2726:
1353:
1420:, Cavalli-Sforza, Menozzi and Piazza provide some of the most detailed and comprehensive estimates of genetic distances between human populations, within and across continents. Their initial database contains 76,676 gene frequencies (using 120 blood polymorphisms), corresponding to 6,633 samples in different locations. By culling and pooling such samples, they restrict their analysis to 491 populations.
20:
1424:
1436:
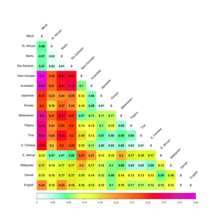
For these 42 populations, Cavalli-Sforza and coauthors report bilateral distances computed from 120 alleles. Among this set of 42 world populations, the greatest genetic distance observed is between Mbuti
Pygmies and Papua New Guineans, where the Fst distance is 0.4573, while the smallest genetic distance (0.0021) is between the Danish and the English.
900:
1439:
When considering more disaggregated data for 26 European populations, the smallest genetic distance (0.0009) is between the Dutch and the Danes, and the largest (0.0667) is between the Lapps and the
Sardinians. The mean genetic distance among the 861 available pairings of the 42 selected populations
1435:
that were at their present location at the end of the 15th century when the great
European migrations began. When studying genetic difference at the world level, the number is reduced to 42 representative populations, aggregating subpopulations characterized by a high level of genetic similarity.
1332:). The average pairwise difference within a population can be calculated as the sum of the pairwise differences divided by the number of pairs. However, this estimator is biased when sample sizes are small or if they vary between populations. Therefore, more elaborate methods are used to compute F
492:
measures the amount of genetic variance that can be explained by population structure. This can also be thought of as the fraction of total diversity that is not a consequence of the average diversity within subpopulations, where diversity is measured by the probability that two randomly selected
1402:
A genetic distance of 0.125 implies that kinship between unrelated individuals of the same ancestry relative to the world population is equivalent to kinship between half siblings in a randomly mating population. This also implies that if a human from a given ancestral population has a mixed
631:
480:
1215:
179:
85:; that is, that the two populations are interbreeding freely. A value of one implies that all genetic variation is explained by the population structure, and that the two populations do not share any genetic diversity.
1141:
In practice, none of the quantities used for the definitions can be easily measured. As a consequence, various estimators have been proposed. A particularly simple estimator applicable to DNA sequence data is:
990:
1124:
895:{\displaystyle F_{ST}={\frac {{\bar {p}}(1-{\bar {p}})-\sum c_{i}p_{i}(1-p_{i})}{{\bar {p}}(1-{\bar {p}})}}={\frac {{\bar {p}}(1-{\bar {p}})-{\overline {p(1-p)}}}{{\bar {p}}(1-{\bar {p}})}}}
358:
1303:
1245:
1330:
1272:
3531:
346:
314:
1129:
This formulation has the advantage that the expected time to coalescence can easily be estimated from genetic data, which led to the development of various estimators for F
3291:
Supplementary Figure 6: Heat map of pairwise FST values among all 1000 Genomes
Project and GME populations identifies three clusters with a low degree of differentiation.
1380:
show values significantly below 1%, indistinguishable from panmixia. Within Europe, the most divergent ethnic groups have been found to have values of the order of 7% (
1049:
282:
1148:
529:
1020:
623:
576:
3461:
207:
102:
3807:
3488:
596:
549:
3223:"European Population Genetic Substructure: Further Definition of Ancestry Informative Markers for Distinguishing among Diverse European Ethnic Groups"
2725:
3209:
3405:
Evolution and the
Genetics of Populations Volume 2: the Theory of Gene Frequencies, pg 294–295, S. Wright, Univ. of Chicago Press, Chicago, 1969
3781:
3639:
3536:
2912:
1403:
half-sibling, that human is closer genetically to an unrelated individual of their ancestral population than to their mixed half-sibling.
1055:
can be interpreted as measuring how much closer two individuals from the same subpopulation are, compared to the total population. If the
2774:
1067:
and T denote the average time to coalescence for individuals from the same subpopulation and the total population, respectively. Then,
1391:
Larger values are found if highly divergent homogenous groups are compared: the highest such value found was at close to 46%, between
216:
can be difficult when the data analyzed are highly polymorphic. In this case, the probability of identity by descent is very low and F
911:
3481:
316:
is the variance in the frequency of the allele among different subpopulations, weighted by the sizes of the subpopulations, and
3576:
1536:
1022:
is the probability of identity by descent of two individuals given that the two individuals are in the same subpopulation, and
2374:(combined from Utah residents of Northern and Western European ancestry from the CEPH collection and Italians from Tuscany),
3581:
1337:
51:
2834:(1998), Advances in molecular ecology (ed. G. Carvalho). NATO Science Series A: Life Sciences, IOS Press, Amsterdam, 39-53.
1452:
3511:
3802:
3474:
2363:
1073:
1336:
in practice. Two of the most widely used procedures are the estimator by Weir & Cockerham (1984), or performing an
3556:
3439:
3309:
475:{\displaystyle F_{ST}={\frac {\sigma _{S}^{2}}{\sigma _{T}^{2}}}={\frac {\sigma _{S}^{2}}{{\bar {p}}(1-{\bar {p}})}}}
220:
can have an arbitrarily low upper bound, which might lead to misinterpretation of the data. Also, strictly speaking F
1051:
is the probability that two individuals from the total population are identical by descent. Using this definition, F
39:
3677:
2748:
3551:
3434:
55:
3729:
3444:
2984:
Weir, B. S.; Cockerham, C. Clark (1984). "Estimating F-Statistics for the
Analysis of Population Structure".
1059:
is small, this interpretation can be made more explicit by linking the probability of identity by descent to
3541:
3270:
2394:
of Ibadan, Nigeria). It reported a value close to 12% between continental populations, and values close to
2378:(combining Han Chinese from Beijing, Chinese from metropolitan Denver and Japanese from Tokyo, Japan) and
1281:
1223:
3526:
1308:
1250:
47:
3768:
3662:
3657:
3561:
89:
69:. Its values range from 0 to 1, with 0 being no differentiation and 1 being complete differentiation.
2788:
3614:
3171:
3109:
2379:
1459:
1275:
319:
287:
225:
77:
This comparison of genetic variability within and between populations is frequently used in applied
3758:
3634:
3521:
3497:
2831:
2740:
1466:
249:
229:
78:
66:
1210:{\displaystyle F_{ST}={\frac {\pi _{\text{Between}}-\pi _{\text{Within}}}{\pi _{\text{Between}}}}}
3748:
3744:
3604:
3334:
3069:
3009:
1543:
1364:
values depend strongly on the choice of populations. Closely related ethnic groups, such as the
3283:
Characterization of
Greater Middle Eastern genetic variation for enhanced disease gene discovery
1025:
258:
1352:
3739:
3734:
3672:
3599:
3591:
3566:
3383:
3326:
3260:
3242:
3199:
3137:
3039:
3017:
3001:
2966:
2908:
2883:
2865:
2846:"Genetics in geographically structured populations: defining, estimating and interpreting FST"
2771:
1373:
1060:
43:
496:
3698:
3609:
3419:
3373:
3365:
3318:
3250:
3234:
3189:
3179:
3127:
3117:
3061:
2993:
2956:
2948:
2873:
2857:
2720:
245:
2832:
Estimation of migration rates and population sizes in geographically structured populations
998:
601:
554:
174:{\displaystyle M={\frac {m}{\mu }}\approx {\frac {1}{4}}\left({\frac {1}{F_{ST}}}-1\right)}
3516:
2778:
1480:
192:
1381:
3175:
3113:
1443:
The following table shows Fst calculated by
Cavalli-Sforza (1994) for some populations:
3763:
3753:
3667:
3546:
3449:
3429:
3378:
3353:
3255:
3222:
3194:
3159:
3132:
3097:
2961:
2936:
2878:
2845:
1522:
1508:
1377:
581:
534:
3796:
3703:
3693:
3649:
3408:
A haplotype map of the human genome, The
International HapMap Consortium, Nature 2005
3338:
3322:
3290:
2391:
2387:
1396:
1056:
485:
59:
3073:
3719:
3571:
3424:
2383:
1473:
1392:
1369:
62:
19:
3184:
3122:
2952:
2902:
96:
can be used to estimate migration rates. Under that model, the migration rate is
3369:
3354:"Empirical Bayes inference of pairwise F(ST) and its distribution in the genome"
2375:
1529:
1494:
2759:
2753:
1423:
3238:
3065:
1385:
3246:
3005:
2869:
2783:
2765:
3724:
2371:
1501:
3387:
3330:
3264:
3203:
3141:
3021:
2887:
2970:
3098:"Empirical Distributions of FST from Large-Scale Human Polymorphism Data"
2395:
82:
3013:
2461:
Intra-European/mediterranean autosomal genetic distances based on SNPs
1487:
3466:
2811:
1427:
The 42 world populations used by the authors and their reported FSTs.
1340:. A list of implementations is available at the end of this article.
2997:
2861:
3158:
Nelis, Mari; et al. (2009-05-08). Fleischer, Robert C. (ed.).
3286:
3034:
Cavalli-Sforza et al. (1994), cited after V. Ginsburgh, S. Weber,
1515:
1365:
2715:
Autosomal genetic distances based on whole exome sequencing (WES)
1278:
between two individuals sampled from different sub-populations (
3052:
Harpending, Henry (2002). "Kinship and
Population Subdivision".
3470:
985:{\displaystyle F_{ST}={\frac {f_{0}-{\bar {f}}}{1-{\bar {f}}}}}
348:
is the variance of the allelic state in the total population, F
284:
is the average frequency of an allele in the total population,
81:. The values range from 0 to 1. A zero value implies complete
2719:
Pairwise Fst values among several populations based on whole
3160:"Genetic Structure of Europeans: A View from the North–East"
2402:
Intercontinental autosomal genetic distances based on SNPs
2937:"Estimation of Levels of Gene Flow from DNA Sequence Data"
3307:: software for the measurement of genetic diversity".
1412:
Autosomal genetic distances based on classical markers
228:
in the mathematical sense, as it does not satisfy the
2370:
between the three major "continental" populations of
1311:
1284:
1253:
1226:
1151:
1076:
1028:
1001:
914:
634:
604:
584:
557:
537:
499:
361:
322:
290:
261:
195:
105:
2935:
Hudson, RR.; Slatkin, M.; Maddison, WP. (Oct 1992).
65:, it is one of the most commonly used statistics in
3712:
3686:
3648:
3623:
3590:
3504:
2398:(smaller than 1%) within continental populations.
1324:
1297:
1266:
1239:
1209:
1119:{\displaystyle F_{ST}\approx 1-{\frac {T_{0}}{T}}}
1118:
1043:
1014:
984:
894:
617:
590:
570:
543:
523:
474:
340:
308:
276:
201:
173:
244:at a given locus are based on 1) the variance of
248:among populations, and on 2) the probability of
3036:The Palgrave Handbook of Economics and Language
1418:The History and Geography of Human Genes (1994)
240:Two of the most commonly used definitions for F
3482:
2904:Probability Models for DNA Sequence Evolution
8:
3489:
3475:
3467:
2844:Holsinger, Kent E.; Bruce S. Weir (2009).
189:is the migration rate per generation, and
3377:
3254:
3221:Tian, Chao; et al. (November 2009).
3193:
3183:
3131:
3121:
2960:
2877:
2358:Autosomal genetic distances based on SNPs
1316:
1310:
1289:
1283:
1258:
1252:
1231:
1225:
1199:
1188:
1175:
1168:
1156:
1150:
1105:
1099:
1081:
1075:
1030:
1029:
1027:
1006:
1000:
968:
967:
948:
947:
938:
931:
919:
913:
875:
874:
854:
853:
824:
807:
806:
786:
785:
782:
762:
761:
741:
740:
729:
710:
700:
676:
675:
655:
654:
651:
639:
633:
609:
603:
583:
562:
556:
536:
498:
455:
454:
434:
433:
426:
421:
415:
404:
399:
389:
384:
378:
366:
360:
332:
327:
321:
300:
295:
289:
263:
262:
260:
194:
149:
140:
125:
112:
104:
3352:Kitada S, Kitakado T, Kishino H (2007).
3153:
3151:
2459:
2400:
1445:
1422:
1351:
18:
3281:Scott, E., Halees, A., Itan, Y. et al.
2823:
23:Fst values between European populations
3782:Index of evolutionary biology articles
1407:Genetic distances in human populations
1276:average number of pairwise differences
1298:{\displaystyle \pi _{\text{Between}}}
1240:{\displaystyle \pi _{\text{Between}}}
209:is the mutation rate per generation.
16:Measure of population differentiation
7:
3808:Mathematical and theoretical biology
1325:{\displaystyle \pi _{\text{Within}}}
1267:{\displaystyle \pi _{\text{Within}}}
2806:
1305:) or from the same sub-population (
3285:. Nat Genet 48, 1071–1076 (2016).
3086:Cavalli-Sforza et al., 1994, p. 24
2901:Richard Durrett (12 August 2008).
1356:FST values in selected populations
46:. It is frequently estimated from
14:
531:. If the allele frequency in the
58:. Developed as a special case of
3323:10.1111/j.1755-0998.2009.02801.x
2724:
3462:BioPerl - Bio::PopGen::PopStats
3303:Crawford, Nicholas G. (2010). "
3287:https://doi.org/10.1038/ng.3592
341:{\displaystyle \sigma _{T}^{2}}
309:{\displaystyle \sigma _{S}^{2}}
52:single-nucleotide polymorphisms
3582:Constructive neutral evolution
1338:Analysis of molecular variance
1035:
973:
953:
886:
880:
865:
859:
842:
830:
818:
812:
797:
791:
773:
767:
752:
746:
735:
716:
687:
681:
666:
660:
518:
506:
493:alleles are different, namely
466:
460:
445:
439:
268:
1:
2772:Microsatellite Analyzer (MSA)
578:and the relative size of the
488:definition illustrates that F
88:For idealized models such as
3532:Fisher's fundamental theorem
3185:10.1371/journal.pone.0005472
3123:10.1371/journal.pone.0049837
2420:Sub-Saharan Africa (Yoruba)
2412:Sub-Saharan Africa (Yoruba)
2364:International HapMap Project
846:
90:Wright's finite island model
3557:Coefficient of relationship
3440:Coefficient of relationship
3370:10.1534/genetics.107.077263
3310:Molecular Ecology Resources
3826:
3054:Population and Environment
2953:10.1093/genetics/132.2.583
2732:Programs for calculating F
1044:{\displaystyle {\bar {f}}}
277:{\displaystyle {\bar {p}}}
40:population differentiation
3777:
3552:Coefficient of inbreeding
3435:Coefficient of inbreeding
3239:10.2119/molmed.2009.00094
2798:Modules for calculating F
2683:
2654:
2625:
2597:
2570:
2545:
2520:
2496:
2491:
2488:
2485:
2482:
2479:
2476:
2473:
2470:
2467:
2465:
2444:
2431:
2419:
2414:
2411:
2408:
2406:
1440:was found to be 0.1338..
3730:Evolutionary game theory
3512:Hardy–Weinberg principle
3445:Hardy-Weinberg principle
2390:of Kinyawa, Kenya and
3542:Shifting balance theory
3066:10.1023/A:1020815420693
2362:A 2012 study based on
524:{\displaystyle 2p(1-p)}
212:The interpretation of F
3527:Linkage disequilibrium
1433:aboriginal populations
1428:
1357:
1326:
1299:
1268:
1241:
1211:
1120:
1045:
1016:
986:
896:
619:
592:
572:
545:
525:
476:
342:
310:
278:
203:
175:
24:
3769:Quantitative genetics
3678:Balding–Nichols model
3663:Population bottleneck
3658:Small population size
3562:Selection coefficient
3096:Elhaik, Eran (2012).
2432:East-Asia (Japanese)
2415:East-Asia (Japanese)
1426:
1355:
1327:
1300:
1269:
1242:
1212:
1121:
1046:
1017:
1015:{\displaystyle f_{0}}
987:
897:
620:
618:{\displaystyle c_{i}}
593:
573:
571:{\displaystyle p_{i}}
546:
526:
477:
343:
311:
279:
204:
176:
22:
3640:Background selection
3627:on genomic variation
3625:Effects of selection
3577:Population structure
3038:, Springer (2016),
2445:East-Asia (Chinese)
2380:Sub-Saharan Africans
1309:
1282:
1251:
1224:
1149:
1074:
1026:
999:
912:
632:
602:
582:
555:
535:
497:
359:
320:
288:
259:
202:{\displaystyle \mu }
193:
103:
48:genetic polymorphism
3803:Population genetics
3759:Population genomics
3635:Genetic hitchhiking
3522:Identity by descent
3498:Population genetics
3176:2009PLoSO...4.5472N
3114:2012PLoSO...749837E
2462:
2403:
2386:of Webuye, Kenya,
431:
409:
394:
337:
305:
250:identity by descent
230:triangle inequality
79:population genetics
67:population genetics
3745:Landscape genetics
3233:(11–12): 371–383.
3227:Molecular Medicine
2777:2015-03-29 at the
2460:
2401:
1429:
1358:
1322:
1295:
1264:
1237:
1207:
1116:
1041:
1012:
982:
892:
615:
588:
568:
541:
521:
472:
417:
395:
380:
338:
323:
306:
291:
274:
246:allele frequencies
199:
171:
38:) is a measure of
25:
3790:
3789:
3740:Genetic genealogy
3735:Fitness landscape
2914:978-0-387-78168-6
2712:
2711:
2458:
2457:
2355:
2354:
2351:
2346:
2341:
2336:
2331:
2326:
2321:
2316:
2311:
2306:
2301:
2296:
2291:
2286:
2281:
2272:
2267:
2262:
2257:
2252:
2247:
2242:
2237:
2232:
2227:
2222:
2217:
2212:
2207:
2196:
2191:
2186:
2181:
2176:
2171:
2166:
2161:
2156:
2151:
2146:
2141:
2136:
2123:
2118:
2113:
2108:
2103:
2098:
2093:
2088:
2083:
2078:
2073:
2068:
2053:
2048:
2043:
2038:
2033:
2028:
2023:
2018:
2013:
2008:
2003:
1986:
1981:
1976:
1971:
1966:
1961:
1956:
1951:
1946:
1941:
1922:
1917:
1912:
1907:
1902:
1897:
1892:
1887:
1882:
1861:
1856:
1851:
1846:
1841:
1836:
1831:
1826:
1803:
1798:
1793:
1788:
1783:
1778:
1773:
1748:
1743:
1738:
1733:
1728:
1723:
1696:
1691:
1686:
1681:
1676:
1647:
1642:
1637:
1632:
1601:
1596:
1591:
1558:
1553:
1546:
1539:
1532:
1525:
1518:
1511:
1504:
1497:
1490:
1483:
1476:
1469:
1462:
1455:
1319:
1292:
1261:
1234:
1205:
1202:
1191:
1178:
1114:
1038:
980:
976:
956:
890:
883:
862:
849:
815:
794:
777:
770:
749:
684:
663:
598:th population is
591:{\displaystyle i}
551:th population is
544:{\displaystyle i}
470:
463:
442:
410:
271:
158:
133:
120:
44:genetic structure
3815:
3699:J. B. S. Haldane
3491:
3484:
3477:
3468:
3420:Genetic distance
3392:
3391:
3381:
3349:
3343:
3342:
3306:
3300:
3294:
3279:
3273:
3268:
3258:
3218:
3212:
3207:
3197:
3187:
3155:
3146:
3145:
3135:
3125:
3093:
3087:
3084:
3078:
3077:
3049:
3043:
3032:
3026:
3025:
2992:(6): 1358–1370.
2981:
2975:
2974:
2964:
2932:
2926:
2925:
2923:
2921:
2898:
2892:
2891:
2881:
2841:
2835:
2828:
2728:
2723:(WES) in 2016:
2721:exome sequencing
2463:
2404:
2366:data estimated F
2349:
2344:
2339:
2334:
2329:
2324:
2319:
2314:
2309:
2304:
2299:
2294:
2289:
2284:
2279:
2270:
2265:
2260:
2255:
2250:
2245:
2240:
2235:
2230:
2225:
2220:
2215:
2210:
2205:
2194:
2189:
2184:
2179:
2174:
2169:
2164:
2159:
2154:
2149:
2144:
2139:
2134:
2121:
2116:
2111:
2106:
2101:
2096:
2091:
2086:
2081:
2076:
2071:
2066:
2051:
2046:
2041:
2036:
2031:
2026:
2021:
2016:
2011:
2006:
2001:
1984:
1979:
1974:
1969:
1964:
1959:
1954:
1949:
1944:
1939:
1920:
1915:
1910:
1905:
1900:
1895:
1890:
1885:
1880:
1859:
1854:
1849:
1844:
1839:
1834:
1829:
1824:
1801:
1796:
1791:
1786:
1781:
1776:
1771:
1746:
1741:
1736:
1731:
1726:
1721:
1694:
1689:
1684:
1679:
1674:
1645:
1640:
1635:
1630:
1599:
1594:
1589:
1556:
1551:
1542:
1535:
1528:
1521:
1514:
1507:
1500:
1493:
1486:
1479:
1472:
1465:
1458:
1451:
1446:
1331:
1329:
1328:
1323:
1321:
1320:
1317:
1304:
1302:
1301:
1296:
1294:
1293:
1290:
1273:
1271:
1270:
1265:
1263:
1262:
1259:
1246:
1244:
1243:
1238:
1236:
1235:
1232:
1216:
1214:
1213:
1208:
1206:
1204:
1203:
1200:
1194:
1193:
1192:
1189:
1180:
1179:
1176:
1169:
1164:
1163:
1125:
1123:
1122:
1117:
1115:
1110:
1109:
1100:
1089:
1088:
1061:coalescent times
1050:
1048:
1047:
1042:
1040:
1039:
1031:
1021:
1019:
1018:
1013:
1011:
1010:
991:
989:
988:
983:
981:
979:
978:
977:
969:
959:
958:
957:
949:
943:
942:
932:
927:
926:
901:
899:
898:
893:
891:
889:
885:
884:
876:
864:
863:
855:
851:
850:
845:
825:
817:
816:
808:
796:
795:
787:
783:
778:
776:
772:
771:
763:
751:
750:
742:
738:
734:
733:
715:
714:
705:
704:
686:
685:
677:
665:
664:
656:
652:
647:
646:
624:
622:
621:
616:
614:
613:
597:
595:
594:
589:
577:
575:
574:
569:
567:
566:
550:
548:
547:
542:
530:
528:
527:
522:
481:
479:
478:
473:
471:
469:
465:
464:
456:
444:
443:
435:
430:
425:
416:
411:
408:
403:
393:
388:
379:
374:
373:
347:
345:
344:
339:
336:
331:
315:
313:
312:
307:
304:
299:
283:
281:
280:
275:
273:
272:
264:
208:
206:
205:
200:
188:
180:
178:
177:
172:
170:
166:
159:
157:
156:
141:
134:
126:
121:
113:
3825:
3824:
3818:
3817:
3816:
3814:
3813:
3812:
3793:
3792:
3791:
3786:
3773:
3708:
3682:
3644:
3628:
3626:
3619:
3586:
3517:Genetic linkage
3500:
3495:
3458:
3416:
3411:
3401:
3399:Further reading
3396:
3395:
3351:
3350:
3346:
3304:
3302:
3301:
3297:
3280:
3276:
3220:
3219:
3215:
3157:
3156:
3149:
3095:
3094:
3090:
3085:
3081:
3051:
3050:
3046:
3033:
3029:
2998:10.2307/2408641
2983:
2982:
2978:
2934:
2933:
2929:
2919:
2917:
2915:
2900:
2899:
2895:
2862:10.1038/nrg2611
2843:
2842:
2838:
2829:
2825:
2820:
2803:
2801:
2779:Wayback Machine
2737:
2735:
2717:
2369:
2360:
1416:In their study
1414:
1409:
1363:
1350:
1347:
1335:
1312:
1307:
1306:
1285:
1280:
1279:
1254:
1249:
1248:
1227:
1222:
1221:
1195:
1184:
1171:
1170:
1152:
1147:
1146:
1139:
1132:
1101:
1077:
1072:
1071:
1066:
1054:
1024:
1023:
1002:
997:
996:
960:
934:
933:
915:
910:
909:
905:Alternatively,
852:
826:
784:
739:
725:
706:
696:
653:
635:
630:
629:
605:
600:
599:
580:
579:
558:
553:
552:
533:
532:
495:
494:
491:
432:
362:
357:
356:
352:is defined as
351:
318:
317:
286:
285:
257:
256:
243:
238:
223:
219:
215:
191:
190:
186:
145:
139:
135:
101:
100:
95:
75:
56:microsatellites
36:
17:
12:
11:
5:
3823:
3822:
3819:
3811:
3810:
3805:
3795:
3794:
3788:
3787:
3785:
3784:
3778:
3775:
3774:
3772:
3771:
3766:
3764:Phylogeography
3761:
3756:
3754:Microevolution
3751:
3742:
3737:
3732:
3727:
3722:
3716:
3714:
3713:Related topics
3710:
3709:
3707:
3706:
3701:
3696:
3690:
3688:
3684:
3683:
3681:
3680:
3675:
3670:
3668:Founder effect
3665:
3660:
3654:
3652:
3646:
3645:
3643:
3642:
3637:
3631:
3629:
3624:
3621:
3620:
3618:
3617:
3612:
3607:
3602:
3596:
3594:
3588:
3587:
3585:
3584:
3579:
3574:
3569:
3564:
3559:
3554:
3549:
3547:Price equation
3544:
3539:
3537:Neutral theory
3534:
3529:
3524:
3519:
3514:
3508:
3506:
3502:
3501:
3496:
3494:
3493:
3486:
3479:
3471:
3465:
3464:
3457:
3456:External links
3454:
3453:
3452:
3450:Wahlund effect
3447:
3442:
3437:
3432:
3430:QST_(genetics)
3427:
3422:
3415:
3412:
3410:
3409:
3406:
3402:
3400:
3397:
3394:
3393:
3344:
3317:(3): 556–557.
3295:
3274:
3213:
3147:
3108:(11): e49837.
3088:
3079:
3060:(2): 141–147.
3044:
3027:
2976:
2927:
2913:
2893:
2856:(9): 639–650.
2836:
2830:Peter Beerli,
2822:
2821:
2819:
2816:
2815:
2814:
2809:
2802:
2799:
2796:
2795:
2794:
2791:
2786:
2781:
2769:
2763:
2757:
2751:
2746:
2743:
2736:
2733:
2730:
2716:
2713:
2710:
2709:
2707:
2705:
2702:
2699:
2696:
2694:
2691:
2688:
2685:
2681:
2680:
2678:
2676:
2673:
2670:
2667:
2666:0.0080-0.0150
2664:
2661:
2659:
2658:0.0030-0.0050
2656:
2652:
2651:
2649:
2647:
2645:
2644:0.0030-0.0037
2642:
2641:0.0070-0.0079
2639:
2638:0.0060-0.0120
2636:
2635:0.0030-0.0036
2633:
2630:
2629:0.0088-0.0120
2627:
2623:
2622:
2620:
2618:
2616:
2614:
2613:0.0015-0.0030
2611:
2610:0.0060-0.0130
2608:
2607:0.0007-0.0010
2605:
2602:
2601:0.0029-0.0080
2599:
2595:
2594:
2592:
2590:
2588:
2586:
2584:
2583:0.0110-0.0170
2581:
2578:
2575:
2574:0.0010-0.0050
2572:
2568:
2567:
2565:
2563:
2561:
2559:
2557:
2555:
2554:0.0050-0.0110
2552:
2550:
2549:0.0130-0.0230
2547:
2543:
2542:
2540:
2538:
2536:
2534:
2532:
2530:
2528:
2525:
2524:0.0064-0.0090
2522:
2518:
2517:
2515:
2513:
2511:
2509:
2507:
2505:
2503:
2501:
2498:
2494:
2493:
2490:
2487:
2484:
2481:
2478:
2475:
2472:
2469:
2466:
2456:
2455:
2452:
2449:
2446:
2442:
2441:
2439:
2436:
2433:
2429:
2428:
2426:
2424:
2421:
2417:
2416:
2413:
2410:
2407:
2367:
2359:
2356:
2353:
2352:
2347:
2342:
2337:
2332:
2327:
2322:
2317:
2312:
2307:
2302:
2297:
2292:
2287:
2282:
2276:
2275:
2273:
2268:
2263:
2258:
2253:
2248:
2243:
2238:
2233:
2228:
2223:
2218:
2213:
2208:
2202:
2201:
2199:
2197:
2192:
2187:
2182:
2177:
2172:
2167:
2162:
2157:
2152:
2147:
2142:
2137:
2131:
2130:
2128:
2126:
2124:
2119:
2114:
2109:
2104:
2099:
2094:
2089:
2084:
2079:
2074:
2069:
2063:
2062:
2060:
2058:
2056:
2054:
2049:
2044:
2039:
2034:
2029:
2024:
2019:
2014:
2009:
2004:
1998:
1997:
1995:
1993:
1991:
1989:
1987:
1982:
1977:
1972:
1967:
1962:
1957:
1952:
1947:
1942:
1936:
1935:
1933:
1931:
1929:
1927:
1925:
1923:
1918:
1913:
1908:
1903:
1898:
1893:
1888:
1883:
1877:
1876:
1874:
1872:
1870:
1868:
1866:
1864:
1862:
1857:
1852:
1847:
1842:
1837:
1832:
1827:
1821:
1820:
1818:
1816:
1814:
1812:
1810:
1808:
1806:
1804:
1799:
1794:
1789:
1784:
1779:
1774:
1768:
1767:
1765:
1763:
1761:
1759:
1757:
1755:
1753:
1751:
1749:
1744:
1739:
1734:
1729:
1724:
1718:
1717:
1715:
1713:
1711:
1709:
1707:
1705:
1703:
1701:
1699:
1697:
1692:
1687:
1682:
1677:
1671:
1670:
1668:
1666:
1664:
1662:
1660:
1658:
1656:
1654:
1652:
1650:
1648:
1643:
1638:
1633:
1627:
1626:
1624:
1622:
1620:
1618:
1616:
1614:
1612:
1610:
1608:
1606:
1604:
1602:
1597:
1592:
1586:
1585:
1583:
1581:
1579:
1577:
1575:
1573:
1571:
1569:
1567:
1565:
1563:
1561:
1559:
1554:
1548:
1547:
1540:
1533:
1526:
1519:
1512:
1505:
1498:
1491:
1484:
1477:
1470:
1463:
1456:
1449:
1431:They focus on
1413:
1410:
1408:
1405:
1361:
1349:
1345:
1342:
1333:
1315:
1288:
1274:represent the
1257:
1230:
1218:
1217:
1198:
1187:
1183:
1174:
1167:
1162:
1159:
1155:
1138:
1135:
1130:
1127:
1126:
1113:
1108:
1104:
1098:
1095:
1092:
1087:
1084:
1080:
1064:
1052:
1037:
1034:
1009:
1005:
993:
992:
975:
972:
966:
963:
955:
952:
946:
941:
937:
930:
925:
922:
918:
903:
902:
888:
882:
879:
873:
870:
867:
861:
858:
848:
844:
841:
838:
835:
832:
829:
823:
820:
814:
811:
805:
802:
799:
793:
790:
781:
775:
769:
766:
760:
757:
754:
748:
745:
737:
732:
728:
724:
721:
718:
713:
709:
703:
699:
695:
692:
689:
683:
680:
674:
671:
668:
662:
659:
650:
645:
642:
638:
612:
608:
587:
565:
561:
540:
520:
517:
514:
511:
508:
505:
502:
489:
483:
482:
468:
462:
459:
453:
450:
447:
441:
438:
429:
424:
420:
414:
407:
402:
398:
392:
387:
383:
377:
372:
369:
365:
349:
335:
330:
326:
303:
298:
294:
270:
267:
241:
237:
234:
221:
217:
213:
198:
183:
182:
169:
165:
162:
155:
152:
148:
144:
138:
132:
129:
124:
119:
116:
111:
108:
93:
74:
73:Interpretation
71:
50:data, such as
34:
29:fixation index
15:
13:
10:
9:
6:
4:
3:
2:
3821:
3820:
3809:
3806:
3804:
3801:
3800:
3798:
3783:
3780:
3779:
3776:
3770:
3767:
3765:
3762:
3760:
3757:
3755:
3752:
3750:
3746:
3743:
3741:
3738:
3736:
3733:
3731:
3728:
3726:
3723:
3721:
3718:
3717:
3715:
3711:
3705:
3704:Sewall Wright
3702:
3700:
3697:
3695:
3692:
3691:
3689:
3685:
3679:
3676:
3674:
3671:
3669:
3666:
3664:
3661:
3659:
3656:
3655:
3653:
3651:
3650:Genetic drift
3647:
3641:
3638:
3636:
3633:
3632:
3630:
3622:
3616:
3613:
3611:
3608:
3606:
3603:
3601:
3598:
3597:
3595:
3593:
3589:
3583:
3580:
3578:
3575:
3573:
3570:
3568:
3565:
3563:
3560:
3558:
3555:
3553:
3550:
3548:
3545:
3543:
3540:
3538:
3535:
3533:
3530:
3528:
3525:
3523:
3520:
3518:
3515:
3513:
3510:
3509:
3507:
3503:
3499:
3492:
3487:
3485:
3480:
3478:
3473:
3472:
3469:
3463:
3460:
3459:
3455:
3451:
3448:
3446:
3443:
3441:
3438:
3436:
3433:
3431:
3428:
3426:
3423:
3421:
3418:
3417:
3413:
3407:
3404:
3403:
3398:
3389:
3385:
3380:
3375:
3371:
3367:
3364:(2): 861–73.
3363:
3359:
3355:
3348:
3345:
3340:
3336:
3332:
3328:
3324:
3320:
3316:
3312:
3311:
3299:
3296:
3292:
3288:
3284:
3278:
3275:
3272:
3266:
3262:
3257:
3252:
3248:
3244:
3240:
3236:
3232:
3228:
3224:
3217:
3214:
3211:
3205:
3201:
3196:
3191:
3186:
3181:
3177:
3173:
3169:
3165:
3161:
3154:
3152:
3148:
3143:
3139:
3134:
3129:
3124:
3119:
3115:
3111:
3107:
3103:
3099:
3092:
3089:
3083:
3080:
3075:
3071:
3067:
3063:
3059:
3055:
3048:
3045:
3041:
3037:
3031:
3028:
3023:
3019:
3015:
3011:
3007:
3003:
2999:
2995:
2991:
2987:
2980:
2977:
2972:
2968:
2963:
2958:
2954:
2950:
2946:
2942:
2938:
2931:
2928:
2916:
2910:
2906:
2905:
2897:
2894:
2889:
2885:
2880:
2875:
2871:
2867:
2863:
2859:
2855:
2851:
2850:Nat Rev Genet
2847:
2840:
2837:
2833:
2827:
2824:
2817:
2813:
2810:
2808:
2805:
2804:
2797:
2792:
2790:
2787:
2785:
2782:
2780:
2776:
2773:
2770:
2767:
2764:
2761:
2758:
2755:
2752:
2750:
2747:
2744:
2742:
2739:
2738:
2731:
2729:
2727:
2722:
2714:
2708:
2706:
2703:
2700:
2697:
2695:
2692:
2689:
2686:
2682:
2679:
2677:
2674:
2671:
2668:
2665:
2662:
2660:
2657:
2653:
2650:
2648:
2646:
2643:
2640:
2637:
2634:
2631:
2628:
2624:
2621:
2619:
2617:
2615:
2612:
2609:
2606:
2603:
2600:
2596:
2593:
2591:
2589:
2587:
2585:
2582:
2579:
2576:
2573:
2569:
2566:
2564:
2562:
2560:
2558:
2556:
2553:
2551:
2548:
2544:
2541:
2539:
2537:
2535:
2533:
2531:
2529:
2526:
2523:
2519:
2516:
2514:
2512:
2510:
2508:
2506:
2504:
2502:
2499:
2497:Palestinians
2495:
2471:Palestinians
2464:
2453:
2450:
2447:
2443:
2440:
2437:
2434:
2430:
2427:
2425:
2422:
2418:
2409:Europe (CEU)
2405:
2399:
2397:
2393:
2389:
2385:
2381:
2377:
2373:
2365:
2357:
2348:
2343:
2338:
2333:
2328:
2323:
2318:
2313:
2308:
2303:
2298:
2293:
2288:
2283:
2278:
2277:
2274:
2269:
2264:
2259:
2254:
2249:
2244:
2239:
2234:
2229:
2224:
2219:
2214:
2209:
2204:
2203:
2200:
2198:
2193:
2188:
2183:
2178:
2173:
2168:
2163:
2158:
2153:
2148:
2143:
2138:
2133:
2132:
2129:
2127:
2125:
2120:
2115:
2110:
2105:
2100:
2095:
2090:
2085:
2080:
2075:
2070:
2065:
2064:
2061:
2059:
2057:
2055:
2050:
2045:
2040:
2035:
2030:
2025:
2020:
2015:
2010:
2005:
2000:
1999:
1996:
1994:
1992:
1990:
1988:
1983:
1978:
1973:
1968:
1963:
1958:
1953:
1948:
1943:
1938:
1937:
1934:
1932:
1930:
1928:
1926:
1924:
1919:
1914:
1909:
1904:
1899:
1894:
1889:
1884:
1879:
1878:
1875:
1873:
1871:
1869:
1867:
1865:
1863:
1858:
1853:
1848:
1843:
1838:
1833:
1828:
1823:
1822:
1819:
1817:
1815:
1813:
1811:
1809:
1807:
1805:
1800:
1795:
1790:
1785:
1780:
1775:
1770:
1769:
1766:
1764:
1762:
1760:
1758:
1756:
1754:
1752:
1750:
1745:
1740:
1735:
1730:
1725:
1720:
1719:
1716:
1714:
1712:
1710:
1708:
1706:
1704:
1702:
1700:
1698:
1693:
1688:
1683:
1678:
1673:
1672:
1669:
1667:
1665:
1663:
1661:
1659:
1657:
1655:
1653:
1651:
1649:
1644:
1639:
1634:
1629:
1628:
1625:
1623:
1621:
1619:
1617:
1615:
1613:
1611:
1609:
1607:
1605:
1603:
1598:
1593:
1588:
1587:
1584:
1582:
1580:
1578:
1576:
1574:
1572:
1570:
1568:
1566:
1564:
1562:
1560:
1555:
1550:
1549:
1545:
1541:
1538:
1534:
1531:
1527:
1524:
1520:
1517:
1513:
1510:
1506:
1503:
1499:
1496:
1492:
1489:
1485:
1482:
1478:
1475:
1471:
1468:
1464:
1461:
1457:
1454:
1450:
1448:
1447:
1444:
1441:
1437:
1434:
1425:
1421:
1419:
1411:
1406:
1404:
1400:
1398:
1394:
1389:
1387:
1383:
1379:
1375:
1371:
1367:
1354:
1343:
1341:
1339:
1313:
1286:
1277:
1255:
1228:
1196:
1185:
1181:
1172:
1165:
1160:
1157:
1153:
1145:
1144:
1143:
1136:
1134:
1111:
1106:
1102:
1096:
1093:
1090:
1085:
1082:
1078:
1070:
1069:
1068:
1062:
1058:
1057:mutation rate
1032:
1007:
1003:
970:
964:
961:
950:
944:
939:
935:
928:
923:
920:
916:
908:
907:
906:
877:
871:
868:
856:
839:
836:
833:
827:
821:
809:
803:
800:
788:
779:
764:
758:
755:
743:
730:
726:
722:
719:
711:
707:
701:
697:
693:
690:
678:
672:
669:
657:
648:
643:
640:
636:
628:
627:
626:
610:
606:
585:
563:
559:
538:
515:
512:
509:
503:
500:
487:
457:
451:
448:
436:
427:
422:
418:
412:
405:
400:
396:
390:
385:
381:
375:
370:
367:
363:
355:
354:
353:
333:
328:
324:
301:
296:
292:
265:
253:
251:
247:
235:
233:
231:
227:
210:
196:
167:
163:
160:
153:
150:
146:
142:
136:
130:
127:
122:
117:
114:
109:
106:
99:
98:
97:
91:
86:
84:
80:
72:
70:
68:
64:
61:
57:
53:
49:
45:
41:
37:
30:
21:
3720:Biogeography
3694:R. A. Fisher
3572:Heritability
3505:Key concepts
3425:F-statistics
3361:
3357:
3347:
3314:
3308:
3298:
3282:
3277:
3230:
3226:
3216:
3170:(5): e5472.
3167:
3163:
3105:
3101:
3091:
3082:
3057:
3053:
3047:
3035:
3030:
2989:
2985:
2979:
2947:(2): 583–9.
2944:
2940:
2930:
2918:. Retrieved
2907:. Springer.
2903:
2896:
2853:
2849:
2839:
2826:
2793:Popoolation2
2718:
2580:0.0040-0055
2361:
1442:
1438:
1432:
1430:
1417:
1415:
1401:
1390:
1359:
1219:
1140:
1128:
994:
904:
484:
254:
239:
211:
184:
87:
76:
63:F-statistics
32:
28:
26:
3673:Coalescence
2768:(R package)
2762:(R package)
2756:(R package)
2382:(combining
2376:East Asians
2206:New Guinean
1590:Nio-Saharan
1537:New Guinean
1460:Nio-Saharan
3797:Categories
3615:Ecological
3605:Artificial
2920:25 October
2818:References
2280:Australian
2135:Melanesian
1940:S. Chinese
1631:W. African
1544:Australian
1530:Melanesian
1509:S. Chinese
1467:W. African
1386:Sardinians
1374:Portuguese
1137:Estimation
236:Definition
3725:Evolution
3592:Selection
3339:205970662
3271:see table
3247:1076-1551
3210:see table
3006:0014-3820
2986:Evolution
2870:1471-0056
2812:BioPython
2760:hierfstat
2754:diveRsity
2626:Russians
2486:Russians
2468:Italians
2372:Europeans
1378:Spaniards
1372:, or the
1348:in humans
1314:π
1287:π
1256:π
1229:π
1197:π
1186:π
1182:−
1173:π
1097:−
1091:≈
1036:¯
974:¯
965:−
954:¯
945:−
881:¯
872:−
860:¯
847:¯
837:−
822:−
813:¯
804:−
792:¯
768:¯
759:−
747:¯
723:−
694:∑
691:−
682:¯
673:−
661:¯
513:−
461:¯
452:−
440:¯
419:σ
397:σ
382:σ
325:σ
293:σ
269:¯
224:is not a
197:μ
161:−
123:≈
118:μ
54:(SNP) or
3749:genomics
3687:Founders
3414:See also
3388:17660541
3358:Genetics
3331:21565057
3265:19707526
3204:19424496
3164:PLOS ONE
3142:23185452
3102:PLOS ONE
3074:15208802
3022:28563791
2941:Genetics
2888:19687804
2784:VCFtools
2775:Archived
2741:Arlequin
2598:Germans
2571:Spanish
2521:Swedish
2483:Germans
2480:Spanish
2474:Swedish
2396:panmixia
1881:Filipino
1722:Japanese
1502:Filipino
1481:Japanese
1376:vs. the
1368:vs. the
486:Wright's
226:distance
83:panmixia
60:Wright's
3600:Natural
3567:Fitness
3379:2034649
3256:2730349
3195:2675054
3172:Bibcode
3133:3504095
3110:Bibcode
3014:2408641
2971:1427045
2962:1205159
2879:4687486
2807:BioPerl
2766:FinePop
2704:0.0108
2701:0.0039
2698:0.0035
2693:0.0084
2690:0.0057
2687:0.0000
2684:Greeks
2675:0.0050
2672:0.0010
2669:0.0010
2663:0.0020
2655:French
2632:0.0202
2604:0.0136
2577:0.0101
2527:0.0191
2500:0.0064
2492:Greeks
2489:French
2067:English
1523:English
1397:Papuans
1291:Between
1233:Between
1220:where
1201:Between
1177:Between
1063:: Let T
625:, then
185:where
42:due to
3610:Sexual
3386:
3376:
3337:
3329:
3263:
3253:
3245:
3202:
3192:
3140:
3130:
3072:
3040:p. 182
3020:
3012:
3004:
2969:
2959:
2911:
2886:
2876:
2868:
2546:Finns
2477:Finns
2454:0.007
2451:0.192
2448:0.110
2438:0.190
2435:0.111
2423:0.153
2392:Yoruba
2388:Maasai
2002:Danish
1772:Korean
1516:Danish
1488:Korean
1318:Within
1260:Within
1190:Within
995:where
3335:S2CID
3305:smogd
3070:S2CID
3010:JSTOR
2789:DnaSP
2749:SMOGD
2745:Fstat
2384:Luhya
1675:Mbuti
1552:Bantu
1474:Mbuti
1453:Bantu
1393:Mbuti
1370:Dutch
1366:Danes
3747:and
3384:PMID
3327:PMID
3261:PMID
3243:ISSN
3200:PMID
3138:PMID
3018:PMID
3002:ISSN
2967:PMID
2922:2012
2909:ISBN
2884:PMID
2866:ISSN
2350:0.00
2345:0.10
2340:0.11
2335:0.15
2330:0.14
2325:0.11
2320:0.13
2315:0.13
2310:0.07
2305:0.06
2300:0.43
2295:0.27
2290:0.36
2285:0.33
2271:0.00
2266:0.07
2261:0.16
2256:0.16
2251:0.15
2246:0.13
2241:0.18
2236:0.14
2231:0.12
2226:0.46
2221:0.28
2216:0.33
2211:0.34
2195:0.00
2190:0.16
2185:0.14
2180:0.06
2175:0.05
2170:0.08
2165:0.11
2160:0.11
2155:0.40
2150:0.27
2145:0.31
2140:0.34
2122:0.00
2117:0.00
2112:0.12
2107:0.11
2102:0.11
2097:0.10
2092:0.12
2087:0.24
2082:0.15
2077:0.18
2072:0.23
2052:0.00
2047:0.13
2042:0.13
2037:0.13
2032:0.09
2027:0.12
2022:0.15
2017:0.15
2012:0.17
2007:0.17
1985:0.00
1980:0.03
1975:0.01
1970:0.05
1965:0.05
1960:0.34
1955:0.20
1950:0.29
1945:0.30
1921:0.00
1916:0.06
1911:0.12
1906:0.10
1901:0.38
1896:0.23
1891:0.30
1886:0.29
1860:0.00
1855:0.06
1850:0.07
1845:0.39
1840:0.25
1835:0.30
1830:0.34
1825:Thai
1802:0.00
1797:0.01
1792:0.30
1787:0.18
1782:0.24
1777:0.27
1747:0.00
1742:0.31
1737:0.23
1732:0.25
1727:0.24
1695:0.00
1690:0.08
1685:0.08
1680:0.07
1646:0.00
1641:0.02
1636:0.02
1600:0.00
1595:0.01
1557:0.00
1495:Thai
1395:and
1384:vs.
1382:Sámi
1247:and
27:The
3374:PMC
3366:doi
3362:177
3319:doi
3251:PMC
3235:doi
3190:PMC
3180:doi
3128:PMC
3118:doi
3062:doi
2994:doi
2957:PMC
2949:doi
2945:132
2874:PMC
2858:doi
1388:).
255:If
92:, F
3799::
3382:.
3372:.
3360:.
3356:.
3333:.
3325:.
3315:10
3313:.
3289:.
3269:,
3259:.
3249:.
3241:.
3231:15
3229:.
3225:.
3208:,
3198:.
3188:.
3178:.
3166:.
3162:.
3150:^
3136:.
3126:.
3116:.
3104:.
3100:.
3068:.
3058:24
3056:.
3016:.
3008:.
3000:.
2990:38
2988:.
2965:.
2955:.
2943:.
2939:.
2882:.
2872:.
2864:.
2854:10
2852:.
2848:.
2800:ST
2734:ST
2368:ST
1399:.
1362:ST
1346:ST
1334:ST
1133:.
1131:ST
1053:ST
490:ST
350:ST
252:.
242:ST
232:.
222:ST
218:ST
214:ST
94:ST
35:ST
3490:e
3483:t
3476:v
3390:.
3368::
3341:.
3321::
3293:.
3267:.
3237::
3206:.
3182::
3174::
3168:4
3144:.
3120::
3112::
3106:7
3076:.
3064::
3042:.
3024:.
2996::
2973:.
2951::
2924:.
2890:.
2860::
1360:F
1344:F
1166:=
1161:T
1158:S
1154:F
1112:T
1107:0
1103:T
1094:1
1086:T
1083:S
1079:F
1065:0
1033:f
1008:0
1004:f
971:f
962:1
951:f
940:0
936:f
929:=
924:T
921:S
917:F
887:)
878:p
869:1
866:(
857:p
843:)
840:p
834:1
831:(
828:p
819:)
810:p
801:1
798:(
789:p
780:=
774:)
765:p
756:1
753:(
744:p
736:)
731:i
727:p
720:1
717:(
712:i
708:p
702:i
698:c
688:)
679:p
670:1
667:(
658:p
649:=
644:T
641:S
637:F
611:i
607:c
586:i
564:i
560:p
539:i
519:)
516:p
510:1
507:(
504:p
501:2
467:)
458:p
449:1
446:(
437:p
428:2
423:S
413:=
406:2
401:T
391:2
386:S
376:=
371:T
368:S
364:F
334:2
329:T
302:2
297:S
266:p
187:m
181:,
168:)
164:1
154:T
151:S
147:F
143:1
137:(
131:4
128:1
115:m
110:=
107:M
33:F
31:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.