
341:
340:
334:
328:
318:
308:
302:
275:
272:
243:
240:
184:
181:
151:
150:
141:
137:
136:
131:
125:
124:
119:
113:
112:
95:
89:
88:
85:
84:
65:
63:
61:Stable release
57:
56:
53:
52:
49:
45:
44:
37:
31:
30:
27:
13:
10:
9:
6:
4:
3:
2:
1101:
1090:
1087:
1085:
1082:
1080:
1077:
1075:
1072:
1070:
1067:
1065:
1062:
1061:
1059:
1050:
1047:
1046:
1042:
1027:
1023:
1017:
1014:
1002:
998:
992:
989:
977:
973:
967:
964:
953:
949:
943:
940:
935:
929:
913:
909:
905:
899:
896:
891:
885:
877:
873:
869:
867:
858:
855:
842:
838:
837:www.ebi.ac.uk
834:
828:
825:
813:
809:
803:
800:
788:
782:
779:
768:
764:
758:
755:
744:
740:
734:
731:
726:
722:
717:
712:
708:
704:
700:
698:
689:
686:
681:
677:
672:
667:
663:
659:
655:
651:
647:
645:
636:
633:
620:
614:
611:
604:
602:
595:
593:
590:
582:
580:
577:
575:
571:
561:
554:
549:
548:Pukka puckers
546:
543:
540:
537:
536:Probe clashes
534:
531:
528:
525:
523:
519:
516:
513:
510:
507:
504:
502:
498:
495:
492:
490:
487:
484:
481:
478:
476:
472:
469:
466:
463:
460:
457:
455:
451:
448:
444:
443:
439:
438:
437:
429:
421:
414:
410:
407:
405:
402:
399:
396:
393:
390:
389:
388:
383:
380:
378:
375:
373:
370:
367:
364:
361:
358:
355:
352:
349:
346:
345:
344:
338:
335:
332:
331:Ideal DNA/RNA
329:
326:
322:
319:
316:
312:
309:
306:
303:
300:
297:
296:
295:
288:
280:
273:
271:
267:
265:
261:
255:
253:
249:
241:
239:
237:
233:
229:
225:
221:
216:
214:
210:
206:
202:
198:
197:free software
193:
191:
182:
180:
178:
174:
170:
166:
162:
158:
149:
145:
142:
138:
135:
132:
130:
126:
123:
120:
118:
114:
111:
107:
103:
99:
96:
94:
90:
86:
70:
66:0.9.4.1
64:
62:
58:
54:
50:
46:
41:
38:
36:
32:
25:
20:
1049:Coot website
1029:. Retrieved
1025:
1016:
1005:. Retrieved
1003:. 1999-03-26
1001:Csb.yale.edu
1000:
991:
980:. Retrieved
978:. 2006-08-09
975:
966:
955:. Retrieved
951:
942:
916:. Retrieved
912:the original
907:
898:
884:cite journal
878:: 2288–2294.
875:
871:
865:
857:
845:. Retrieved
841:the original
836:
827:
816:. Retrieved
814:. 2014-10-23
811:
802:
791:. Retrieved
781:
770:. Retrieved
766:
757:
746:. Retrieved
742:
733:
706:
702:
696:
688:
653:
649:
643:
635:
623:. Retrieved
613:
599:
586:
578:
566:
547:
541:
535:
529:
520:
514:
508:
499:
493:
488:
482:
473:
467:
461:
452:
440:
434:
408:
403:
397:
391:
386:
381:
376:
371:
365:
359:
353:
347:
342:
337:Find ligands
336:
330:
320:
310:
304:
298:
293:
268:
256:
245:
217:
194:
186:
160:
156:
155:The program
154:
35:Developer(s)
392:Find waters
325:beta strand
315:alpha helix
220:Paul Emsley
40:Paul Emsley
1058:Categories
1031:2017-02-27
1007:2017-02-27
982:2017-02-27
957:2017-02-27
918:17 January
847:17 January
818:2017-02-27
793:2017-02-27
789:. JISCMail
772:2017-02-27
748:2017-02-27
605:References
572:, and the
976:Sdsc.edu
928:cite web
725:15572765
680:20383002
524:analysis
454:Kleywegt
242:Features
195:Coot is
183:Overview
671:2852313
625:4 March
522:Rotamer
501:Peptide
140:Website
129:License
98:Windows
77: (
739:"Coot"
723:
678:
668:
477:peaks
252:mmcif
102:Linux
934:link
920:2022
890:link
849:2022
721:PMID
676:PMID
627:2021
570:FFTW
456:plot
260:ccp4
213:Fink
209:CCP4
157:Coot
117:Type
110:Unix
106:OS X
51:2002
17:Coot
876:D60
711:doi
707:D60
666:PMC
658:doi
654:D66
264:cns
262:or
248:pdb
238:).
1060::
1024:.
999:.
974:.
950:.
930:}}
926:{{
906:.
886:}}
882:{{
874:.
870:.
835:.
810:.
765:.
741:.
719:.
705:.
701:.
674:.
664:.
652:.
648:.
576:.
250:,
179:.
108:,
104:,
100:,
1034:.
1010:.
985:.
960:.
936:)
922:.
892:)
868:"
864:"
851:.
821:.
796:.
775:.
751:.
727:.
713::
699:"
695:"
682:.
660::
646:"
642:"
629:.
517:-
511:-
222:(
159:(
81:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.