943:
smallest numbers of genes of a known genome sequence for a eukaryote, and has only three chromosomes in its genome. Many of the genes responsible for cell division and cellular organization in fission yeast cell are also found in the human's genome. Cell cycle regulation and division are crucial for growth and development of any cell. Fission yeast's conserved genes has been heavily studied and the reason for many recent biomedical developments. Fission yeast is also a practical model system to observe cell division because fission yeast's are cylindrically shaped single celled eukaryotes that divide and reproduce by medial fission. This can easily be seen using microscopy. Fission yeast also have an extremely short generation time, 2 to 4 hours, which also makes it an easy model system to observe and grow in the laboratory
Fission yeast's simplicity in genomic structure yet similarities with mammalian genome, ease of ability to manipulate, and ability to be used for drug analysis is why fission yeast is making many contributions to biomedicine and cellular biology research, and a model system for genetic analysis.
1017:
of 57 strains of fission yeast that each differed by ≥1,900 SNPs, and all detected 57 strains of fission yeast were prototrophic (able to grow on the same minimal medium as the reference strain). A number of studies on S.pombe genome support the idea that the genetic diversity of fission yeast strains is slightly less than budding yeast. Indeed, only limited variations of S.pombe occur in proliferation in different environments. In addition, the amount of phenotypic variation segregating in fission yeast is less than that seen, in S. cerevisiae. Since most strains of fission yeast were isolated from brewed beverages, there is no ecological or historical context to this dispersal.
797:
785:
731:
886:
be required for the change of the cell type. Studies of the mating-type switching system lead to a discovery and characterization of a site-specific replication termination site RTS1, a site-specific replication pause site MPS1, and a novel type of chromosomal imprint, marking one of the sister chromatids at the mating-type locus mat1. In addition, work on the silenced donor region has led to great advances in understanding formation and maintenance of heterochromatin.
1069:
points such as these, mutagens are created and replicated, resulting in multitudes of cellular issues including cell death or tumorigenesis seen in cancerous cells. Paul Nurse, Leland
Hartwell, and Tim Hunt were awarded the Nobel Prize in Physiology or Medicine in 2001. They discovered key conserved checkpoints that are crucial for a cell to divide properly. These findings have been linked to cancer and diseased cells and are a notable finding for biomedicine.
57:
773:
817:
758:
979:
be 34, 65, and 110 kb. This is 300–100 times longer than the centromeres of budding yeast. An extremely high level of conservation (97%) is also seen over 1,780-bp region in the DGS regions of the centromere. This elongation of centromeres and its conservative sequences makes fission yeast a practical model system to use to observe cell division and in humans because of their likeness.
834:
1104:
However, there are limitations with using fission yeast as a model system: its multidrug resistance. "The MDR response involves overexpression of two types of drug efflux pumps, the ATP-binding cassette (ABC) family... and the major facilitator superfamily". Paul Nurse and some of his colleagues have
1072:
Researchers using fission yeast as a model system also look at organelle dynamics and responses and the possible correlations between yeast cells and mammalian cells. Mitochondria diseases, and various organelle systems such as the Golgi apparatus and endoplasmic reticulum, can be further understood,
1016:
Biodiversity and evolutionary study of fission yeast was carried out on 161 strains of
Schizosaccharomyces pombe collected from 20 countries. Modeling of the evolutionary rate showed that all strains derived from a common ancestor that has lived since ~2,300 years ago. The study also identified a set
885:
Fission yeast switches mating type by a replication-coupled recombination event, which takes place during S phase of the cell cycle. Fission yeast uses intrinsic asymmetry of the DNA replication process to switch the mating type; it was the first system where the direction of replication was shown to
978:
s having a large number of introns gives opportunities for an increase of range of protein types produced from alternative splicing and genes that code for comparable genes in human. 81% of the three centromeres in fission yeast have been sequenced. The lengths of the three centromeres were found to
854:
as measured by increased cell division time and increased probability of cell death. Finally, wee1 mutant fission yeast cells are smaller than wild-type cells, but take just as long to go through the cell cycle. This is possible because small yeast cells grow slower, that is, their added total mass
1025:
DNA replication in yeast has been increasingly studied by many researchers. Further understanding of DNA replication, gene expression, and conserved mechanisms in yeast can provide researchers with information on how these systems operate in mammalian cells in general and human cells in particular.
942:
Fission yeast has become a notable model system to study basic principles of a cell that can be used to understand more complex organisms like mammals and in particular humans. This single cell eukaryote is nonpathogenic and easily grown and manipulated in the lab. Fission yeast contains one of the
753:
The central events of cell reproduction are chromosome duplication, which takes place in S (Synthetic) phase, followed by chromosome segregation and nuclear division (mitosis) and cell division (cytokinesis), which are collectively called M (Mitotic) phase. G1 is the gap between M and S phases, and
1129:
Virtually any genetics experiment or technique can, therefore, be applied to this model system such as: tetrad dissection, mutagens analysis, transformations, and microscopy techniques such as FRAP and FRET. New models, such as Tug-Of-War (gTOW), are also being used to analyze yeast robustness and
1068:
One of the safety precautions that the cell takes to ensure precise cell division occurs is the cell-cycle checkpoint. These checkpoints ensure any mutagens are eliminated. This is done often by relay signals that stimulate ubiquitination of the targets and delay cytokinesis. Without mitotic check
970:
Forty-three percent of the genes in the Genome
Project were found to contain introns in 4,739 genes. Fission yeast does not have as many duplicated genes compared to budding yeast, only containing 5%, making fission yeast a great model genome to observe and gives researchers the ability to create
963:
centromeric (40kb) and telomeric (260kb) regions. After the initial sequencing of the fission yeast's genome, other previous non-sequenced regions of the genes have been sequenced. Structural and functional analysis of these gene regions can be found on large scale fission yeast databases such as
1125:
is normally a haploid cell but, when put under stressful conditions, usually nitrogen deficiency, two cells will conjugate to form a diploid that later form four spores within a tetrad ascus. This process is easily visible and observable under any microscope and allows us to look at meiosis in a
1112:
For example, Doxorubicin, a very common chemotherapeutic antibiotic, has many adverse side-effects. Researchers are looking for ways to further understand how doxorubicin works by observing the genes linked to resistance by using fission yeast as a model system. Links between doxorubicin adverse
876:
gradient does not reach the cortical nodes (red dots), and therefore Cdr2 and Cdr1 remain active in the nodes. Cdr2 and Cdr1 inhibit Wee1, preventing phosphorylation of Cdk1 and thereby leading to activation of CDK and mitotic entry. (This simplified diagram omits several other regulators of CDK
1055:
Cytokinesis is one of the components of cell division that is often observed in fission yeast. Well-conserved components of cytokinesis are observed in fission yeast and allow us to look at various genomic scenarios and pinpoint mutations. Cytokinesis is a permanent step and very crucial to the
765:
Fission yeast governs mitosis by mechanisms that are similar to those in multicellular animals. It normally proliferates in a haploid state. When starved, cells of opposite mating types (P and M) fuse to form a diploid zygote that immediately enters meiosis to generate four haploid spores. When
523:
is unique because of the deposition of α-(1,3)-glucan or pseudonigeran in the cell wall in addition to the better known β-glucans and the virtual lack of chitin. Species of this genus also differ in mannan composition, which shows terminal d-galactose sugars in the side-chains of their mannans.
845:
mutation causes entry into mitosis at an abnormally small size, resulting in a shorter G2. G1 is lengthened, suggesting that progression through Start (beginning of cell cycle) is responsive to growth when the G2/M control is lost. Furthermore, cells in poor nutrient conditions grow slowly and
962:
s genome was fully sequenced in 2002, the sixth eukaryotic genome to be sequenced as part of the Genome
Project. An estimated 4,979 genes were discovered within three chromosomes containing about 14Mb of DNA. This DNA is contained within 3 different chromosomes in the nucleus with gaps in the
518:
belongs to the division
Ascomycota, which represents the largest and most diverse group of fungi. Free-living ascomycetes are commonly found in tree exudates, on plant roots and in surrounding soil, on ripe and rotting fruits, and in association with insect vectors that transport them between
954:
is often used to study cell division and growth because of conserved genomic regions also seen in humans including: heterochromatin proteins, large origins of replication, large centromeres, conserved cellular checkpoints, telomere function, gene splicing, and many other cellular processes.
1060:
as a model system. The contractile ring is highly conserved in both fission yeast and human cytokinesis. Mutations in cytokinesis can result in many malfunctions of the cell including cell death and development of cancerous cells. This is a complex process in human cell division, but in
477:
was first discovered in 1893 when a group working in a
Brewery Association Laboratory in Germany was looking at sediment found in millet beer imported from East Africa that gave it an acidic taste. The term schizo, meaning "split" or "fission", had previously been used to describe other
519:
substrates. Many of these associations are symbiotic or saprophytic, although numerous ascomycetes (and their basidiomycete cousins) represent important plant pathogens that target myriad plant species, including commercial crops. Among the ascomycetous yeast genera, the fission yeast
3293:
Brown, William R. A.; Liti, Gianni; Rosa, Carlos; James, Steve; Roberts, Ian; Robert, Vincent; Jolly, Neil; Tang, Wen; Baumann, Peter; Green, Carter; Schlegel, Kristina; Young, Jonathan; Hirchaud, Fabienne; Leek, Spencer; Thomas, Geraint; Blomberg, Anders; Warringer, Jonas (2011).
824:
The general features of cytokinesis are shown here. The site of cell division is determined before anaphase. The anaphase spindle (in green on the figure) is then positioned so that the segregated chromosomes are on opposite sides of the predetermined cleavage plane.
922:, an enzyme that removes uracil from the DNA backbone and initiates base excision repair. On the basis of this finding, it was proposed that base excision repair of either a uracil base, an abasic site, or a single-strand nick is sufficient to initiate
913:
and formation of meiotic spores. This finding suggests that meiosis, and particularly meiotic recombination, may be an adaptation for repairing DNA damage. Supporting this view is the finding that single base lesions of the type dU:dG in the DNA of
862:
protein kinase (green) is localized to the cell cortex, with the highest concentration at the cell tips. The cell-cycle regulators Cdr2, Cdr1 and Wee1 are present in cortical nodes in the middle of the cell (blue and red dots). a, In small cells, the
44:
562:
are both extensively studied; these two species diverged approximately 300 to 600 million years before present, and are significant tools in molecular and cellular biology. Some of the technical discriminants between these two species are:
402:). This has fully unlocked the power of this organism, with many genes orthologous to human genes identified - 70% to date, including many genes involved in human disease. In 2006, sub-cellular localization of almost all the proteins in
1113:
side-effects and chromosome metabolism and membrane transport were seen. Metabolic models for drug targeting are now being used in biotechnology, and further advances are expected in the future using the fission yeast model system.
502:, Switzerland. The culture used by Urs Leupold contained (besides others) cells with the mating types h90 (strain 968), h- (strain 972), and h+ (strain 975). Subsequent to this, there have been two large efforts to isolate
749:
is a rod-shaped cell, approximately 3 μm in diameter, that grows entirely by elongation at the ends. After mitosis, division occurs by the formation of a septum, or cell plate, that cleaves the cell at its midpoint.
871:
inhibits Cdr2, preventing Cdr2 and Cdr1 from inhibiting Wee1, and allowing Wee1 to phosphorylate Cdk1, thus inactivating cyclin-dependent kinase (CDK) activity and preventing entry into mitosis. b, In long cells, the
3635:
Senoo T, Kawano S, Ikeda S (March 2017). "DNA base excision repair and nucleotide excision repair synergistically contribute to survival of stationary-phase cells of the fission yeast
Schizosaccharomyces pombe".
846:
therefore take longer to double in size and divide. Low nutrient levels also reset the growth threshold so that cell progresses through the cell cycle at a smaller size. Upon exposure to stressful conditions
431:
have been isolated. These have been collected from a variety of locations including Europe, North and South
America, and Asia. The majority of these strains have been collected from cultivated fruits such as
4292:
Grishchuk AL, Kohli J. Five RecA-like proteins of
Schizosaccharomyces pombe are involved in meiotic recombination. Genetics. 2003 Nov;165(3):1031-43. doi: 10.1093/genetics/165.3.1031. PMID: 14668362; PMCID:
754:
G2 is the gap between S and M phases. In the fission yeast, the G2 phase is particularly extended, and cytokinesis (daughter-cell segregation) does not happen until a new S (Synthetic) phase is launched.
3679:
Cadou A, Couturier A, Le Goff C, Xie L, Paulson JR, Le Goff X (March 2013). "The Kin1 kinase and the calcineurin phosphatase cooperate to link actin ring assembly and septum synthesis in fission yeast".
1493:
Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, et al. (July 2006). "ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe".
796:
1205:
Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, et al. (June 2008). "Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution".
745:
The fission yeast is a single-celled fungus with simple, fully characterized genome and a rapid growth rate. It has long been used in brewing, baking, and molecular genetics.
1130:
observe gene expression. Making knock-in and knock-out genes is fairly easy and with the fission yeast's genome being sequenced this task is very accessible and well known.
506:
from fruit, nectar, or fermentations: one by Florenzano et al. in the vineyards of western Sicily, and the other by Gomes et al. (2002) in four regions of southeast Brazil.
4379:
3723:
Balazs A, Batta G, Miklos I, Acs-Szabo L, Vazquez de Aldana CR, Sipiczki M (March 2012). "Conserved regulators of the cell separation process in Schizosaccharomyces".
833:
4198:
Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, et al. (September 2012). "Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae".
4688:
4727:
784:
1109:
strains sensitive to chemical inhibitors and common probes to see whether it is possible to use fission yeast as a model system of chemical drug research.
1896:
Moseley, James B.; Mayeux, Adeline; Paoletti, Anne; Nurse, Paul (2009). "A spatial gradient coordinates cell size and mitotic entry in fission yeast".
4428:
1955:
Martin, Sophie G.; Berthelot-Grosjean, Martine (2009-06-11). "Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle".
4662:
1183:
1643:"The evolution of aerobic fermentation in Schizosaccharomyces pombe was associated with regulatory reprogramming but not nucleosome reorganization"
2266:"Activation of an alternative, rec12 (spo11)-independent pathway of fission yeast meiotic recombination in the absence of a DNA flap endonuclease"
1121:
Fission yeast is easily accessible, easily grown and manipulated to make mutants, and able to be maintained at either a haploid or diploid state.
706:
has large, repetitive centromeres (40–100 kb) more similar to mammalian centromeres, and degenerate replication origins of at least 1kb.
4459:
4355:
1823:
1026:
Other stages, such as cellular growth and aging, are also observed in yeast in order to understand these mechanisms in more complex systems.
369:
360:, a fission yeast researcher, successfully merged the independent schools of fission yeast genetics and cell cycle research. Together with
2129:
2112:
498:, after he isolated it in 1924 from French wine (most probably rancid) at the Federal Experimental Station of Vini- and Horticulture in
816:
3834:
2653:"Cellular robustness conferred by genetic crosstalk underlies resistance against chemotherapeutic drug doxorubicin in fission yeast"
529:
4793:
1282:
genetics. In: Hall MN, Linder P. eds. The Early Days of Yeast Genetics. New York. Cold Spring Harbor Laboratory Press. p 125–128.
1139:
482:. The addition of the word pombe was due to its isolation from East African beer, as pombe means "beer" in Swahili. The standard
2313:
Forsburg SL (June 2005). "The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe: models for cell biology research".
56:
2601:"Analyzing fission yeast multidrug resistance mechanisms to develop a genetically tractable model system for chemical biology"
772:
657:
remains in the G2 phase of the cell cycle for an extended period (as a consequence, G2-M transition is under tight control).
4338:
Green, M. D. Sabatinos, S. A. Forsburg, S. L. (2009). "Microscopy Techniques to Examine DNA Replication in Fission Yeast".
4714:
4421:
1065:
simpler experiments can yield results that can then be applied for research in higher-order model systems such as humans.
4601:
4092:"Relationships between cell cycle regulator gene copy numbers and protein expression levels in Schizosaccharomyces pombe"
3047:"SAC-ing mitotic errors: how the spindle assembly checkpoint (SAC) plays defense against chromosome mis-segregation"
2065:
Klar, Amar J.S. (2007-12-01). "Lessons Learned from Studies of Fission Yeast Mating-Type Switching and Silencing".
1571:
Florenzano G, Balloni W, Materassi R (1977). "Contributo alla ecologia dei lieviti Schizosaccharomyces sulle uve".
1049:
997:
a great system to use to study human genes and disease pathways, especially cell cycle and DNA checkpoint systems.
486:
strains were isolated by Urs Leupold in 1946 and 1947 from a culture that he obtained from the yeast collection in
4732:
4466:
3978:
407:
3817:
Das M, Chiron S, Verde F (2010). "Microtubule-dependent spatial organization of mitochondria in fission yeast".
4479:
4414:
3188:"Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete-killing efficiency"
2456:
1085:
558:
118:
3921:"Mutations disrupting histone methylation have different effects on replication timing in S. pombe centromere"
3495:"The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42"
4798:
4523:
1037:
399:
108:
730:
4783:
4563:
4516:
934:, causes DNA damages such as single-strand nicks or gaps, and that these stimulate meiotic recombination.
919:
536:
can degrade L-malic acid, one of the dominant organic acids in wine, which makes them diverse among other
3592:
4311:
1056:
wellbeing of the cell. Contractile ring formation in particular is heavily studied by researchers using
923:
897:
is a facultative sexual microorganism that can undergo mating when nutrients are limiting. Exposure of
479:
151:
98:
719:
4742:
4654:
4623:
4502:
4207:
4103:
3932:
2664:
2506:
2023:
1964:
1905:
1703:
1214:
1045:
718:
gene products (proteins and RNAs) participate in many cellular processes common across all life. The
4401:
4055:"Mitochondria as protean organelles: membrane processes that influence mitochondrial shape in yeast"
3593:"Schizosaccharomyces pombe grows exponentially during the division cycle with no rate change points"
1692:"The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils?"
1005:
4778:
4530:
4509:
2547:"Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p"
462:
319:
These cells maintain their shape by growing exclusively through the cell tips and divide by medial
128:
4006:"Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates"
1416:
Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, et al. (January 2012).
4373:
4231:
3871:
3799:
3705:
3661:
3524:
2142:
2047:
1996:
1937:
1518:
1238:
722:
provides a categorical high level overview of the biological role of all S. pombe gene products.
571:
441:
176:
51:
2996:
McDowall MD, Harris MA, Lock A, Rutherford K, Staines DM, Bähler J, et al. (January 2015).
2747:"An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System"
4693:
1096:, and all five RecA homologs appear to be required for normal levels of meiotic recombination.
1073:
by observing fission yeast's chromosome dynamics and protein expression levels and regulation.
308:, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding
4750:
4610:
4490:
4361:
4351:
4272:
4223:
4180:
4131:
4072:
4035:
3960:
3901:
3860:"Cell-cycle regulated transcription associates with DNA replication timing in yeast and human"
3840:
3830:
3791:
3750:
3697:
3653:
3617:
3573:
3516:
3475:
3426:
3382:
3333:
3275:
3219:
3186:
Núñez MAB, Sabbarini IM, Eickbush MT, Liang Y, Lange JJ, Kent AM, Zanders SE (February 2020).
3168:
3124:
3095:"Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives"
3068:
3027:
2978:
2926:
2877:
2825:
2776:
2727:
2692:
2630:
2576:
2524:
2428:
2374:
2322:
2295:
2246:
2194:
2134:
2090:
2082:
2039:
1988:
1980:
1929:
1921:
1878:
1829:
1819:
1797:
1780:
1731:
1672:
1623:
1553:
1510:
1447:
1393:
1344:
1309:
1230:
931:
858:
A spatial gradient is thought to coordinate cell size and mitotic entry in fission yeast. The
702:
of 125 bp, and sequence-defined replication origins of about the same size. On the converse,
351:
285:
1364:"A Brief History of Schizosaccharomyces pombe Research: A Perspective Over the Past 70 Years"
4755:
4546:
4343:
4262:
4215:
4170:
4162:
4121:
4111:
4062:
4025:
4017:
3950:
3940:
3891:
3881:
3822:
3781:
3740:
3732:
3689:
3645:
3607:
3563:
3555:
3506:
3465:
3457:
3416:
3374:
3364:
3323:
3315:
3265:
3257:
3209:
3199:
3158:
3114:
3106:
3058:
3017:
3009:
2968:
2960:
2916:
2908:
2867:
2859:
2815:
2807:
2766:
2758:
2719:
2682:
2672:
2620:
2612:
2566:
2558:
2514:
2418:
2410:
2364:
2356:
2285:
2277:
2236:
2228:
2184:
2176:
2124:
2074:
2031:
1972:
1913:
1868:
1860:
1847:
Coelho M, Dereli A, Haese A, Kühn S, Malinovska L, DeSantis ME, et al. (October 2013).
1804:. "Epigenetics in Saccharomyces cerevisiae." Epigenetics. 1. Cold Spring Harbor Press, 2007.
1770:
1762:
1721:
1711:
1662:
1654:
1613:
1603:
1545:
1502:
1437:
1429:
1383:
1375:
1336:
1301:
1222:
902:
383:
343:
2493:
Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, et al. (February 2002).
2217:"Alternative induction of meiotic recombination from single-base lesions of DNA deaminases"
2078:
4788:
4324:
1801:
421:
313:
3353:"Specification of DNA replication origins and genomic base composition in fission yeasts"
1292:
Mitchison JM (October 1957). "The growth of single cells. I. Schizosaccharomyces pombe".
4303:
4211:
4107:
3936:
2668:
2510:
2215:
Pauklin S, Burkert JS, Martin J, Osman F, Weller S, Boulton SJ, et al. (May 2009).
2027:
1968:
1909:
1707:
1218:
761:
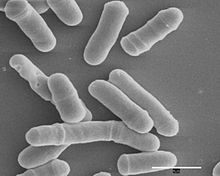
Calcofluor white confocal fluorescence image series demonstrating fission yeast division
660:
Both species share genes with higher eukaryotes that they do not share with each other.
653:
for an extended period (as a consequence, G1-S transition is tightly controlled), while
4649:
4541:
4438:
4175:
4150:
4126:
4091:
4030:
4005:
3955:
3920:
3896:
3859:
3568:
3543:
3470:
3445:
3328:
3295:
3270:
3245:
3214:
3187:
3119:
3094:
3022:
2997:
2973:
2948:
2921:
2896:
2872:
2847:
2846:
Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H (January 2012).
2820:
2795:
2771:
2746:
2687:
2652:
2625:
2600:
2571:
2546:
2423:
2398:
2369:
2344:
2290:
2265:
2241:
2216:
1873:
1848:
1775:
1750:
1667:
1642:
1442:
1417:
1388:
1363:
757:
320:
297:
281:
3826:
3421:
3404:
2723:
2189:
2164:
1726:
1691:
1618:
1591:
1549:
4772:
4267:
4250:
3163:
3146:
1305:
1242:
1041:
4406:
3803:
3709:
3665:
3528:
3446:"Overexpression limits of fission yeast cell-cycle regulators in vivo and in silico"
2949:"Dicentric chromosomes: unique models to study centromere function and inactivation"
2848:"Rif1 is a global regulator of timing of replication origin firing in fission yeast"
2494:
2180:
2146:
1749:
Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE (August 2010).
1608:
1536:
Teoh AL, Heard G, Cox J (September 2004). "Yeast ecology of Kombucha fermentation".
1522:
4615:
4453:
4235:
3145:
Rinaldi T, Dallabona C, Ferrero I, Frontali L, Bolotin-Fukuhara M (December 2010).
2616:
2051:
2000:
1941:
766:
conditions improve, these spores germinate to produce proliferating haploid cells.
361:
331:
289:
167:
499:
4347:
4116:
3945:
3204:
2677:
2651:
Tay Z, Eng RJ, Sajiki K, Lim KK, Tang MY, Yanagida M, Chen ES (24 January 2013).
1849:"Fission yeast does not age under favorable conditions, but does so after stress"
323:
to produce two daughter cells of equal size, which makes them a powerful tool in
4719:
4675:
4595:
4473:
2811:
2762:
2545:
Das M, Wiley DJ, Medina S, Vincent HA, Larrea M, Oriolo A, Verde F (June 2007).
2281:
2232:
1379:
699:
685:
347:
346:
word for beer. It was first developed as an experimental model in the 1950s: by
335:
4586:
3886:
2165:"Sexual reproduction as a response to H2O2 damage in Schizosaccharomyces pombe"
1696:
Proceedings of the National Academy of Sciences of the United States of America
4166:
4021:
3736:
3559:
3511:
3494:
3369:
3352:
2964:
2912:
1864:
1144:
906:
734:
650:
600:
460:
is the major fermenter or a contaminant in such brews. The natural ecology of
445:
417:
357:
324:
301:
88:
2796:"A mammalian-like DNA damage response of fission yeast to nucleoside analogs"
2086:
1984:
1925:
1088:
of DNA double-strand breaks. Five RecA-like proteins have been described in
17:
4636:
4536:
4067:
4054:
3612:
2562:
2130:
10.1002/(SICI)1097-0061(199812)14:16<1529::AID-YEA357>3.0.CO;2-0
1833:
1716:
1658:
632:
416:
has also become an important organism in studying the cellular responses to
391:
293:
68:
4365:
4276:
4227:
4184:
4135:
4076:
4039:
3964:
3905:
3844:
3795:
3754:
3701:
3657:
3621:
3577:
3520:
3479:
3430:
3386:
3337:
3279:
3223:
3172:
3128:
3072:
3031:
2982:
2930:
2881:
2863:
2829:
2780:
2731:
2696:
2634:
2580:
2528:
2432:
2378:
2326:
2299:
2250:
2094:
2043:
1992:
1933:
1882:
1784:
1735:
1676:
1627:
1557:
1514:
1479:
1465:
1451:
1397:
1340:
1327:
Mitchison JM (April 1990). "The fission yeast, Schizosaccharomyces pombe".
1313:
1234:
990:
490:, The Netherlands. It was deposited there by A. Osterwalder under the name
43:
3693:
3319:
3013:
2198:
2138:
1766:
1348:
986:
4701:
4580:
4442:
3770:"Mid1/anillin and the spatial regulation of cytokinesis in fission yeast"
1433:
636:
453:
365:
4219:
3461:
3378:
2014:
Sawin KE (June 2009). "Cell cycle: Cell division brought down to size".
1976:
1917:
1226:
930:
indicated that faulty processing of DNA replication intermediates, i.e.
4680:
4667:
3745:
3649:
1690:
Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H (October 2004).
1256:
Leupold U (1950). "Die Vererbung von Homothallie und Heterothallie bei
1154:
1093:
982:
964:
622:
614:
395:
273:
3405:"Only connect: linking meiotic DNA replication to chromosome dynamics"
3304:
Shows Limited Phenotypic Variation but Extensive Karyotypic Diversity"
2519:
1174:
837:
Cell-cycle length of the fission yeast depends on nutrient conditions.
4706:
3786:
3769:
3444:
Moriya H, Chino A, Kapuy O, Csikász-Nagy A, Novák B (December 2011).
3110:
3063:
3046:
2897:"Neocentromeres and epigenetically inherited features of centromeres"
2360:
1033:
910:
851:
586:
437:
433:
387:
379:
305:
78:
4557:
3261:
2460:
2035:
1506:
4641:
3246:"The genomic and phenotypic diversity of Schizosaccharomyces pombe"
2414:
2264:
Farah JA, Cromie G, Davis L, Steiner WW, Smith GR (December 2005).
841:
In fission yeast, where growth governs progression through G2/M, a
4496:
3876:
2338:
2336:
1149:
756:
487:
309:
277:
4149:
Raychaudhuri S, Young BP, Espenshade PJ, Loewen C (August 2012).
2399:"Featured organism: Schizosaccharomyces pombe, the fission yeast"
4342:. Methods in Molecular Biology. Vol. 521. pp. 463–82.
1081:
873:
868:
864:
859:
842:
665:
4628:
4561:
4410:
2794:
Sabatinos SA, Mastro TL, Green MD, Forsburg SL (January 2013).
1052:. Defects in these repair processes lead to reduced survival.
918:
stimulate meiotic recombination. This recombination requires
1418:"PomBase: a comprehensive online resource for fission yeast"
832:
815:
668:
genes like those in vertebrates, while this is missing from
4396:
3821:. Methods in Cell Biology. Vol. 97. pp. 203–21.
2599:
Kawashima SA, Takemoto A, Nurse P, Kapoor TM (July 2012).
1126:
simpler model system to see how this phenomenon operates.
3239:
3237:
3235:
3233:
3147:"Mitochondrial diseases and the role of the yeast models"
2594:
2592:
2590:
867:
gradient reaches most of the cortical nodes (blue dots).
3493:
Das M, Wiley DJ, Chen X, Shah K, Verde F (August 2009).
676:
also has greatly simplified heterochromatin compared to
1044:. Most such damages can ordinarily be repaired by DNA
4151:"Regulation of lipid metabolism: a tale of two yeasts"
2998:"PomBase 2015: updates to the fission yeast database"
2392:
2390:
2388:
386:, becoming the sixth model eukaryotic organism whose
3351:
Mojardín L, Vázquez E, Antequera F (November 2013).
855:
per unit time is smaller than that of normal cells.
4570:
3088:
3086:
3084:
3082:
2942:
2940:
1751:"Evolution of CST function in telomere maintenance"
304:in diameter and 7 to 14 micrometres in length. Its
2947:Stimpson KM, Matheny JE, Sullivan BA (July 2012).
2495:"The genome sequence of Schizosaccharomyces pombe"
382:was published in 2002, by a consortium led by the
4090:Chino A, Makanae K, Moriya H (3 September 2013).
2841:
2839:
2710:Forsburg SL (September 1999). "The best yeast?".
4288:
4286:
3398:
3396:
2488:
2486:
2484:
2482:
2480:
2478:
300:are rod-shaped. Cells typically measure 3 to 4
3140:
3138:
2745:Hoffman CS, Wood V, Fantes PA (October 2015).
2158:
2156:
2106:
2104:
1411:
1409:
1407:
985:reports over 69% of protein coding genes have
452:is also known to be present in fermented tea,
4422:
4402:MicrobeWiki page on Schizosaccharomyces pombe
2646:
2644:
1032:stationary phase cells undergo chronological
790:The particular cell cycle of a fission yeast.
8:
4378:: CS1 maint: multiple names: authors list (
3544:"Cellular aging: symmetry evades senescence"
578:has approximately 5,070 open reading frames.
394:. S. pombe researchers are supported by the
2540:
2538:
2450:
2448:
2446:
2444:
2442:
901:to hydrogen peroxide, an agent that causes
4558:
4429:
4415:
4407:
4004:Zhao J, Lendahl U, Nistér M (March 2013).
3093:Lee IJ, Coffman VC, Wu JQ (October 2012).
2210:
2208:
1538:International Journal of Food Microbiology
42:
31:
4266:
4174:
4125:
4115:
4066:
4029:
3954:
3944:
3895:
3885:
3875:
3785:
3744:
3611:
3567:
3510:
3469:
3420:
3368:
3327:
3269:
3244:Jeffares, Daniel C.; et al. (2015).
3213:
3203:
3162:
3118:
3062:
3021:
2972:
2920:
2871:
2819:
2770:
2686:
2676:
2624:
2570:
2518:
2422:
2368:
2289:
2240:
2188:
2128:
1872:
1774:
1725:
1715:
1666:
1617:
1607:
1590:Gómez EB, Bailis JM, Forsburg SL (2002).
1441:
1387:
806:in bright and dark field light microscopy
3296:"A Geographically Diverse Collection of
2315:Gravitational and Space Biology Bulletin
1184:Global Biodiversity Information Facility
1084:and RecA-like proteins are required for
1008:and drive suppressors called wtf genes.
729:
711:S. pombe pathways and cellular processes
1592:"Fission yeast enters a joyful new era"
1166:
768:
4371:
4320:
4309:
3768:Rincon SA, Paoletti A (October 2012).
2343:Forsburg SL, Rhind N (February 2006).
2079:10.1146/annurev.genet.39.073103.094316
330:Fission yeast was isolated in 1893 by
3919:Li PC, Green MD, Forsburg SL (2013).
971:more functional research approaches.
926:in S. pombe. Other experiments with
456:. It is not clear at present whether
427:Approximately 160 natural strains of
370:Nobel Prize in Physiology or Medicine
280:used in traditional brewing and as a
7:
4743:3adedc6f-6aa6-47b6-b49d-df9a4e503285
4010:Cellular and Molecular Life Sciences
3983:The Official Site of the Nobel Prize
1816:The Cell Cycle Principles of Control
4397:PomBase - The Pombe Genome Database
2403:Comparative and Functional Genomics
2163:Bernstein C, Johns V (April 1989).
1362:Fantes PA, Hoffman CS (June 2016).
778:General features of the cell cycle.
372:for work on cell cycle regulation.
3051:Cell Motility and the Cytoskeleton
2895:Burrack LS, Berman J (July 2012).
1262:C R Trav Lab Carlsberg Ser Physiol
202:Schizosaccharomyces acidodevoratus
25:
4059:General Physiology and Biophysics
2345:"Basic methods for fission yeast"
1550:10.1016/j.ijfoodmicro.2003.12.020
1278:Leupold U. (1993) The origins of
820:Cytokinesis of the fission yeast.
532:in the presence of excess sugar.
4268:10.1111/j.1600-0854.2011.01316.x
3164:10.1111/j.1567-1364.2010.00685.x
1140:DNA damage (naturally occurring)
795:
783:
771:
210:Schizosaccharomyces liquefaciens
55:
4249:Suda Y, Nakano A (April 2012).
4155:Current Opinion in Cell Biology
3979:"Sir Paul Nurse - Biographical"
3045:Kadura S, Sazer S (July 2005).
2181:10.1128/jb.171.4.1893-1897.1989
1647:Molecular Biology and Evolution
1609:10.1186/gb-2002-3-6-reports4017
544:Comparison with budding yeast (
2617:10.1016/j.chembiol.2012.06.008
581:Despite similar gene numbers,
350:for studying genetics, and by
1:
3827:10.1016/S0091-679X(10)97012-X
3422:10.1016/S1097-2765(02)00508-7
2724:10.1016/s0168-9525(99)01798-9
2551:Molecular Biology of the Cell
1818:. London: New Science Press.
991:associated with human disease
354:for studying the cell cycle.
4348:10.1007/978-1-60327-815-7_26
4304:"Trans-NIH.pombe Initiative"
4117:10.1371/journal.pone.0073319
3946:10.1371/journal.pone.0061464
3357:Journal of Molecular Biology
3308:G3: Genes, Genomes, Genetics
3205:10.1371/journal.pgen.1008350
2678:10.1371/journal.pone.0055041
1306:10.1016/0014-4827(57)90005-8
466:yeasts is not well-studied.
218:Schizosaccharomyces mellacei
4251:"The yeast Golgi apparatus"
3725:Fungal Genetics and Biology
3542:Moseley JB (October 2013).
3008:(Database issue): D656-61.
2812:10.1534/genetics.112.145730
2763:10.1534/genetics.115.181503
2282:10.1534/genetics.105.046821
2233:10.1534/genetics.109.101683
2113:"Fusion of a fission yeast"
1641:Lin Z, Li WH (April 2011).
1380:10.1534/genetics.116.189407
4815:
4061:. 30 Spec No (5): S13-24.
3887:10.1186/gb-2013-14-10-r111
3638:Cell Biology International
3403:Forsburg SL (April 2002).
1428:(Database issue): D695-9.
1294:Experimental Cell Research
1050:nucleotide excision repair
989:and over 500 of these are
649:is in the G1 phase of the
4602:Schizosaccharomyces pombe
4572:Schizosaccharomyces pombe
4449:
4167:10.1016/j.ceb.2012.05.006
4022:10.1007/s00018-012-1066-6
3737:10.1016/j.fgb.2012.01.003
3560:10.1016/j.cub.2013.08.013
3512:10.1016/j.cub.2009.06.057
3450:Molecular Systems Biology
3370:10.1016/j.jmb.2013.09.023
2965:10.1007/s10577-012-9302-3
2913:10.1007/s10577-012-9296-x
2111:Davey J (December 1998).
2067:Annual Review of Genetics
1865:10.1016/j.cub.2013.07.084
1814:Morgan, David O. (2007).
1280:Schizosaccharomyces pombe
1258:Schizosaccharomyces pombe
1177:Schizosaccharomyces pombe
952:Schizosaccharomyces pombe
895:Schizosaccharomyces pombe
554:Schizosaccharomyces pombe
475:Schizosaccharomyces pombe
414:Schizosaccharomyces pombe
408:green fluorescent protein
265:Schizosaccharomyces pombe
247:Schizosaccharomyces pombe
235:Schizosaccharomyces pombe
226:Schizosaccharomyces pombe
182:
175:
161:Schizosaccharomyces pombe
157:
150:
52:Scientific classification
50:
41:
36:Schizosaccharomyces pombe
34:
570:has approximately 5,600
559:Saccharomyces cerevisiae
546:Saccharomyces cerevisiae
119:Schizosaccharomycetaceae
4794:Fungi described in 1893
4068:10.4149/gpb_2011_SI1_13
3613:10.1111/1567-1364.12072
2852:Genes & Development
2605:Chemistry & Biology
2563:10.1091/mbc.E06-09-0883
2169:Journal of Bacteriology
1717:10.1073/pnas.0403984101
1117:Experimental approaches
1038:reactive oxygen species
890:Responses to DNA damage
400:model organism database
109:Schizosaccharomycetales
4319:Cite journal requires
3002:Nucleic Acids Research
2864:10.1101/gad.178491.111
1422:Nucleic Acids Research
1341:10.1002/bies.950120409
1086:recombinational repair
920:uracil-DNA glycosylase
838:
821:
762:
742:
440:, or from the various
292:. It is a unicellular
186:Saccharomyces mellacei
3819:Microtubules: In vivo
3694:10.1111/boc.201200042
3320:10.1534/g3.111.001123
1767:10.4161/cc.9.16.12547
1659:10.1093/molbev/msq324
1094:meiotic recombination
1077:Meiotic recombination
1036:due to production of
905:leading to oxidative
881:Mating-type switching
836:
819:
760:
733:
720:fission yeast GO slim
480:Schizosaccharomycetes
368:, Nurse won the 2001
99:Schizosaccharomycetes
4053:Abelovska L (2011).
1495:Nature Biotechnology
1046:base excision repair
530:aerobic fermentation
444:, such as Brazilian
410:as a molecular tag.
406:was published using
375:The sequence of the
254:Sakag. & Y.Otani
242:Sakag. & Y.Otani
4510:Medicago truncatula
4220:10.1038/nature11354
4212:2012Natur.489..585B
4108:2013PLoSO...873319C
3937:2013PLoSO...861464L
3682:Biology of the Cell
3462:10.1038/msb.2011.91
3298:Schizosaccharomyces
3151:FEMS Yeast Research
3014:10.1093/nar/gku1040
2953:Chromosome Research
2901:Chromosome Research
2669:2013PLoSO...855041T
2511:2002Natur.415..871W
2028:2009Natur.459..782S
1977:10.1038/nature08054
1969:2009Natur.459..852M
1918:10.1038/nature08074
1910:2009Natur.459..857M
1708:2004PNAS..10115386D
1227:10.1038/nature07002
1219:2008Natur.453.1239W
1092:that are linked to
1021:Cell cycle analysis
909:, strongly induces
804:Schizosaccharomyces
802:Division stages of
684:has well-developed
585:has only about 250
572:open reading frames
521:Schizosaccharomyces
463:Schizosaccharomyces
442:alcoholic beverages
420:and the process of
338:. The species name
194:Saccharomyces pombe
130:Schizosaccharomyces
3858:Fraser HB (2013).
3650:10.1002/cbin.10722
2712:Trends in Genetics
1798:Grunstein, Michael
1602:(6): REPORTS4017.
1434:10.1093/nar/gkr853
839:
822:
763:
743:
552:The yeast species
514:The fission yeast
334:from East African
4766:
4765:
4751:Open Tree of Life
4564:Taxon identifiers
4555:
4554:
4357:978-1-60327-814-0
3591:Cooper S (2013).
2520:10.1038/nature724
1963:(7248): 852–856.
1904:(7248): 857–860.
1825:978-0-19-920610-0
1213:(7199): 1239–43.
1105:recently created
1012:Genetic diversity
938:As a model system
932:Okazaki fragments
593:has nearly 5,000.
352:Murdoch Mitchison
312:and at least 450
261:
260:
255:
243:
222:
221:(A.Jörg.) Lindner
214:
206:
198:
197:(Lindner) A.Jörg.
190:
16:(Redirected from
4806:
4759:
4758:
4746:
4745:
4736:
4735:
4723:
4722:
4720:NBNSYS0000016749
4710:
4709:
4697:
4696:
4684:
4683:
4671:
4670:
4658:
4657:
4645:
4644:
4632:
4631:
4619:
4618:
4606:
4605:
4604:
4591:
4590:
4589:
4559:
4431:
4424:
4417:
4408:
4384:
4383:
4377:
4369:
4335:
4329:
4328:
4322:
4317:
4315:
4307:
4300:
4294:
4290:
4281:
4280:
4270:
4246:
4240:
4239:
4195:
4189:
4188:
4178:
4146:
4140:
4139:
4129:
4119:
4087:
4081:
4080:
4070:
4050:
4044:
4043:
4033:
4001:
3995:
3994:
3992:
3990:
3975:
3969:
3968:
3958:
3948:
3916:
3910:
3909:
3899:
3889:
3879:
3855:
3849:
3848:
3814:
3808:
3807:
3789:
3787:10.1002/cm.21056
3765:
3759:
3758:
3748:
3720:
3714:
3713:
3676:
3670:
3669:
3632:
3626:
3625:
3615:
3597:
3588:
3582:
3581:
3571:
3539:
3533:
3532:
3514:
3490:
3484:
3483:
3473:
3441:
3435:
3434:
3424:
3400:
3391:
3390:
3372:
3348:
3342:
3341:
3331:
3290:
3284:
3283:
3273:
3241:
3228:
3227:
3217:
3207:
3183:
3177:
3176:
3166:
3142:
3133:
3132:
3122:
3111:10.1002/cm.21052
3090:
3077:
3076:
3066:
3064:10.1002/cm.20072
3042:
3036:
3035:
3025:
2993:
2987:
2986:
2976:
2944:
2935:
2934:
2924:
2892:
2886:
2885:
2875:
2843:
2834:
2833:
2823:
2791:
2785:
2784:
2774:
2742:
2736:
2735:
2707:
2701:
2700:
2690:
2680:
2648:
2639:
2638:
2628:
2596:
2585:
2584:
2574:
2542:
2533:
2532:
2522:
2505:(6874): 871–80.
2490:
2473:
2472:
2470:
2468:
2459:. Archived from
2452:
2437:
2436:
2426:
2397:Wixon J (2002).
2394:
2383:
2382:
2372:
2361:10.1002/yea.1347
2340:
2331:
2330:
2310:
2304:
2303:
2293:
2261:
2255:
2254:
2244:
2212:
2203:
2202:
2192:
2160:
2151:
2150:
2132:
2108:
2099:
2098:
2062:
2056:
2055:
2011:
2005:
2004:
1952:
1946:
1945:
1893:
1887:
1886:
1876:
1844:
1838:
1837:
1811:
1805:
1795:
1789:
1788:
1778:
1746:
1740:
1739:
1729:
1719:
1702:(43): 15386–91.
1687:
1681:
1680:
1670:
1638:
1632:
1631:
1621:
1611:
1587:
1581:
1580:
1568:
1562:
1561:
1533:
1527:
1526:
1490:
1484:
1483:
1476:
1470:
1469:
1462:
1456:
1455:
1445:
1413:
1402:
1401:
1391:
1359:
1353:
1352:
1324:
1318:
1317:
1289:
1283:
1276:
1270:
1269:
1253:
1247:
1246:
1202:
1196:
1195:
1193:
1191:
1171:
977:
961:
903:oxidative stress
799:
787:
775:
698:has small point
384:Sanger Institute
253:
241:
220:
212:
204:
196:
188:
163:
60:
59:
46:
32:
27:Species of yeast
21:
4814:
4813:
4809:
4808:
4807:
4805:
4804:
4803:
4769:
4768:
4767:
4762:
4754:
4749:
4741:
4739:
4731:
4726:
4718:
4713:
4705:
4700:
4692:
4687:
4679:
4674:
4666:
4661:
4653:
4648:
4640:
4635:
4627:
4622:
4614:
4609:
4600:
4599:
4594:
4585:
4584:
4579:
4566:
4556:
4551:
4445:
4439:model organisms
4435:
4393:
4388:
4387:
4370:
4358:
4340:DNA Replication
4337:
4336:
4332:
4318:
4308:
4302:
4301:
4297:
4291:
4284:
4248:
4247:
4243:
4206:(7417): 585–9.
4197:
4196:
4192:
4148:
4147:
4143:
4089:
4088:
4084:
4052:
4051:
4047:
4003:
4002:
3998:
3988:
3986:
3977:
3976:
3972:
3918:
3917:
3913:
3857:
3856:
3852:
3837:
3816:
3815:
3811:
3767:
3766:
3762:
3722:
3721:
3717:
3678:
3677:
3673:
3634:
3633:
3629:
3595:
3590:
3589:
3585:
3548:Current Biology
3541:
3540:
3536:
3499:Current Biology
3492:
3491:
3487:
3443:
3442:
3438:
3402:
3401:
3394:
3363:(23): 4706–13.
3350:
3349:
3345:
3292:
3291:
3287:
3262:10.1038/ng.3215
3250:Nature Genetics
3243:
3242:
3231:
3198:(2): e1008350.
3185:
3184:
3180:
3144:
3143:
3136:
3092:
3091:
3080:
3044:
3043:
3039:
2995:
2994:
2990:
2946:
2945:
2938:
2894:
2893:
2889:
2845:
2844:
2837:
2793:
2792:
2788:
2744:
2743:
2739:
2709:
2708:
2704:
2650:
2649:
2642:
2598:
2597:
2588:
2557:(6): 2090–101.
2544:
2543:
2536:
2492:
2491:
2476:
2466:
2464:
2454:
2453:
2440:
2396:
2395:
2386:
2342:
2341:
2334:
2312:
2311:
2307:
2276:(4): 1499–511.
2263:
2262:
2258:
2214:
2213:
2206:
2162:
2161:
2154:
2123:(16): 1529–66.
2110:
2109:
2102:
2064:
2063:
2059:
2036:10.1038/459782a
2022:(7248): 782–3.
2013:
2012:
2008:
1954:
1953:
1949:
1895:
1894:
1890:
1859:(19): 1844–52.
1853:Current Biology
1846:
1845:
1841:
1826:
1813:
1812:
1808:
1796:
1792:
1761:(16): 3157–65.
1748:
1747:
1743:
1689:
1688:
1684:
1640:
1639:
1635:
1589:
1588:
1584:
1570:
1569:
1565:
1535:
1534:
1530:
1507:10.1038/nbt1222
1492:
1491:
1487:
1478:
1477:
1473:
1464:
1463:
1459:
1415:
1414:
1405:
1361:
1360:
1356:
1326:
1325:
1321:
1291:
1290:
1286:
1277:
1273:
1255:
1254:
1250:
1204:
1203:
1199:
1189:
1187:
1173:
1172:
1168:
1163:
1136:
1119:
1102:
1100:Biomedical tool
1079:
1023:
1014:
1006:meiotic drivers
987:human orthologs
975:
959:
949:
940:
892:
883:
831:
814:
807:
800:
791:
788:
779:
776:
728:
713:
550:
512:
472:
422:DNA replication
390:has been fully
314:non-coding RNAs
268:, also called "
171:
165:
159:
146:
54:
28:
23:
22:
15:
12:
11:
5:
4812:
4810:
4802:
4801:
4799:Fungus species
4796:
4791:
4786:
4781:
4771:
4770:
4764:
4763:
4761:
4760:
4747:
4737:
4724:
4711:
4698:
4685:
4672:
4659:
4646:
4633:
4620:
4607:
4592:
4576:
4574:
4568:
4567:
4562:
4553:
4552:
4550:
4549:
4544:
4539:
4534:
4527:
4520:
4513:
4506:
4499:
4494:
4487:
4482:
4477:
4470:
4463:
4456:
4450:
4447:
4446:
4436:
4434:
4433:
4426:
4419:
4411:
4405:
4404:
4399:
4392:
4391:External links
4389:
4386:
4385:
4356:
4330:
4321:|journal=
4295:
4282:
4241:
4190:
4141:
4082:
4045:
3996:
3970:
3911:
3864:Genome Biology
3850:
3835:
3809:
3780:(10): 764–77.
3760:
3715:
3671:
3644:(3): 276–286.
3627:
3600:FEMS Yeast Res
3583:
3554:(19): R871-3.
3534:
3505:(15): 1314–9.
3485:
3436:
3409:Molecular Cell
3392:
3343:
3314:(7): 615–626.
3285:
3256:(3): 235–241.
3229:
3178:
3157:(8): 1006–22.
3134:
3105:(10): 751–63.
3078:
3037:
2988:
2959:(5): 595–605.
2936:
2887:
2835:
2786:
2737:
2702:
2640:
2611:(7): 893–901.
2586:
2534:
2474:
2463:on 6 June 2015
2438:
2415:10.1002/cfg.92
2409:(2): 194–204.
2384:
2332:
2305:
2256:
2204:
2152:
2100:
2073:(1): 213–236.
2057:
2006:
1947:
1888:
1839:
1824:
1806:
1790:
1741:
1682:
1653:(4): 1407–13.
1633:
1596:Genome Biology
1582:
1563:
1528:
1485:
1471:
1457:
1403:
1354:
1319:
1284:
1271:
1248:
1197:
1165:
1164:
1162:
1159:
1158:
1157:
1152:
1147:
1142:
1135:
1132:
1118:
1115:
1101:
1098:
1078:
1075:
1022:
1019:
1013:
1010:
1000:The genome of
948:
945:
939:
936:
891:
888:
882:
879:
850:cells undergo
830:
827:
813:
810:
809:
808:
801:
794:
792:
789:
782:
780:
777:
770:
727:
724:
712:
709:
708:
707:
693:
680:. Conversely,
666:RNAi machinery
658:
644:
641:S. cerevisiae
639:complex while
626:
608:
594:
579:
549:
542:
511:
508:
471:
468:
282:model organism
259:
258:
257:
256:
251:ogasawaraensis
244:
232:
230:acidodevoratus
223:
215:
207:
199:
191:
180:
179:
173:
172:
166:
155:
154:
148:
147:
140:
138:
134:
133:
126:
122:
121:
116:
112:
111:
106:
102:
101:
96:
92:
91:
86:
82:
81:
76:
72:
71:
66:
62:
61:
48:
47:
39:
38:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4811:
4800:
4797:
4795:
4792:
4790:
4787:
4785:
4784:Fungal models
4782:
4780:
4777:
4776:
4774:
4757:
4752:
4748:
4744:
4738:
4734:
4729:
4725:
4721:
4716:
4712:
4708:
4703:
4699:
4695:
4690:
4686:
4682:
4677:
4673:
4669:
4664:
4660:
4656:
4651:
4647:
4643:
4638:
4634:
4630:
4625:
4621:
4617:
4612:
4608:
4603:
4597:
4593:
4588:
4582:
4578:
4577:
4575:
4573:
4569:
4565:
4560:
4548:
4545:
4543:
4540:
4538:
4535:
4533:
4532:
4528:
4526:
4525:
4521:
4519:
4518:
4514:
4512:
4511:
4507:
4505:
4504:
4500:
4498:
4495:
4493:
4492:
4488:
4486:
4485:Fission yeast
4483:
4481:
4480:Budding yeast
4478:
4476:
4475:
4471:
4469:
4468:
4467:Chlamydomonas
4464:
4462:
4461:
4457:
4455:
4452:
4451:
4448:
4444:
4440:
4432:
4427:
4425:
4420:
4418:
4413:
4412:
4409:
4403:
4400:
4398:
4395:
4394:
4390:
4381:
4375:
4367:
4363:
4359:
4353:
4349:
4345:
4341:
4334:
4331:
4326:
4313:
4305:
4299:
4296:
4289:
4287:
4283:
4278:
4274:
4269:
4264:
4261:(4): 505–10.
4260:
4256:
4252:
4245:
4242:
4237:
4233:
4229:
4225:
4221:
4217:
4213:
4209:
4205:
4201:
4194:
4191:
4186:
4182:
4177:
4172:
4168:
4164:
4160:
4156:
4152:
4145:
4142:
4137:
4133:
4128:
4123:
4118:
4113:
4109:
4105:
4102:(9): e73319.
4101:
4097:
4093:
4086:
4083:
4078:
4074:
4069:
4064:
4060:
4056:
4049:
4046:
4041:
4037:
4032:
4027:
4023:
4019:
4016:(6): 951–76.
4015:
4011:
4007:
4000:
3997:
3984:
3980:
3974:
3971:
3966:
3962:
3957:
3952:
3947:
3942:
3938:
3934:
3931:(5): e61464.
3930:
3926:
3922:
3915:
3912:
3907:
3903:
3898:
3893:
3888:
3883:
3878:
3873:
3869:
3865:
3861:
3854:
3851:
3846:
3842:
3838:
3836:9780123813497
3832:
3828:
3824:
3820:
3813:
3810:
3805:
3801:
3797:
3793:
3788:
3783:
3779:
3775:
3771:
3764:
3761:
3756:
3752:
3747:
3742:
3738:
3734:
3731:(3): 235–49.
3730:
3726:
3719:
3716:
3711:
3707:
3703:
3699:
3695:
3691:
3688:(3): 129–48.
3687:
3683:
3675:
3672:
3667:
3663:
3659:
3655:
3651:
3647:
3643:
3639:
3631:
3628:
3623:
3619:
3614:
3609:
3605:
3601:
3594:
3587:
3584:
3579:
3575:
3570:
3565:
3561:
3557:
3553:
3549:
3545:
3538:
3535:
3530:
3526:
3522:
3518:
3513:
3508:
3504:
3500:
3496:
3489:
3486:
3481:
3477:
3472:
3467:
3463:
3459:
3455:
3451:
3447:
3440:
3437:
3432:
3428:
3423:
3418:
3415:(4): 703–11.
3414:
3410:
3406:
3399:
3397:
3393:
3388:
3384:
3380:
3376:
3371:
3366:
3362:
3358:
3354:
3347:
3344:
3339:
3335:
3330:
3325:
3321:
3317:
3313:
3309:
3305:
3303:
3299:
3289:
3286:
3281:
3277:
3272:
3267:
3263:
3259:
3255:
3251:
3247:
3240:
3238:
3236:
3234:
3230:
3225:
3221:
3216:
3211:
3206:
3201:
3197:
3193:
3192:PLOS Genetics
3189:
3182:
3179:
3174:
3170:
3165:
3160:
3156:
3152:
3148:
3141:
3139:
3135:
3130:
3126:
3121:
3116:
3112:
3108:
3104:
3100:
3096:
3089:
3087:
3085:
3083:
3079:
3074:
3070:
3065:
3060:
3057:(3): 145–60.
3056:
3052:
3048:
3041:
3038:
3033:
3029:
3024:
3019:
3015:
3011:
3007:
3003:
2999:
2992:
2989:
2984:
2980:
2975:
2970:
2966:
2962:
2958:
2954:
2950:
2943:
2941:
2937:
2932:
2928:
2923:
2918:
2914:
2910:
2907:(5): 607–19.
2906:
2902:
2898:
2891:
2888:
2883:
2879:
2874:
2869:
2865:
2861:
2858:(2): 137–50.
2857:
2853:
2849:
2842:
2840:
2836:
2831:
2827:
2822:
2817:
2813:
2809:
2806:(1): 143–57.
2805:
2801:
2797:
2790:
2787:
2782:
2778:
2773:
2768:
2764:
2760:
2757:(2): 403–23.
2756:
2752:
2748:
2741:
2738:
2733:
2729:
2725:
2721:
2717:
2713:
2706:
2703:
2698:
2694:
2689:
2684:
2679:
2674:
2670:
2666:
2663:(1): e55041.
2662:
2658:
2654:
2647:
2645:
2641:
2636:
2632:
2627:
2622:
2618:
2614:
2610:
2606:
2602:
2595:
2593:
2591:
2587:
2582:
2578:
2573:
2568:
2564:
2560:
2556:
2552:
2548:
2541:
2539:
2535:
2530:
2526:
2521:
2516:
2512:
2508:
2504:
2500:
2496:
2489:
2487:
2485:
2483:
2481:
2479:
2475:
2462:
2458:
2455:Forsburg SL.
2451:
2449:
2447:
2445:
2443:
2439:
2434:
2430:
2425:
2420:
2416:
2412:
2408:
2404:
2400:
2393:
2391:
2389:
2385:
2380:
2376:
2371:
2366:
2362:
2358:
2355:(3): 173–83.
2354:
2350:
2346:
2339:
2337:
2333:
2328:
2324:
2320:
2316:
2309:
2306:
2301:
2297:
2292:
2287:
2283:
2279:
2275:
2271:
2267:
2260:
2257:
2252:
2248:
2243:
2238:
2234:
2230:
2226:
2222:
2218:
2211:
2209:
2205:
2200:
2196:
2191:
2186:
2182:
2178:
2175:(4): 1893–7.
2174:
2170:
2166:
2159:
2157:
2153:
2148:
2144:
2140:
2136:
2131:
2126:
2122:
2118:
2114:
2107:
2105:
2101:
2096:
2092:
2088:
2084:
2080:
2076:
2072:
2068:
2061:
2058:
2053:
2049:
2045:
2041:
2037:
2033:
2029:
2025:
2021:
2017:
2010:
2007:
2002:
1998:
1994:
1990:
1986:
1982:
1978:
1974:
1970:
1966:
1962:
1958:
1951:
1948:
1943:
1939:
1935:
1931:
1927:
1923:
1919:
1915:
1911:
1907:
1903:
1899:
1892:
1889:
1884:
1880:
1875:
1870:
1866:
1862:
1858:
1854:
1850:
1843:
1840:
1835:
1831:
1827:
1821:
1817:
1810:
1807:
1803:
1799:
1794:
1791:
1786:
1782:
1777:
1772:
1768:
1764:
1760:
1756:
1752:
1745:
1742:
1737:
1733:
1728:
1723:
1718:
1713:
1709:
1705:
1701:
1697:
1693:
1686:
1683:
1678:
1674:
1669:
1664:
1660:
1656:
1652:
1648:
1644:
1637:
1634:
1629:
1625:
1620:
1615:
1610:
1605:
1601:
1597:
1593:
1586:
1583:
1578:
1574:
1567:
1564:
1559:
1555:
1551:
1547:
1544:(2): 119–26.
1543:
1539:
1532:
1529:
1524:
1520:
1516:
1512:
1508:
1504:
1500:
1496:
1489:
1486:
1481:
1475:
1472:
1467:
1461:
1458:
1453:
1449:
1444:
1439:
1435:
1431:
1427:
1423:
1419:
1412:
1410:
1408:
1404:
1399:
1395:
1390:
1385:
1381:
1377:
1373:
1369:
1365:
1358:
1355:
1350:
1346:
1342:
1338:
1335:(4): 189–91.
1334:
1330:
1323:
1320:
1315:
1311:
1307:
1303:
1300:(2): 244–62.
1299:
1295:
1288:
1285:
1281:
1275:
1272:
1267:
1263:
1259:
1252:
1249:
1244:
1240:
1236:
1232:
1228:
1224:
1220:
1216:
1212:
1208:
1201:
1198:
1186:
1185:
1180:
1178:
1170:
1167:
1160:
1156:
1153:
1151:
1148:
1146:
1143:
1141:
1138:
1137:
1133:
1131:
1127:
1124:
1116:
1114:
1110:
1108:
1099:
1097:
1095:
1091:
1087:
1083:
1076:
1074:
1070:
1066:
1064:
1059:
1053:
1051:
1047:
1043:
1039:
1035:
1031:
1027:
1020:
1018:
1011:
1009:
1007:
1003:
998:
996:
993:. This makes
992:
988:
984:
980:
974:
968:
966:
958:
953:
946:
944:
937:
935:
933:
929:
925:
924:recombination
921:
917:
912:
908:
904:
900:
896:
889:
887:
880:
878:
875:
870:
866:
861:
856:
853:
849:
844:
835:
828:
826:
818:
811:
805:
798:
793:
786:
781:
774:
769:
767:
759:
755:
751:
748:
740:
736:
732:
725:
723:
721:
717:
710:
705:
701:
697:
696:S. cerevisiae
694:
691:
687:
683:
682:S. cerevisiae
679:
675:
674:S. cerevisiae
671:
670:S. cerevisiae
667:
663:
659:
656:
652:
648:
647:S. cerevisiae
645:
642:
638:
634:
630:
627:
624:
620:
616:
612:
611:S. cerevisiae
609:
606:
602:
598:
597:S. cerevisiae
595:
592:
588:
584:
583:S. cerevisiae
580:
577:
573:
569:
568:S. cerevisiae
566:
565:
564:
561:
560:
555:
547:
543:
541:
539:
538:Saccharomyces
535:
531:
527:
522:
517:
509:
507:
505:
501:
497:
493:
489:
485:
481:
476:
469:
467:
465:
464:
459:
455:
451:
447:
443:
439:
435:
430:
425:
423:
419:
415:
411:
409:
405:
401:
397:
393:
389:
385:
381:
378:
373:
371:
367:
363:
359:
355:
353:
349:
345:
341:
337:
333:
328:
326:
322:
317:
315:
311:
307:
303:
299:
295:
291:
287:
283:
279:
275:
271:
270:fission yeast
267:
266:
252:
248:
245:
240:
236:
233:
231:
227:
224:
219:
216:
211:
208:
203:
200:
195:
192:
187:
184:
183:
181:
178:
174:
169:
164:
162:
156:
153:
152:Binomial name
149:
145:
144:
143:S. pombe
139:
136:
135:
132:
131:
127:
124:
123:
120:
117:
114:
113:
110:
107:
104:
103:
100:
97:
94:
93:
90:
87:
84:
83:
80:
77:
74:
73:
70:
67:
64:
63:
58:
53:
49:
45:
40:
37:
33:
30:
19:
18:Fission yeast
4571:
4529:
4522:
4515:
4508:
4501:
4489:
4484:
4472:
4465:
4458:
4454:Lambda phage
4339:
4333:
4312:cite journal
4298:
4258:
4254:
4244:
4203:
4199:
4193:
4161:(4): 502–8.
4158:
4154:
4144:
4099:
4095:
4085:
4058:
4048:
4013:
4009:
3999:
3987:. Retrieved
3982:
3973:
3928:
3924:
3914:
3870:(10): R111.
3867:
3863:
3853:
3818:
3812:
3777:
3774:Cytoskeleton
3773:
3763:
3728:
3724:
3718:
3685:
3681:
3674:
3641:
3637:
3630:
3606:(7): 650–8.
3603:
3599:
3586:
3551:
3547:
3537:
3502:
3498:
3488:
3453:
3449:
3439:
3412:
3408:
3379:10261/104754
3360:
3356:
3346:
3311:
3307:
3301:
3297:
3288:
3253:
3249:
3195:
3191:
3181:
3154:
3150:
3102:
3099:Cytoskeleton
3098:
3054:
3050:
3040:
3005:
3001:
2991:
2956:
2952:
2904:
2900:
2890:
2855:
2851:
2803:
2799:
2789:
2754:
2750:
2740:
2718:(9): 340–4.
2715:
2711:
2705:
2660:
2656:
2608:
2604:
2554:
2550:
2502:
2498:
2465:. Retrieved
2461:the original
2406:
2402:
2352:
2348:
2318:
2314:
2308:
2273:
2269:
2259:
2227:(1): 41–54.
2224:
2220:
2172:
2168:
2120:
2116:
2070:
2066:
2060:
2019:
2015:
2009:
1960:
1956:
1950:
1901:
1897:
1891:
1856:
1852:
1842:
1815:
1809:
1802:Susan Gasser
1793:
1758:
1754:
1744:
1699:
1695:
1685:
1650:
1646:
1636:
1599:
1595:
1585:
1576:
1572:
1566:
1541:
1537:
1531:
1501:(7): 841–7.
1498:
1494:
1488:
1474:
1460:
1425:
1421:
1374:(2): 621–9.
1371:
1367:
1357:
1332:
1328:
1322:
1297:
1293:
1287:
1279:
1274:
1265:
1261:
1257:
1251:
1210:
1206:
1200:
1188:. Retrieved
1182:
1176:
1169:
1128:
1122:
1120:
1111:
1106:
1103:
1089:
1080:
1071:
1067:
1062:
1057:
1054:
1029:
1028:
1024:
1015:
1001:
999:
994:
981:
972:
969:
956:
951:
950:
941:
927:
915:
898:
894:
893:
884:
857:
847:
840:
829:Size control
823:
803:
764:
752:
746:
744:
738:
715:
714:
703:
695:
689:
681:
677:
673:
669:
661:
654:
646:
640:
628:
618:
610:
604:
596:
590:
582:
575:
567:
557:
553:
551:
545:
537:
533:
525:
520:
515:
513:
503:
496:liquefaciens
495:
491:
483:
474:
473:
461:
457:
449:
428:
426:
413:
412:
403:
376:
374:
362:Lee Hartwell
356:
339:
332:Paul Lindner
329:
318:
290:cell biology
269:
264:
263:
262:
250:
246:
238:
234:
229:
225:
217:
209:
201:
193:
185:
160:
158:
142:
141:
129:
35:
29:
4676:iNaturalist
4596:Wikispecies
4503:Arabidopsis
4474:Tetrahymena
3746:10261/51389
1042:DNA damages
1040:that cause
877:activity.)
812:Cytokinesis
686:peroxisomes
621:is usually
601:chromosomes
348:Urs Leupold
336:millet beer
302:micrometres
4779:Ascomycota
4773:Categories
4524:Drosophila
4517:C. elegans
4491:Neurospora
4293:PMC1462848
3989:7 February
3456:(1): 556.
2467:7 February
2457:"PombeNet"
2321:(2): 3–9.
1755:Cell Cycle
1268:: 381–480.
1161:References
1145:DNA repair
907:DNA damage
735:Centrosome
726:Life cycle
700:centromere
651:cell cycle
418:DNA damage
358:Paul Nurse
327:research.
325:cell cycle
205:Tschalenko
89:Ascomycota
85:Division:
4537:Zebrafish
4374:cite book
3877:1308.1985
2087:0066-4197
1985:1476-4687
1926:1476-4687
1480:"PomBase"
1466:"PomBase"
1329:BioEssays
1243:205213499
1190:14 August
1004:contains
692:does not.
643:does not.
633:shelterin
613:is often
540:strains.
500:Wädenswil
392:sequenced
294:eukaryote
286:molecular
239:iotoensis
137:Species:
75:Kingdom:
69:Eukaryota
4702:MycoBank
4694:10390016
4650:Fungorum
4587:Q2236682
4581:Wikidata
4443:genetics
4366:19563123
4277:22132734
4228:22940862
4185:22694927
4136:24019917
4096:PLOS ONE
4077:21869447
4040:22806564
3965:23658693
3925:PLOS ONE
3906:24098959
3845:20719273
3804:22906028
3796:22888038
3755:22300943
3710:21404821
3702:23294323
3666:39454427
3658:28032397
3622:23981297
3578:24112980
3529:12744756
3521:19646873
3480:22146300
3431:11983163
3387:24095860
3338:22384373
3302:Isolates
3280:25665008
3224:32032353
3173:20946356
3129:22887981
3073:15887295
3032:25361970
2983:22801777
2931:22723125
2882:22279046
2830:23150603
2800:Genetics
2781:26447128
2751:Genetics
2732:10461200
2697:23365689
2657:PLOS ONE
2635:22840777
2581:17377067
2529:11859360
2433:18628834
2379:16498704
2327:16038088
2300:16118186
2270:Genetics
2251:19237686
2221:Genetics
2147:44652765
2095:17614787
2044:19516326
1993:19474792
1934:19474789
1883:24035542
1834:70173205
1785:20697207
1736:15494441
1677:21127171
1628:12093372
1579:: 38–44.
1558:15282124
1523:10397608
1515:16823372
1452:22039153
1398:27270696
1368:Genetics
1314:13480293
1235:18488015
1134:See also
1123:S. pombe
1107:S. pombe
1090:S. pombe
1063:S. pombe
1058:S. pombe
1030:S. pombe
1002:S. pombe
995:S. pombe
973:S. pombe
957:S. pombe
928:S. pombe
916:S. pombe
899:S. pombe
848:S. pombe
747:S. pombe
739:S. pombe
716:S. pombe
704:S. pombe
690:S. pombe
688:, while
678:S. pombe
662:S. pombe
655:S. pombe
637:telomere
629:S. pombe
619:S. pombe
605:S. pombe
591:S. pombe
589:, while
576:S. pombe
534:S. pombe
528:undergo
526:S. pombe
516:S. pombe
504:S. pombe
492:S. pombe
484:S. pombe
458:S. pombe
454:kombucha
450:S. pombe
429:S. pombe
404:S. pombe
377:S. pombe
366:Tim Hunt
296:, whose
272:", is a
177:Synonyms
115:Family:
65:Domain:
4681:1023705
4668:2586935
4531:Xenopus
4460:E. coli
4306:. 2002.
4255:Traffic
4236:4344457
4208:Bibcode
4176:4339016
4127:3760898
4104:Bibcode
4031:3578726
3956:3641051
3933:Bibcode
3897:3983658
3569:4276399
3471:3737731
3329:3276172
3271:4645456
3215:7032740
3120:5322539
3023:4383888
2974:3557915
2922:3409321
2873:3273838
2821:3527242
2772:4596657
2688:3554685
2665:Bibcode
2626:3589755
2572:1877093
2507:Bibcode
2424:2447254
2370:5074380
2291:1456079
2242:2674839
2199:2703462
2139:9885154
2052:4402226
2024:Bibcode
2001:4412402
1965:Bibcode
1942:4330336
1906:Bibcode
1874:4620659
1776:3041159
1704:Bibcode
1668:3058771
1443:3245111
1389:4896181
1349:2185750
1215:Bibcode
1155:PomBase
983:PomBase
965:PomBase
623:haploid
615:diploid
599:has 16
587:introns
510:Ecology
470:History
446:Cachaça
396:PomBase
344:Swahili
342:is the
321:fission
274:species
213:Osterw.
189:A.Jörg.
168:Lindner
125:Genus:
105:Order:
95:Class:
4789:Yeasts
4756:990004
4740:NZOR:
4707:212377
4655:212377
4642:SSACPO
4629:187820
4437:Major
4364:
4354:
4275:
4234:
4226:
4200:Nature
4183:
4173:
4134:
4124:
4075:
4038:
4028:
3985:. 2001
3963:
3953:
3904:
3894:
3843:
3833:
3802:
3794:
3753:
3708:
3700:
3664:
3656:
3620:
3576:
3566:
3527:
3519:
3478:
3468:
3429:
3385:
3336:
3326:
3300:pombe
3278:
3268:
3222:
3212:
3171:
3127:
3117:
3071:
3030:
3020:
2981:
2971:
2929:
2919:
2880:
2870:
2828:
2818:
2779:
2769:
2730:
2695:
2685:
2633:
2623:
2579:
2569:
2527:
2499:Nature
2431:
2421:
2377:
2367:
2325:
2298:
2288:
2249:
2239:
2197:
2190:209837
2187:
2145:
2137:
2093:
2085:
2050:
2042:
2016:Nature
1999:
1991:
1983:
1957:Nature
1940:
1932:
1924:
1898:Nature
1881:
1871:
1832:
1822:
1800:, and
1783:
1773:
1734:
1727:524432
1724:
1675:
1665:
1626:
1619:139370
1616:
1556:
1521:
1513:
1450:
1440:
1396:
1386:
1347:
1312:
1241:
1233:
1207:Nature
947:Genome
911:mating
635:-like
631:has a
617:while
607:has 3.
438:grapes
434:apples
388:genome
380:genome
306:genome
170:(1893)
4689:IRMNG
4616:4VCRL
4547:Mouse
4497:Maize
4232:S2CID
3872:arXiv
3800:S2CID
3706:S2CID
3662:S2CID
3596:(PDF)
3525:S2CID
2349:Yeast
2143:S2CID
2117:Yeast
2048:S2CID
1997:S2CID
1938:S2CID
1573:Vitis
1519:S2CID
1239:S2CID
1150:Yeast
1034:aging
976:'
960:'
852:aging
494:var.
488:Delft
398:MOD (
340:pombe
310:genes
298:cells
278:yeast
249:var.
237:var.
228:var.
79:Fungi
4733:4896
4728:NCBI
4663:GBIF
4637:EPPO
4380:link
4362:PMID
4352:ISBN
4325:help
4273:PMID
4224:PMID
4181:PMID
4132:PMID
4073:PMID
4036:PMID
3991:2016
3961:PMID
3902:PMID
3841:PMID
3831:ISBN
3792:PMID
3751:PMID
3698:PMID
3654:PMID
3618:PMID
3574:PMID
3517:PMID
3476:PMID
3427:PMID
3383:PMID
3334:PMID
3276:PMID
3220:PMID
3169:PMID
3125:PMID
3069:PMID
3028:PMID
2979:PMID
2927:PMID
2878:PMID
2826:PMID
2777:PMID
2728:PMID
2693:PMID
2631:PMID
2577:PMID
2525:PMID
2469:2016
2429:PMID
2375:PMID
2323:PMID
2296:PMID
2247:PMID
2195:PMID
2135:PMID
2091:PMID
2083:ISSN
2040:PMID
1989:PMID
1981:ISSN
1930:PMID
1922:ISSN
1879:PMID
1830:OCLC
1820:ISBN
1781:PMID
1732:PMID
1673:PMID
1624:PMID
1554:PMID
1511:PMID
1448:PMID
1394:PMID
1345:PMID
1310:PMID
1231:PMID
1192:2021
1082:RecA
1048:and
874:Pom1
869:Pom1
865:Pom1
860:Pom1
843:wee1
664:has
556:and
436:and
364:and
288:and
4715:NBN
4624:EoL
4611:CoL
4542:Rat
4441:in
4344:doi
4263:doi
4216:doi
4204:489
4171:PMC
4163:doi
4122:PMC
4112:doi
4063:doi
4026:PMC
4018:doi
3951:PMC
3941:doi
3892:PMC
3882:doi
3823:doi
3782:doi
3741:hdl
3733:doi
3690:doi
3686:105
3646:doi
3608:doi
3564:PMC
3556:doi
3507:doi
3466:PMC
3458:doi
3417:doi
3375:hdl
3365:doi
3361:425
3324:PMC
3316:doi
3266:PMC
3258:doi
3210:PMC
3200:doi
3159:doi
3115:PMC
3107:doi
3059:doi
3018:PMC
3010:doi
2969:PMC
2961:doi
2917:PMC
2909:doi
2868:PMC
2860:doi
2816:PMC
2808:doi
2804:193
2767:PMC
2759:doi
2755:201
2720:doi
2683:PMC
2673:doi
2621:PMC
2613:doi
2567:PMC
2559:doi
2515:doi
2503:415
2419:PMC
2411:doi
2365:PMC
2357:doi
2286:PMC
2278:doi
2274:171
2237:PMC
2229:doi
2225:182
2185:PMC
2177:doi
2173:171
2125:doi
2075:doi
2032:doi
2020:459
1973:doi
1961:459
1914:doi
1902:459
1869:PMC
1861:doi
1771:PMC
1763:doi
1722:PMC
1712:doi
1700:101
1663:PMC
1655:doi
1614:PMC
1604:doi
1546:doi
1503:doi
1438:PMC
1430:doi
1384:PMC
1376:doi
1372:203
1337:doi
1302:doi
1260:".
1223:doi
1211:453
737:of
284:in
276:of
4775::
4753::
4730::
4717::
4704::
4691::
4678::
4665::
4652::
4639::
4626::
4613::
4598::
4583::
4376:}}
4372:{{
4360:.
4350:.
4316::
4314:}}
4310:{{
4285:^
4271:.
4259:13
4257:.
4253:.
4230:.
4222:.
4214:.
4202:.
4179:.
4169:.
4159:24
4157:.
4153:.
4130:.
4120:.
4110:.
4098:.
4094:.
4071:.
4057:.
4034:.
4024:.
4014:70
4012:.
4008:.
3981:.
3959:.
3949:.
3939:.
3927:.
3923:.
3900:.
3890:.
3880:.
3868:14
3866:.
3862:.
3839:.
3829:.
3798:.
3790:.
3778:69
3776:.
3772:.
3749:.
3739:.
3729:49
3727:.
3704:.
3696:.
3684:.
3660:.
3652:.
3642:41
3640:.
3616:.
3604:13
3602:.
3598:.
3572:.
3562:.
3552:23
3550:.
3546:.
3523:.
3515:.
3503:19
3501:.
3497:.
3474:.
3464:.
3452:.
3448:.
3425:.
3411:.
3407:.
3395:^
3381:.
3373:.
3359:.
3355:.
3332:.
3322:.
3310:.
3306:.
3274:.
3264:.
3254:47
3252:.
3248:.
3232:^
3218:.
3208:.
3196:16
3194:.
3190:.
3167:.
3155:10
3153:.
3149:.
3137:^
3123:.
3113:.
3103:69
3101:.
3097:.
3081:^
3067:.
3055:61
3053:.
3049:.
3026:.
3016:.
3006:43
3004:.
3000:.
2977:.
2967:.
2957:20
2955:.
2951:.
2939:^
2925:.
2915:.
2905:20
2903:.
2899:.
2876:.
2866:.
2856:26
2854:.
2850:.
2838:^
2824:.
2814:.
2802:.
2798:.
2775:.
2765:.
2753:.
2749:.
2726:.
2716:15
2714:.
2691:.
2681:.
2671:.
2659:.
2655:.
2643:^
2629:.
2619:.
2609:19
2607:.
2603:.
2589:^
2575:.
2565:.
2555:18
2553:.
2549:.
2537:^
2523:.
2513:.
2501:.
2497:.
2477:^
2441:^
2427:.
2417:.
2405:.
2401:.
2387:^
2373:.
2363:.
2353:23
2351:.
2347:.
2335:^
2319:18
2317:.
2294:.
2284:.
2272:.
2268:.
2245:.
2235:.
2223:.
2219:.
2207:^
2193:.
2183:.
2171:.
2167:.
2155:^
2141:.
2133:.
2121:14
2119:.
2115:.
2103:^
2089:.
2081:.
2071:41
2069:.
2046:.
2038:.
2030:.
2018:.
1995:.
1987:.
1979:.
1971:.
1959:.
1936:.
1928:.
1920:.
1912:.
1900:.
1877:.
1867:.
1857:23
1855:.
1851:.
1828:.
1779:.
1769:.
1757:.
1753:.
1730:.
1720:.
1710:.
1698:.
1694:.
1671:.
1661:.
1651:28
1649:.
1645:.
1622:.
1612:.
1598:.
1594:.
1577:16
1575:.
1552:.
1542:95
1540:.
1517:.
1509:.
1499:24
1497:.
1446:.
1436:.
1426:40
1424:.
1420:.
1406:^
1392:.
1382:.
1370:.
1366:.
1343:.
1333:12
1331:.
1308:.
1298:13
1296:.
1266:24
1264:.
1237:.
1229:.
1221:.
1209:.
1181:.
967:.
672:.
603:,
574:;
448:.
424:.
316:.
4430:e
4423:t
4416:v
4382:)
4368:.
4346::
4327:)
4323:(
4279:.
4265::
4238:.
4218::
4210::
4187:.
4165::
4138:.
4114::
4106::
4100:8
4079:.
4065::
4042:.
4020::
3993:.
3967:.
3943::
3935::
3929:8
3908:.
3884::
3874::
3847:.
3825::
3806:.
3784::
3757:.
3743::
3735::
3712:.
3692::
3668:.
3648::
3624:.
3610::
3580:.
3558::
3531:.
3509::
3482:.
3460::
3454:7
3433:.
3419::
3413:9
3389:.
3377::
3367::
3340:.
3318::
3312:1
3282:.
3260::
3226:.
3202::
3175:.
3161::
3131:.
3109::
3075:.
3061::
3034:.
3012::
2985:.
2963::
2933:.
2911::
2884:.
2862::
2832:.
2810::
2783:.
2761::
2734:.
2722::
2699:.
2675::
2667::
2661:8
2637:.
2615::
2583:.
2561::
2531:.
2517::
2509::
2471:.
2435:.
2413::
2407:3
2381:.
2359::
2329:.
2302:.
2280::
2253:.
2231::
2201:.
2179::
2149:.
2127::
2097:.
2077::
2054:.
2034::
2026::
2003:.
1975::
1967::
1944:.
1916::
1908::
1885:.
1863::
1836:.
1787:.
1765::
1759:9
1738:.
1714::
1706::
1679:.
1657::
1630:.
1606::
1600:3
1560:.
1548::
1525:.
1505::
1482:.
1468:.
1454:.
1432::
1400:.
1378::
1351:.
1339::
1316:.
1304::
1245:.
1225::
1217::
1194:.
1179:"
1175:"
741:.
625:.
548:)
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.