361:
397:
resulting in a two electron reduced state with one electron each on the FAD and cluster domains. Then, the bound ubiquinone is reduced to ubiquinol, at least transiently forming the singly reduced semiubiquinone. The second pathway involves the donation of electrons from ETF to the iron cluster, followed by internal transitions between the two electron centers. After equilibration, the rest of the pathway follows as above.
2645:
29:
421:
deficiency. More severe cases involve congenital defects and full metabolic crisis. Genetically, it is an autosomal recessive disorder, making its occurrence fairly rare. Most affected patients are the result of single point mutations around the FAD ubiquinone interface. Milder forms of the disorder
1107:
Olsen RK, Olpin SE, Andresen BS, Miedzybrodzka ZH, Pourfarzam M, Merinero B, Frerman FE, Beresford MW, Dean JC, Cornelius N, Andersen O, Oldfors A, Holme E, Gregersen N, Turnbull DM, Morris AA (Aug 2007). "ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation
396:
The exact mechanism for the reduction is unknown, although there are two hypothesized pathways. The first pathway is the transferral of electrons from one electron reduced ETF one at a time to the lower potential FAD center. One electron is transferred from the reduced FAD to the iron cluster,
972:
Turnbull DM, Bartlett K, Eyre JA, Gardner-Medwin D, Johnson MA, Fisher J, Watmough NJ (Oct 1988). "Lipid storage myopathy due to glutaric aciduria type II: treatment of a potentially fatal myopathy".
794:
Watmough NJ, Loehr JP, Drake SK, Frerman FE (Feb 1991). "Tryptophan fluorescence in electron-transfer flavoprotein:ubiquinone oxidoreductase: fluorescence quenching by a brominated pseudosubstrate".
1228:
1072:
Goodman SI, Binard RJ, Woontner MR, Frerman FE (2002). "Glutaric acidemia type II: gene structure and mutations of the electron transfer flavoprotein:ubiquinone oxidoreductase (ETF:QO) gene".
929:
Singla M, Guzman G, Griffin AJ, Bharati S (Mar 2008). "Cardiomyopathy in multiple Acyl-CoA dehydrogenase deficiency: a clinico-pathological correlation and review of literature".
409:(also known as MADD for multiple acyl-CoA dehydrogenase deficiency), in which there is an improper buildup of fats and proteins in the body. Complications can involve
426:
therapy and are coined riboflavin-responsive MADD (RR-MADD), although due to the varying mutations causing the disease treatment and symptoms can vary considerably.
2212:
1221:
368:
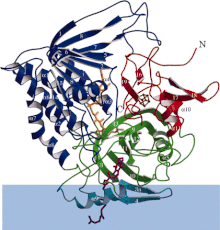
ETF-QO consists of one structural domain with three functional domains packed in close proximity: a FAD domain, a 4Fe4S cluster domain, and a UQ-binding domain.
1740:
1501:
63:
1145:"Multiple acyl-coenzyme A dehydrogenation disorder responsive to riboflavin: substrate oxidation, flavin metabolism, and flavoenzyme activities in fibroblasts"
2242:
1214:
384:
components. Ubiquinone binding is achieved through a deep hydrophobic binding pocket which is a different mode than other UQ-binding proteins such as
1472:
1017:"ETFDH mutations, CoQ10 levels, and respiratory chain activities in patients with riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency"
831:"Deficiency of Electron Transfer Flavoprotein or Electron Transfer Flavoprotein:Ubiquinone Oxidoreductase in Glutaric Acidemia Type II Fibroblasts"
759:
Beckmann JD, Frerman FE (Jul 1985). "Reaction of electron-transfer flavoprotein with electron-transfer flavoprotein-ubiquinone oxidoreductase".
2666:
1604:
2261:
2205:
1197:
87:
2284:
1354:
1273:
277:
222:
2371:
2238:
36:
of electron-transferring-flavoprotein dehydrogenase with each functional domain differentially colored. Blue band is membrane area.
653:"Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool"
75:
1735:
388:. Although ETF-QO is an integral membrane protein, it does not traverse the entire membrane unlike other UQ-binding proteins.
68:
2198:
1511:
1341:
1329:
1278:
2527:
2671:
2138:
1350:
1337:
234:
369:
511:
He M, Rutledge SL, Kelly DR, Palmer CA, Murdoch G, Majumder N, Nicholls RD, Pei Z, Watkins PA, Vockley J (Jul 2007).
2256:
1645:
601:
Vianey-Liaud C, Divry P, Gregersen N, Mathieu M (1987). "The inborn errors of mitochondrial fatty acid oxidation".
2512:
2628:
2615:
2602:
2589:
2576:
2563:
2550:
2304:
2271:
2234:
2009:
1931:
1686:
1438:
1015:
Liang WC, Ohkuma A, Hayashi YK, López LC, Hirano M, Nonaka I, Noguchi S, Chen LH, Jong YJ, Nishino I (Mar 2009).
435:
385:
2522:
372:
is in an extended conformation and is buried deeply within its functional domain. Multiple hydrogen bonds and a
2476:
2419:
2225:
2029:
1750:
1691:
1597:
1433:
1393:
1268:
440:
406:
257:
238:
215:
140:
2024:
2424:
2338:
2333:
2251:
2123:
1872:
1867:
1853:
1828:
1458:
1374:
1285:
380:
intermediate. The 4Fe4S cluster is also stabilized by extensive hydrogen bonding around the cluster and its
276:
to oxidative phosphorylation in the mitochondria. Specifically, it catalyzes the transfer of electrons from
712:"Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein: ubiquinone oxidoreductase"
360:
2158:
2092:
1978:
1973:
1678:
1417:
1366:
1333:
287:
80:
2445:
2364:
2163:
2128:
2084:
1848:
1707:
1311:
280:(ETF) to ubiquinone, reducing it to ubiquinol. The entire sequence of transfer reactions is as follows:
2517:
2102:
1807:
1783:
1745:
1482:
1412:
1300:
842:
664:
445:
226:
157:
319:
as the electron donor and ubiquinone-1 as the electron acceptor. The enzyme can also be assayed via
2676:
2481:
2047:
1666:
1590:
1562:
246:
1192:
2414:
2170:
1843:
1812:
1788:
1775:
1453:
1054:
997:
954:
626:
320:
417:, with other symptoms such as general weakness, liver enlargement, increased heart failure, and
2681:
2289:
2042:
1838:
1682:
1640:
1635:
1241:
1166:
1125:
1089:
1046:
989:
946:
911:
870:
811:
776:
741:
692:
618:
583:
542:
493:
312:
99:
2460:
2455:
2429:
2357:
2065:
1536:
1531:
1404:
1258:
1156:
1117:
1081:
1036:
1028:
981:
938:
901:
860:
850:
803:
768:
731:
723:
682:
672:
610:
573:
532:
524:
483:
152:
2507:
2491:
2404:
2294:
2019:
1901:
254:
181:
145:
1206:
1041:
1016:
846:
668:
471:
2650:
2545:
2486:
2221:
2096:
2014:
2004:
1953:
1941:
1914:
985:
736:
711:
687:
652:
537:
512:
161:
33:
1085:
906:
889:
865:
830:
2660:
2450:
2409:
1968:
1655:
1161:
1144:
513:"A new genetic disorder in mitochondrial fatty acid beta-oxidation: ACAD9 deficiency"
488:
1582:
1058:
1001:
2399:
1948:
1918:
1723:
1695:
1477:
1463:
1385:
890:"Abnormal myocardial lipid composition in an infant with type II glutaric aciduria"
630:
414:
316:
242:
2190:
958:
578:
561:
116:
2623:
2558:
2394:
2074:
1936:
377:
373:
291:
273:
269:
191:
2644:
657:
Proceedings of the
National Academy of Sciences of the United States of America
2640:
2175:
1897:
1818:
1799:
1731:
1674:
1617:
1613:
1321:
1237:
1032:
942:
450:
423:
230:
123:
2597:
2571:
2070:
1877:
1621:
1263:
855:
677:
418:
306:
1129:
1121:
1093:
1050:
950:
696:
587:
546:
497:
376:
modulate the redox potential of FAD and can possibly stabilize the anionic
1170:
993:
915:
874:
815:
780:
745:
622:
1295:
410:
381:
283:
128:
807:
772:
241:. The enzyme is found in both prokaryotes and eukaryotes and contains a
2308:
2133:
2118:
614:
218:
1187:
727:
104:
28:
2610:
2380:
2275:
1244:
111:
528:
2584:
1546:
1541:
359:
250:
1526:
1521:
1516:
1506:
1496:
1289:
1201:
562:"The electron transfer flavoprotein: ubiquinone oxidoreductases"
92:
2353:
2194:
1586:
1210:
2349:
1202:
MULTIPLE ACYL-CoA DEHYDROGENASE DEFICIENCY; MADD - 231680
302:
The overall reaction catalyzed by ETF-QO is as follows:
212:
electron transfer flavoprotein-ubiquinone oxidoreductase
253:
gene. Deficiency in ETF dehydrogenase causes the human
1815:(amino acid→pyruvate, acetyl CoA, or TCA intermediate)
1143:
Rhead W, Roettger V, Marshall T, Amendt B (Feb 1993).
405:
Deficiency of ETF-QO results in a disorder known as
2536:
2500:
2469:
2438:
2387:
2326:
2303:
2270:
2233:
2151:
2111:
2083:
2064:
1993:
1886:
1866:
1827:
1798:
1774:
1765:
1722:
1706:
1665:
1654:
1628:
1555:
1446:
1430:
1402:
1383:
1364:
1319:
1310:
1251:
888:Galloway JH, Cartwright IJ, Bennett MJ (Mar 1987).
566:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
187:
177:
172:
151:
139:
134:
122:
110:
98:
86:
74:
62:
54:
46:
41:
21:
472:"Acyl-CoA dehydrogenases. A mechanistic overview"
2318:Electron-transferring-flavoprotein dehydrogenase
1568:Electron-transferring-flavoprotein dehydrogenase
204:Electron-transferring-flavoprotein dehydrogenase
22:Electron-transferring-flavoprotein dehydrogenase
1473:Complex III/Coenzyme Q - cytochrome c reductase
835:Proceedings of the National Academy of Sciences
710:Ramsay RR, Steenkamp DJ, Husain M (Feb 1987).
646:
644:
642:
640:
323:of ETF semiquinone. Both reactions are below:
2365:
2206:
1598:
1222:
221:) is an enzyme that transfers electrons from
8:
2372:
2358:
2350:
2213:
2199:
2191:
2080:
1997:
1890:
1883:
1795:
1771:
1662:
1605:
1591:
1583:
1443:
1316:
1229:
1215:
1207:
974:Developmental Medicine and Child Neurology
258:multiple acyl-CoA dehydrogenase deficiency
169:
27:
1160:
1040:
905:
864:
854:
735:
686:
676:
577:
536:
487:
651:Zhang J, Frerman FE, Kim JJ (Oct 2006).
305:ETF-QO(red) + ubiquinone ↔ ETF-QO(ox) +
829:Frerman, F. E.; Goodman, S. I. (1985).
462:
603:Journal of Inherited Metabolic Disease
311:Enzymatic activity is usually assayed
18:
7:
1715:Electron acceptors other than oxygen
560:Watmough NJ, Frerman FE (Dec 2010).
2262:Methylenetetrahydrofolate reductase
1198:Online Mendelian Inheritance in Man
2285:Dihydrobenzophenanthridine oxidase
1459:Complex II/Succinate dehydrogenase
1355:Pyruvate dehydrogenase phosphatase
986:10.1111/j.1469-8749.1988.tb04806.x
517:American Journal of Human Genetics
278:electron transferring flavoprotein
249:. In humans, it is encoded by the
223:electron-transferring flavoprotein
14:
1074:Molecular Genetics and Metabolism
2643:
1162:10.1203/00006450-199302000-00008
489:10.1046/j.1432-1033.2003.03946.x
476:European Journal of Biochemistry
1736:Substrate-level phosphorylation
1483:Complex IV/Cytochrome c oxidase
470:Ghisla S, Thorpe C (Feb 2004).
1330:Pyruvate dehydrogenase complex
268:ETQ-QO links the oxidation of
1:
2139:Reverse cholesterol transport
1351:Pyruvate dehydrogenase kinase
1086:10.1016/S1096-7192(02)00138-5
907:10.1016/S0022-2275(20)38707-1
1791:(protein→peptide→amino acid)
1454:Complex I/NADH dehydrogenase
579:10.1016/j.bbabio.2010.10.007
235:inner mitochondrial membrane
2667:Genes on human chromosome 4
2698:
2257:Saccharopine dehydrogenase
2176:Phospagen system (ATP-PCr)
1646:Primary nutritional groups
1274:Oxoglutarate dehydrogenase
1193:BRENDA entry on EC 1.5.5.1
1188:ENZYME entry on EC 1.5.5.1
386:succinate-Q oxidoreductase
2528:Michaelis–Menten kinetics
2038:
2010:Anoxygenic photosynthesis
2000:
1964:
1932:Pentose phosphate pathway
1927:
1910:
1893:
1687:Oxidative phosphorylation
1439:oxidative phosphorylation
1033:10.1016/j.nmd.2009.01.008
943:10.1007/s00246-007-9119-6
894:Journal of Lipid Research
436:Oxidative phosphorylation
407:glutaric acidemia type II
364:ETF-QO Functional Domains
168:
26:
2420:Diffusion-limited enzyme
2030:Entner-Doudoroff pathway
1692:electron transport chain
1679:Pyruvate decarboxylation
1434:electron transport chain
1394:Methylmalonyl-CoA mutase
1269:Isocitrate dehydrogenase
441:Electron transport chain
422:have been responsive to
239:electron transport chain
2339:Cytokinin dehydrogenase
2334:Sarcosine dehydrogenase
2252:Dihydrofolate reductase
2124:Sphingolipid metabolism
2025:DeLey-Doudoroff pathway
1873:carbohydrate catabolism
1868:Carbohydrate metabolism
1854:Purine nucleotide cycle
1375:Glutamate dehydrogenase
1286:Succinate dehydrogenase
1279:Succinyl CoA synthetase
1021:Neuromuscular Disorders
856:10.1073/pnas.82.13.4517
716:The Biochemical Journal
678:10.1073/pnas.0604567103
2093:Fatty acid degradation
1813:Amino acid degradation
1418:Aspartate transaminase
365:
313:spectrophotometrically
290:→ ETF → ETF-QO → UQ →
288:Acyl-CoA dehydrogenase
2513:Eadie–Hofstee diagram
2446:Allosteric regulation
2129:Eicosanoid metabolism
2085:Fatty acid metabolism
1849:Pyrimidine metabolism
1708:Anaerobic respiration
401:Clinical significance
374:positive helix dipole
363:
2672:Cellular respiration
2523:Lineweaver–Burk plot
2103:Fatty acid synthesis
1808:Amino acid synthesis
1413:Pyruvate carboxylase
1301:Malate dehydrogenase
1122:10.1093/brain/awm135
931:Pediatric Cardiology
609:(Suppl 1): 159–200.
446:Microbial metabolism
237:. It is part of the
227:mitochondrial matrix
1667:Aerobic respiration
1563:Alternative oxidase
1367:α-ketoglutaric acid
847:1985PNAS...82.4517F
808:10.1021/bi00219a023
773:10.1021/bi00336a017
669:2006PNAS..10316212Z
2482:Enzyme superfamily
2415:Enzyme promiscuity
2171:Ethanol metabolism
2119:Steroid metabolism
1844:Nucleotide salvage
1776:Protein metabolism
1149:Pediatric Research
615:10.1007/bf01812855
366:
338:+ Oct-2-enoyl-CoA
321:disproportionation
298:Catalyzed reaction
2638:
2637:
2347:
2346:
2290:Sarcosine oxidase
2188:
2187:
2184:
2183:
2147:
2146:
2060:
2059:
2056:
2055:
2043:Xylose metabolism
1989:
1988:
1862:
1861:
1839:Purine metabolism
1784:Protein synthesis
1761:
1760:
1683:Citric acid cycle
1641:Metabolic network
1636:Metabolic pathway
1580:
1579:
1576:
1575:
1426:
1425:
1242:Citric acid cycle
1116:(Pt 8): 2045–54.
841:(13): 4517–4520.
728:10.1042/bj2410883
315:by reaction with
208:ETF dehydrogenase
201:
200:
197:
196:
2689:
2653:
2648:
2647:
2518:Hanes–Woolf plot
2461:Enzyme activator
2456:Enzyme inhibitor
2430:Enzyme catalysis
2374:
2367:
2360:
2351:
2215:
2208:
2201:
2192:
2159:Metal metabolism
2081:
2066:Lipid metabolism
1998:
1891:
1884:
1796:
1772:
1698:
1663:
1607:
1600:
1593:
1584:
1444:
1405:oxaloacetic acid
1317:
1259:Citrate synthase
1231:
1224:
1217:
1208:
1175:
1174:
1164:
1140:
1134:
1133:
1104:
1098:
1097:
1069:
1063:
1062:
1044:
1012:
1006:
1005:
969:
963:
962:
926:
920:
919:
909:
885:
879:
878:
868:
858:
826:
820:
819:
791:
785:
784:
756:
750:
749:
739:
707:
701:
700:
690:
680:
648:
635:
634:
598:
592:
591:
581:
557:
551:
550:
540:
508:
502:
501:
491:
467:
326:Octanoyl-CoA + Q
170:
31:
19:
2697:
2696:
2692:
2691:
2690:
2688:
2687:
2686:
2657:
2656:
2649:
2642:
2639:
2634:
2546:Oxidoreductases
2532:
2508:Enzyme kinetics
2496:
2492:List of enzymes
2465:
2434:
2405:Catalytic triad
2383:
2378:
2348:
2343:
2322:
2299:
2295:Proline oxidase
2266:
2229:
2222:Oxidoreductases
2219:
2189:
2180:
2164:Iron metabolism
2143:
2107:
2068:
2052:
2034:
2020:Carbon fixation
1985:
1960:
1923:
1906:
1902:Gluconeogenesis
1875:
1870:
1858:
1830:
1823:
1794:
1767:
1757:
1718:
1702:
1690:
1657:
1650:
1624:
1611:
1581:
1572:
1551:
1493:
1467:
1437:
1432:
1422:
1398:
1379:
1360:
1306:
1247:
1235:
1184:
1179:
1178:
1142:
1141:
1137:
1106:
1105:
1101:
1071:
1070:
1066:
1014:
1013:
1009:
971:
970:
966:
928:
927:
923:
887:
886:
882:
828:
827:
823:
793:
792:
788:
758:
757:
753:
709:
708:
704:
663:(44): 16212–7.
650:
649:
638:
600:
599:
595:
559:
558:
554:
510:
509:
505:
469:
468:
464:
459:
432:
403:
394:
358:
352:
348:
344:
337:
333:
329:
300:
266:
255:genetic disease
37:
17:
12:
11:
5:
2695:
2693:
2685:
2684:
2679:
2674:
2669:
2659:
2658:
2655:
2654:
2651:Biology portal
2636:
2635:
2633:
2632:
2619:
2606:
2593:
2580:
2567:
2554:
2540:
2538:
2534:
2533:
2531:
2530:
2525:
2520:
2515:
2510:
2504:
2502:
2498:
2497:
2495:
2494:
2489:
2484:
2479:
2473:
2471:
2470:Classification
2467:
2466:
2464:
2463:
2458:
2453:
2448:
2442:
2440:
2436:
2435:
2433:
2432:
2427:
2422:
2417:
2412:
2407:
2402:
2397:
2391:
2389:
2385:
2384:
2379:
2377:
2376:
2369:
2362:
2354:
2345:
2344:
2342:
2341:
2336:
2330:
2328:
2324:
2323:
2321:
2320:
2314:
2312:
2301:
2300:
2298:
2297:
2292:
2287:
2281:
2279:
2268:
2267:
2265:
2264:
2259:
2254:
2248:
2246:
2231:
2230:
2220:
2218:
2217:
2210:
2203:
2195:
2186:
2185:
2182:
2181:
2179:
2178:
2173:
2168:
2167:
2166:
2155:
2153:
2149:
2148:
2145:
2144:
2142:
2141:
2136:
2131:
2126:
2121:
2115:
2113:
2109:
2108:
2106:
2105:
2100:
2097:Beta oxidation
2089:
2087:
2078:
2062:
2061:
2058:
2057:
2054:
2053:
2051:
2050:
2045:
2039:
2036:
2035:
2033:
2032:
2027:
2022:
2017:
2015:Chemosynthesis
2012:
2007:
2005:Photosynthesis
2001:
1995:
1991:
1990:
1987:
1986:
1984:
1983:
1982:
1981:
1976:
1965:
1962:
1961:
1959:
1958:
1957:
1956:
1954:Leloir pathway
1946:
1945:
1944:
1942:Polyol pathway
1934:
1928:
1925:
1924:
1922:
1921:
1915:Glycogenolysis
1911:
1908:
1907:
1905:
1904:
1894:
1888:
1881:
1864:
1863:
1860:
1859:
1857:
1856:
1851:
1846:
1841:
1835:
1833:
1825:
1824:
1822:
1821:
1816:
1810:
1804:
1802:
1793:
1792:
1786:
1780:
1778:
1769:
1763:
1762:
1759:
1758:
1756:
1755:
1754:
1753:
1748:
1743:
1728:
1726:
1720:
1719:
1717:
1716:
1712:
1710:
1704:
1703:
1701:
1700:
1671:
1669:
1660:
1652:
1651:
1649:
1648:
1643:
1638:
1632:
1630:
1626:
1625:
1612:
1610:
1609:
1602:
1595:
1587:
1578:
1577:
1574:
1573:
1571:
1570:
1565:
1559:
1557:
1553:
1552:
1550:
1549:
1544:
1539:
1534:
1529:
1524:
1519:
1514:
1509:
1504:
1499:
1491:
1486:
1485:
1480:
1475:
1470:
1465:
1461:
1456:
1450:
1448:
1441:
1428:
1427:
1424:
1423:
1421:
1420:
1415:
1409:
1407:
1400:
1399:
1397:
1396:
1390:
1388:
1381:
1380:
1378:
1377:
1371:
1369:
1362:
1361:
1359:
1358:
1349:(regulated by
1346:
1345:
1326:
1324:
1314:
1308:
1307:
1305:
1304:
1298:
1293:
1282:
1281:
1276:
1271:
1266:
1261:
1255:
1253:
1249:
1248:
1236:
1234:
1233:
1226:
1219:
1211:
1205:
1204:
1195:
1190:
1183:
1182:External links
1180:
1177:
1176:
1135:
1099:
1080:(1–2): 86–90.
1064:
1007:
964:
921:
880:
821:
802:(5): 1317–23.
786:
767:(15): 3922–5.
751:
702:
636:
593:
572:(12): 1910–6.
552:
529:10.1086/519219
503:
482:(3): 494–508.
461:
460:
458:
455:
454:
453:
448:
443:
438:
431:
428:
402:
399:
393:
390:
357:
354:
350:
346:
342:
335:
331:
327:
299:
296:
265:
262:
199:
198:
195:
194:
189:
185:
184:
179:
175:
174:
166:
165:
155:
149:
148:
143:
137:
136:
132:
131:
126:
120:
119:
114:
108:
107:
102:
96:
95:
90:
84:
83:
78:
72:
71:
66:
60:
59:
56:
52:
51:
48:
44:
43:
39:
38:
34:Ribbon diagram
32:
24:
23:
16:Protein family
15:
13:
10:
9:
6:
4:
3:
2:
2694:
2683:
2680:
2678:
2675:
2673:
2670:
2668:
2665:
2664:
2662:
2652:
2646:
2641:
2630:
2626:
2625:
2620:
2617:
2613:
2612:
2607:
2604:
2600:
2599:
2594:
2591:
2587:
2586:
2581:
2578:
2574:
2573:
2568:
2565:
2561:
2560:
2555:
2552:
2548:
2547:
2542:
2541:
2539:
2535:
2529:
2526:
2524:
2521:
2519:
2516:
2514:
2511:
2509:
2506:
2505:
2503:
2499:
2493:
2490:
2488:
2487:Enzyme family
2485:
2483:
2480:
2478:
2475:
2474:
2472:
2468:
2462:
2459:
2457:
2454:
2452:
2451:Cooperativity
2449:
2447:
2444:
2443:
2441:
2437:
2431:
2428:
2426:
2423:
2421:
2418:
2416:
2413:
2411:
2410:Oxyanion hole
2408:
2406:
2403:
2401:
2398:
2396:
2393:
2392:
2390:
2386:
2382:
2375:
2370:
2368:
2363:
2361:
2356:
2355:
2352:
2340:
2337:
2335:
2332:
2331:
2329:
2325:
2319:
2316:
2315:
2313:
2310:
2306:
2302:
2296:
2293:
2291:
2288:
2286:
2283:
2282:
2280:
2277:
2273:
2269:
2263:
2260:
2258:
2255:
2253:
2250:
2249:
2247:
2244:
2240:
2236:
2232:
2227:
2223:
2216:
2211:
2209:
2204:
2202:
2197:
2196:
2193:
2177:
2174:
2172:
2169:
2165:
2162:
2161:
2160:
2157:
2156:
2154:
2150:
2140:
2137:
2135:
2132:
2130:
2127:
2125:
2122:
2120:
2117:
2116:
2114:
2110:
2104:
2101:
2098:
2094:
2091:
2090:
2088:
2086:
2082:
2079:
2076:
2072:
2067:
2063:
2049:
2048:Radiotrophism
2046:
2044:
2041:
2040:
2037:
2031:
2028:
2026:
2023:
2021:
2018:
2016:
2013:
2011:
2008:
2006:
2003:
2002:
1999:
1996:
1992:
1980:
1977:
1975:
1972:
1971:
1970:
1969:Glycosylation
1967:
1966:
1963:
1955:
1952:
1951:
1950:
1947:
1943:
1940:
1939:
1938:
1935:
1933:
1930:
1929:
1926:
1920:
1916:
1913:
1912:
1909:
1903:
1899:
1896:
1895:
1892:
1889:
1885:
1882:
1879:
1874:
1869:
1865:
1855:
1852:
1850:
1847:
1845:
1842:
1840:
1837:
1836:
1834:
1832:
1826:
1820:
1817:
1814:
1811:
1809:
1806:
1805:
1803:
1801:
1797:
1790:
1787:
1785:
1782:
1781:
1779:
1777:
1773:
1770:
1764:
1752:
1749:
1747:
1744:
1742:
1739:
1738:
1737:
1733:
1730:
1729:
1727:
1725:
1721:
1714:
1713:
1711:
1709:
1705:
1697:
1693:
1688:
1684:
1680:
1676:
1673:
1672:
1670:
1668:
1664:
1661:
1659:
1653:
1647:
1644:
1642:
1639:
1637:
1634:
1633:
1631:
1627:
1623:
1619:
1615:
1608:
1603:
1601:
1596:
1594:
1589:
1588:
1585:
1569:
1566:
1564:
1561:
1560:
1558:
1554:
1548:
1545:
1543:
1540:
1538:
1535:
1533:
1530:
1528:
1525:
1523:
1520:
1518:
1515:
1513:
1510:
1508:
1505:
1503:
1500:
1498:
1494:
1488:
1487:
1484:
1481:
1479:
1476:
1474:
1471:
1469:
1462:
1460:
1457:
1455:
1452:
1451:
1449:
1445:
1442:
1440:
1435:
1431:Mitochondrial
1429:
1419:
1416:
1414:
1411:
1410:
1408:
1406:
1401:
1395:
1392:
1391:
1389:
1387:
1382:
1376:
1373:
1372:
1370:
1368:
1363:
1356:
1352:
1348:
1347:
1343:
1339:
1335:
1331:
1328:
1327:
1325:
1323:
1318:
1315:
1313:
1309:
1302:
1299:
1297:
1294:
1291:
1287:
1284:
1283:
1280:
1277:
1275:
1272:
1270:
1267:
1265:
1262:
1260:
1257:
1256:
1254:
1250:
1246:
1243:
1239:
1232:
1227:
1225:
1220:
1218:
1213:
1212:
1209:
1203:
1199:
1196:
1194:
1191:
1189:
1186:
1185:
1181:
1172:
1168:
1163:
1158:
1155:(2): 129–35.
1154:
1150:
1146:
1139:
1136:
1131:
1127:
1123:
1119:
1115:
1111:
1108:deficiency".
1103:
1100:
1095:
1091:
1087:
1083:
1079:
1075:
1068:
1065:
1060:
1056:
1052:
1048:
1043:
1038:
1034:
1030:
1026:
1022:
1018:
1011:
1008:
1003:
999:
995:
991:
987:
983:
980:(5): 667–72.
979:
975:
968:
965:
960:
956:
952:
948:
944:
940:
937:(2): 446–51.
936:
932:
925:
922:
917:
913:
908:
903:
900:(3): 279–84.
899:
895:
891:
884:
881:
876:
872:
867:
862:
857:
852:
848:
844:
840:
836:
832:
825:
822:
817:
813:
809:
805:
801:
797:
790:
787:
782:
778:
774:
770:
766:
762:
755:
752:
747:
743:
738:
733:
729:
725:
722:(3): 883–92.
721:
717:
713:
706:
703:
698:
694:
689:
684:
679:
674:
670:
666:
662:
658:
654:
647:
645:
643:
641:
637:
632:
628:
624:
620:
616:
612:
608:
604:
597:
594:
589:
585:
580:
575:
571:
567:
563:
556:
553:
548:
544:
539:
534:
530:
526:
523:(1): 87–103.
522:
518:
514:
507:
504:
499:
495:
490:
485:
481:
477:
473:
466:
463:
456:
452:
449:
447:
444:
442:
439:
437:
434:
433:
429:
427:
425:
420:
416:
412:
408:
400:
398:
391:
389:
387:
383:
379:
375:
371:
362:
355:
353:
339:
324:
322:
318:
314:
309:
308:
303:
297:
295:
293:
289:
285:
281:
279:
275:
271:
263:
261:
259:
256:
252:
248:
244:
240:
236:
232:
228:
224:
220:
217:
213:
209:
205:
193:
190:
186:
183:
180:
176:
171:
167:
164:
163:
159:
156:
154:
150:
147:
144:
142:
138:
133:
130:
127:
125:
121:
118:
115:
113:
109:
106:
103:
101:
97:
94:
91:
89:
85:
82:
79:
77:
73:
70:
67:
65:
61:
57:
53:
49:
45:
40:
35:
30:
25:
20:
2624:Translocases
2621:
2608:
2595:
2582:
2569:
2559:Transferases
2556:
2543:
2400:Binding site
2317:
1949:Galactolysis
1919:Glycogenesis
1724:Fermentation
1696:ATP synthase
1567:
1489:
1478:Cytochrome c
1386:succinyl-CoA
1152:
1148:
1138:
1113:
1109:
1102:
1077:
1073:
1067:
1027:(3): 212–6.
1024:
1020:
1010:
977:
973:
967:
934:
930:
924:
897:
893:
883:
838:
834:
824:
799:
796:Biochemistry
795:
789:
764:
761:Biochemistry
760:
754:
719:
715:
705:
660:
656:
606:
602:
596:
569:
565:
555:
520:
516:
506:
479:
475:
465:
415:hypoglycemia
404:
395:
367:
340:
325:
317:octanoyl-CoA
310:
304:
301:
282:
267:
247:FE-S cluster
233:pool in the
211:
207:
203:
202:
160:
55:Alt. symbols
2395:Active site
2075:lipogenesis
1937:Fructolysis
1751:Lactic acid
1495:synthesis:
1312:Anaplerotic
378:semiquinone
292:Complex III
274:amino acids
270:fatty acids
182:Swiss-model
42:Identifiers
2677:Metabolism
2661:Categories
2598:Isomerases
2572:Hydrolases
2439:Regulation
1898:Glycolysis
1831:metabolism
1829:Nucleotide
1819:Urea cycle
1800:Amino acid
1789:Catabolism
1732:Glycolysis
1675:Glycolysis
1658:metabolism
1618:catabolism
1614:Metabolism
1490:Coenzyme Q
1464:Coenzyme Q
1322:acetyl-CoA
1238:Metabolism
457:References
451:Metabolism
424:riboflavin
231:ubiquinone
178:Structures
173:Search for
135:Other data
2477:EC number
2224:: CH-NH (
2071:lipolysis
1878:anabolism
1622:anabolism
1264:Aconitase
419:carnitine
392:Mechanism
356:Structure
307:ubiquinol
272:and some
229:, to the
141:EC number
117:NM_004453
64:NCBI gene
2682:EC 1.5.5
2501:Kinetics
2425:Cofactor
2388:Activity
2311:acceptor
2278:acceptor
2245:acceptor
1994:Nonhuman
1979:O-linked
1974:N-linked
1766:Specific
1296:Fumarase
1200:(OMIM):
1130:17584774
1094:12359134
1059:28328495
1051:19249206
1042:10409523
1002:33989343
951:17912479
697:17050691
588:20937244
547:17564966
498:14728676
430:See also
411:acidosis
382:cysteine
284:Acyl-CoA
264:Function
192:InterPro
2611:Ligases
2381:Enzymes
2309:quinone
2134:Ketosis
1746:Ethanol
1629:General
1468:(CoQ10)
1447:Primary
1303:and ETC
1245:enzymes
1171:8433888
994:3229565
916:3572253
875:2989828
843:Bibcode
816:1991113
781:2996585
746:3593226
737:1147643
688:1637562
665:Bibcode
631:9771779
623:3119938
538:1950923
225:in the
219:1.5.5.1
188:Domains
162:q4q32.1
146:1.5.5.1
124:UniProt
2585:Lyases
2327:1.5.99
2276:oxygen
1656:Energy
1537:COQ10B
1532:COQ10A
1169:
1128:
1092:
1057:
1049:
1039:
1000:
992:
959:370626
957:
949:
914:
873:
866:391133
863:
814:
779:
744:
734:
695:
685:
629:
621:
586:
545:
535:
496:
243:flavin
158:Chr. 4
129:Q16134
112:RefSeq
93:231675
58:ETF-QO
47:Symbol
2537:Types
2305:1.5.5
2272:1.5.3
2235:1.5.1
2152:Other
2112:Other
1887:Human
1768:paths
1556:Other
1547:PDSS2
1542:PDSS1
1252:Cycle
1110:Brain
1055:S2CID
998:S2CID
955:S2CID
627:S2CID
349:+ ETF
345:↔ ETF
341:2 ETF
251:ETFDH
153:Locus
2629:list
2622:EC7
2616:list
2609:EC6
2603:list
2596:EC5
2590:list
2583:EC4
2577:list
2570:EC3
2564:list
2557:EC2
2551:list
2544:EC1
2243:NADP
2228:1.5)
1876:and
1527:COQ9
1522:COQ7
1517:COQ6
1512:COQ5
1507:COQ4
1502:COQ3
1497:COQ2
1353:and
1290:SDHA
1167:PMID
1126:PMID
1090:PMID
1047:PMID
990:PMID
947:PMID
912:PMID
871:PMID
812:PMID
777:PMID
742:PMID
693:PMID
619:PMID
584:PMID
570:1797
543:PMID
494:PMID
245:and
105:2GMH
88:OMIM
81:3483
76:HGNC
69:2110
50:ETFD
2241:or
2239:NAD
1900:⇄
1741:ABE
1403:to
1384:to
1365:to
1320:to
1157:doi
1118:doi
1114:130
1082:doi
1037:PMC
1029:doi
982:doi
939:doi
902:doi
861:PMC
851:doi
804:doi
769:doi
732:PMC
724:doi
720:241
683:PMC
673:doi
661:103
611:doi
574:doi
533:PMC
525:doi
484:doi
480:271
413:or
370:FAD
330:↔ Q
210:or
100:PDB
2663::
2307::
2274::
2237::
2226:EC
2073:,
1917:⇄
1734:→
1694:+
1685:→
1681:→
1677:→
1620:,
1616:,
1492:10
1466:10
1342:E3
1340:,
1338:E2
1336:,
1334:E1
1240::
1165:.
1153:33
1151:.
1147:.
1124:.
1112:.
1088:.
1078:77
1076:.
1053:.
1045:.
1035:.
1025:19
1023:.
1019:.
996:.
988:.
978:30
976:.
953:.
945:.
935:29
933:.
910:.
898:28
896:.
892:.
869:.
859:.
849:.
839:82
837:.
833:.
810:.
800:30
798:.
775:.
765:24
763:.
740:.
730:.
718:.
714:.
691:.
681:.
671:.
659:.
655:.
639:^
625:.
617:.
607:10
605:.
582:.
568:.
564:.
541:.
531:.
521:81
519:.
515:.
492:.
478:.
474:.
351:2-
347:ox
343:1-
294:.
286:→
260:.
216:EC
214:,
2631:)
2627:(
2618:)
2614:(
2605:)
2601:(
2592:)
2588:(
2579:)
2575:(
2566:)
2562:(
2553:)
2549:(
2373:e
2366:t
2359:v
2214:e
2207:t
2200:v
2099:)
2095:(
2077:)
2069:(
1880:)
1871:(
1699:)
1689:(
1606:e
1599:t
1592:v
1436:/
1357:)
1344:)
1332:(
1292:)
1288:(
1230:e
1223:t
1216:v
1173:.
1159::
1132:.
1120::
1096:.
1084::
1061:.
1031::
1004:.
984::
961:.
941::
918:.
904::
877:.
853::
845::
818:.
806::
783:.
771::
748:.
726::
699:.
675::
667::
633:.
613::
590:.
576::
549:.
527::
500:.
486::
336:2
334:H
332:1
328:1
206:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.