278:
33:
294:
437:
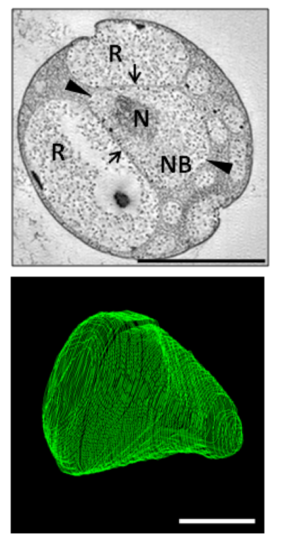
303:. This reconstruction suggests a closed membrane around the nuclear body. The top panel labels the nuclear body (NB), nuclear DNA (N), and riboplasm (R). The nuclear body appears surrounded by a membrane that is a single layer in some places (arrowheads) and a double membrane in others (arrows). Top panel scale bar = 1µm, bottom panel scale bar = 500nm.
651:-dependent, likely receptor-mediated import of extracellular proteins has been observed under laboratory conditions, although it is of unknown functional significance. This may suggest that Planctomycetota and eukaryote endocytosis mechanism share a common evolutionary origin, that the two processes may be an example of
481:
It has been reported that the budding process involves transfer of naked DNA to the daughter cell, after which it is then surrounded by a nucleoid membrane. However, three-dimensional reconstructions indicate that DNA is never surrounded by a closed membrane in a newly created bud, but instead is
376:
membranes are continuous and do not enclose distinct cellular compartments. However, this study has been criticized for not detecting or modeling some commonly reported structural features, and a 2014 study using similar methodology was interpreted as supporting the earlier hypothesis of closed
473:
phase during which budding occurs, although these are less distinct than in other
Planctomycetota whose life cycles have been studied. Observations of individual cells in culture found that approximately 12 hours were required for bud maturation and separation, followed by an asymmetrical
365:; the lineage that gave rise to the PVC superphylum is speculated to be related to an "intermediate" state between prokaryotes and eukaryotes. The question of how the PVC superfamily membranes are organized and how they relate to eukaryotes is an active and controversial area of research.
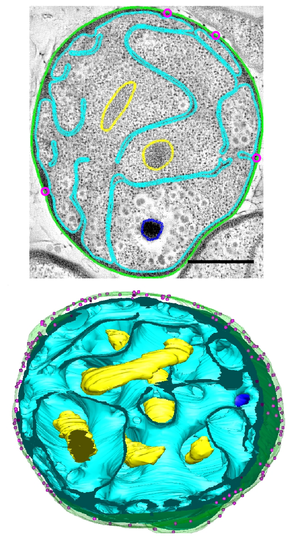
287:. This reconstruction suggests continuous, non-enclosed membranes. The outer membrane is shown in green, the inner membrane in cyan, the DNA in yellow, a poly-phosphate granule in blue, and membrane cavitation in pink. Scale bar = 500nm.
444:
in the process of dividing by budding. Labels indicate the nucleoid (N) of the mother (larger) and daughter (smaller) cells, and the nucleoid envelope (NE) of the daughter cell, which is described as not yet fully formed. Scale bar =
663:; of the three possibilities, the latter is considered unlikely due to statistical features of the bacterial genes associated with the process. There is disagreement over the possibility that proteins with
2105:
701:
Santarella-Mellwig, Rachel; Franke, Josef; Jaedicke, Andreas; Gorjanacz, Matyas; Bauer, Ulrike; Budd, Aidan; Mattaj, Iain W.; Devos, Damien P.; Schmid, Sandra L. (19 January 2010).
404:. More recent reports have found the exterior to more closely chemically resemble typical Gram-negative features. Compositional analysis of the membrane has been reported to find
2079:
1397:"Global and targeted lipid analysis of Gemmata obscuriglobus reveals the presence of lipopolysaccharide, a signature of the classical Gram-negative outer membrane"
1444:
Jeske, O; Schüler, M; Schumann, P; Schneider, A; Boedeker, C; Jogler, M; Bollschweiler, D; Rohde, M; Mayer, C; Engelhardt, H; Spring, S; Jogler, C (12 May 2015).
2118:
592:
have been reported to occur in spatially segregated locations within the cell, which is otherwise characteristic of eukaryotic but not prokaryotic cells.
353:
and other members of the PVC group possess closed internal membranes and therefore have a unique "cell plan" is considered important in understanding the
318:
system, including deep invaginations of its membrane that historically have been described as closed internal membranes that may surround the bacterium's
277:
293:
2066:
2092:
1880:
Lonhienne, T. G. A.; Sagulenko, E.; Webb, R. I.; Lee, K.-C.; Franke, J.; Devos, D. P.; Nouwens, A.; Carroll, B. J.; Fuerst, J. A. (21 June 2010).
810:
682:
may also be involved in its capacity for membrane invaginations and endocytosis because sterols are known to facilitate membrane deformation.
612:, a process critical to the maintenance of eukaryotic cell membranes and ubiquitous in eukaryotes. The sterols identified in the bacterium,
1563:"Genome-Wide Influence of Indel Substitutions on Evolution of Bacteria of the PVC Superphylum, Revealed Using a Novel Computational Method"
2164:
538:
in protein-coding genes of the PVC grouping identified a number of biochemical pathways with unusually high numbers of indels in the
436:
703:"The Compartmentalized Bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum Have Membrane Coat-Like Proteins"
530:. It possesses unusual genetic infrastructure, lacking a key component of most bacterial cell division processes, the protein
2097:
1712:"Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus"
806:
32:
763:
Franzmann, P. D.; Skerman, V. B. D. (May 1984). "Gemmata obscuriglobus, a new genus and species of the budding bacteria".
1612:"Electron tomography of the nucleoid of Gemmata obscuriglobus reveals complex liquid crystalline cholesteric structure"
1323:"Towards understanding the molecular mechanism of the endocytosis-like process in the bacterium Gemmata obscuriglobus"
1215:
Devos, DP (February 2014). "Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin".
1123:
McInerney, JO; Martin, WF; Koonin, EV; Allen, JF; Galperin, MY; Lane, N; Archibald, JM; Embley, TM (November 2011).
1833:"Phylogenomics of Sterol Synthesis: Insights into the Origin, Evolution, and Diversity of a Key Eukaryotic Feature"
1271:
Devos, Damien P. (January 2014). "PVC bacteria: variation of, but not exception to, the Gram-negative cell plan".
511:
2123:
1364:"16S ribosomal RNA- and cell wall analysis of Gemmata obscuriglobus, a new member of the order Planctomycetales"
2159:
660:
643:
was the first bacterium shown to possess a mechanism for protein import into the cell, analogous to eukaryotic
581:
413:
215:
occur in freshwater habitats and was first described in 1984, and is the only described species in its genus.
329:. The concept of membrane-bound genetic material has been described as a "cell plan" unique to a proposed PVC
1995:
648:
585:
161:
478:
in which mother cells were quicker to begin a new budding cycle than were newly budded daughter cells.
2053:
1785:
1723:
1457:
1016:
957:
652:
629:
222:
1774:"Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus"
1104:
Fuerst, John A. (2010). "Beyond
Prokaryotes and Eukaryotes: Planctomycetes and Cell Organization".
470:
40:
1976:
397:
and instead possessed a proteinaceous exterior layer, later described as possibly analogous to an
1956:
1250:
1197:
788:
664:
405:
52:
420:
identified the presence of a PG cell wall following the typical Gram-negative structure by both
2084:
299:
Electron micrograph (top) and three-dimensional reconstruction of the nuclear body (bottom) of
2131:
2040:
1948:
1913:
1862:
1813:
1751:
1692:
1643:
1610:
Yee, Benjamin; Sagulenko, Evgeny; Morgan, Garry P.; Webb, Richard I.; Fuerst, John A. (2012).
1592:
1543:
1483:
1426:
1344:
1298:
1242:
1189:
1154:
1086:
1044:
985:
919:
861:
780:
734:
543:
461:
more commonly observed in bacterial species. It is relatively slow-growing, with an estimated
458:
338:
229:, often considered to represent large differences in internal organization compared with most
1063:
2136:
1940:
1903:
1893:
1852:
1844:
1803:
1793:
1741:
1731:
1682:
1674:
1633:
1623:
1582:
1574:
1533:
1523:
1473:
1465:
1416:
1408:
1375:
1334:
1288:
1280:
1232:
1224:
1181:
1144:
1136:
1078:
1034:
1024:
975:
965:
909:
899:
851:
772:
724:
714:
372:
reconstructions of whole cells reported in 2013 suggest that contrary to historical belief,
357:
of membrane-bound compartments, which are often considered a distinguishing feature between
194:
1172:
Fuerst, JA (October 2013). "The PVC superphylum: exceptions to the bacterial definition?".
486:
after the "neck" between the mother and daughter cell membranes, initially as narrow as 30
462:
334:
208:
76:
264:
Dense, compact DNA and a deeply invaginated membrane are characteristics of the species.
1789:
1727:
1461:
1020:
961:
1908:
1881:
1857:
1832:
1746:
1711:
1687:
1662:
1638:
1611:
1587:
1562:
1538:
1507:
1478:
1445:
1421:
1396:
1380:
1363:
1149:
1124:
1064:"The planctomycetes: emerging models for microbial ecology, evolution and cell biology"
980:
941:
914:
887:
729:
702:
567:
559:
90:
1808:
1773:
283:
Electron micrograph (top) and whole-cell three-dimensional reconstruction (bottom) of
2153:
2110:
1395:
Mahat, Rajendra; Seebart, Corrine; Basile, Franco; Ward, Naomi L. (19 October 2015).
1039:
1004:
425:
390:
346:
315:
198:
190:
1960:
1663:"Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus"
1254:
1201:
792:
2045:
621:
466:
326:
2018:
1339:
1322:
970:
904:
888:"Three-dimensional reconstruction of bacteria with a complex endomembrane system"
719:
554:
One or more nucleoid-like regions of densely compact DNA is commonly observed in
1082:
644:
421:
342:
330:
118:
1009:
Proceedings of the
National Academy of Sciences of the United States of America
1284:
1228:
1185:
886:
Santarella-Mellwig, R; Pruggnaller, S; Roos, N; Mattaj, IW; Devos, DP (2013).
856:
839:
613:
527:
362:
323:
249:
230:
104:
1628:
1898:
1798:
1736:
1029:
571:
563:
487:
483:
475:
394:
358:
354:
201:
1952:
1931:
Jermy, Andrew (August 2010). "Evolution: Bacterial endocytosis uncovered".
1917:
1866:
1817:
1755:
1696:
1647:
1596:
1547:
1528:
1487:
1430:
1348:
1302:
1246:
1193:
1158:
1140:
1048:
989:
923:
865:
738:
1710:
Gottshall, E. Y.; Seebart, C.; Gatlin, J. C.; Ward, N. L. (14 July 2014).
1090:
784:
2012:
1848:
1578:
675:
668:
519:
369:
253:
63:
1944:
1882:"Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus"
1678:
1412:
1293:
1237:
940:
Sagulenko, E; Morgan, GP; Webb, RI; Yee, B; Lee, KC; Fuerst, JA (2014).
620:, are relatively simple compared to eukaryotic sterols; as indicated by
2071:
2032:
2027:
1469:
776:
617:
454:
401:
398:
1661:
Lieber, A; Leis, A; Kushmaro, A; Minsky, A; Medalia, O (March 2009).
679:
609:
515:
507:
226:
205:
1989:
1362:
Stackebrandt, Erko; Wehmeyer, Uta; Liesack, Werner (December 1986).
1005:"Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus"
570:. The structure of the nucleoid has been implicated in the unusual
248:
is a large, roughly spherical bacterium with a cell diameter of 1–2
632:
was among the most primitive known at the time it was identified.
535:
435:
2058:
1561:
Kamneva, O. K.; Liberles, D. A.; Ward, N. L. (3 November 2010).
589:
531:
523:
1993:
1125:"Planctomycetes and eukaryotes: a case of analogy not homology"
469:. Its life cycle consists of a motile or "swarmer" phase and a
319:
1327:
Biochimica et
Biophysica Acta (BBA) - Molecular Cell Research
562:
has been reported, with some structural similarities to the
1772:
Pearson, A.; Budin, M.; Brocks, J. J. (5 December 2003).
314:
is its highly complex and morphologically distinctive
1506:
Lee, Kuo-Chang; Webb, Rick I; Fuerst, John A (2009).
1446:"Planctomycetes do possess a peptidoglycan cell wall"
490:, widens sufficiently to accommodate condensed DNA.
416:. A 2015 study of several Planctomycetota including
412:, consistent with typical features of Gram-negative
2002:
1321:Fuerst, John A.; Sagulenko, Evgeny (August 2014).
608:is one of the few prokaryotes known to synthesize
946:support cell compartmentalisation in a bacterium"
696:
694:
518:by the standards of other PVC bacteria, around 9
482:free to diffuse from the mother to daughter cell
389:found that it lacked a traditional Gram-negative
659:acquired its endocytotic infrastructure through
1886:Proceedings of the National Academy of Sciences
1831:Desmond, E.; Gribaldo, S. (10 September 2009).
1778:Proceedings of the National Academy of Sciences
1716:Proceedings of the National Academy of Sciences
1266:
1264:
558:cells. Complex internal structure resembling a
1316:
1314:
1312:
8:
881:
879:
877:
875:
935:
933:
758:
756:
754:
752:
750:
748:
1990:
1767:
1765:
1512:with respect to cell compartmentalization"
1003:Fuerst, JA; Webb, RI (15 September 1991).
31:
20:
1907:
1897:
1856:
1807:
1797:
1745:
1735:
1686:
1637:
1627:
1586:
1537:
1527:
1477:
1420:
1379:
1338:
1292:
1236:
1148:
1038:
1028:
979:
969:
913:
903:
855:
728:
718:
678:. The bacterium's ability to synthesize
47:internal morphology. Scale bar = 500nm.
43:of representative examples illustrating
1985:- the Bacterial Diversity Metadatabase
833:
831:
829:
827:
811:Integrated Taxonomic Information System
690:
1501:
1499:
1497:
252:. It is motile and possesses multiple
1508:"The cell cycle of the planctomycete
942:"Structural studies of planctomycete
7:
225:and for the unusual features in its
838:Devos, Damien P. (September 2013).
310:Among the most notable features of
1381:10.1111/j.1574-6968.1986.tb01810.x
381:Membrane and cell wall composition
349:bacteria. The question of whether
345:and distinct from the rest of the
14:
465:of around 13 hours based on bulk
499:Genomic content and organization
292:
276:
221:is exceptional for its unusual
1:
1837:Genome Biology and Evolution
1567:Genome Biology and Evolution
1340:10.1016/j.bbamcr.2013.10.002
971:10.1371/journal.pone.0091344
905:10.1371/journal.pbio.1001565
720:10.1371/journal.pbio.1000281
514:. The bacterium has a large
1933:Nature Reviews Microbiology
1083:10.1099/13500872-141-7-1493
522:, and contains about 8,000
449:Like most Planctomycetota,
2181:
2165:Bacteria described in 1985
385:Early characterization of
177:Franzmann and Skerman 1985
140:Franzmann and Skerman 1985
1616:Frontiers in Microbiology
1368:FEMS Microbiology Letters
1285:10.1016/j.tim.2013.10.008
1229:10.1007/s10482-013-0087-y
1186:10.1007/s10482-013-9986-1
857:10.1016/j.cub.2013.07.013
512:J. Craig Venter Institute
260:Internal cell composition
241:Cell shape and appendages
167:
160:
53:Scientific classification
51:
39:
30:
23:
1629:10.3389/fmicb.2012.00326
1062:Fuerst, JA (July 1995).
661:horizontal gene transfer
236:Structure and Morphology
1899:10.1073/pnas.1001085107
1799:10.1073/pnas.2536559100
1737:10.1073/pnas.1409187111
1667:Journal of Bacteriology
1401:Journal of Bacteriology
1217:Antonie van Leeuwenhoek
1174:Antonie van Leeuwenhoek
1030:10.1073/pnas.88.18.8184
840:"Gemmata obscuriglobus"
807:"Gemmata obscuriglobus"
765:Antonie van Leeuwenhoek
671:are represented in the
494:Genetic Characteristics
440:Electron micrograph of
377:internal compartments.
1529:10.1186/1471-2121-10-4
1273:Trends in Microbiology
1141:10.1002/bies.201100045
566:of eukaryotes such as
446:
2111:gemmata-obscuriglobus
2004:Gemmata obscuriglobus
1979:Gemmata obscuriglobus
1510:Gemmata obscuriglobus
1450:Nature Communications
944:Gemmata obscuriglobus
510:was sequenced by the
439:
301:Gemmata obscuriglobus
285:Gemmata obscuriglobus
186:Gemmata obscuriglobus
171:Gemmata obscuriglobus
153:G. obscuriglobus
25:Gemmata obscuriglobus
653:convergent evolution
630:biosynthetic pathway
41:Electron micrographs
1945:10.1038/nrmicro2408
1892:(29): 12883–12888.
1790:2003PNAS..10015352P
1784:(26): 15352–15357.
1728:2014PNAS..11111067G
1722:(30): 11067–11072.
1679:10.1128/jb.01513-08
1462:2015NatCo...6.7116J
1413:10.1128/JB.00517-15
1021:1991PNAS...88.8184F
962:2014PLoSO...991344S
16:Species of bacteria
1849:10.1093/gbe/evp036
1579:10.1093/gbe/evq071
1470:10.1038/ncomms8116
1407:(2): JB.00517–15.
777:10.1007/BF02342136
544:ribosomal proteins
542:genome, including
447:
406:lipopolysaccharide
368:Three-dimensional
268:Membrane structure
2147:
2146:
2132:Open Tree of Life
1996:Taxon identifiers
850:(17): R705–R707.
339:Verrucomicrobiota
182:
181:
2172:
2140:
2139:
2127:
2126:
2114:
2113:
2101:
2100:
2088:
2087:
2075:
2074:
2062:
2061:
2049:
2048:
2036:
2035:
2023:
2022:
2021:
1991:
1965:
1964:
1928:
1922:
1921:
1911:
1901:
1877:
1871:
1870:
1860:
1828:
1822:
1821:
1811:
1801:
1769:
1760:
1759:
1749:
1739:
1707:
1701:
1700:
1690:
1658:
1652:
1651:
1641:
1631:
1607:
1601:
1600:
1590:
1558:
1552:
1551:
1541:
1531:
1516:BMC Cell Biology
1503:
1492:
1491:
1481:
1441:
1435:
1434:
1424:
1392:
1386:
1385:
1383:
1359:
1353:
1352:
1342:
1333:(8): 1732–1738.
1318:
1307:
1306:
1296:
1268:
1259:
1258:
1240:
1212:
1206:
1205:
1169:
1163:
1162:
1152:
1120:
1114:
1113:
1106:Nature Education
1101:
1095:
1094:
1068:
1059:
1053:
1052:
1042:
1032:
1000:
994:
993:
983:
973:
937:
928:
927:
917:
907:
883:
870:
869:
859:
835:
822:
821:
819:
817:
803:
797:
796:
760:
743:
742:
732:
722:
698:
673:G. obscuriglobus
657:G. obscuriglobus
641:G. obscuriglobus
626:G. obscuriglobus
606:G. obscuriglobus
601:Sterol synthesis
576:G. obscuriglobus
556:G. obscuriglobus
540:G. obscuriglobus
505:G. obscuriglobus
457:rather than the
451:G. obscuriglobus
442:G. obscuriglobus
418:G. obscuriglobus
410:G. obscuriglobus
387:G. obscuriglobus
374:G. obscuriglobus
351:G. obscuriglobus
333:composed of the
322:by analogy to a
312:G. obscuriglobus
296:
280:
246:G. obscuriglobus
219:G. obscuriglobus
213:G. obscuriglobus
189:is a species of
173:
141:
45:G. obscuriglobus
35:
21:
2180:
2179:
2175:
2174:
2173:
2171:
2170:
2169:
2160:Planctomycetota
2150:
2149:
2148:
2143:
2135:
2130:
2122:
2117:
2109:
2104:
2096:
2091:
2083:
2078:
2070:
2065:
2057:
2052:
2044:
2039:
2031:
2026:
2017:
2016:
2011:
1998:
1977:Type strain of
1973:
1968:
1930:
1929:
1925:
1879:
1878:
1874:
1830:
1829:
1825:
1771:
1770:
1763:
1709:
1708:
1704:
1660:
1659:
1655:
1609:
1608:
1604:
1560:
1559:
1555:
1505:
1504:
1495:
1443:
1442:
1438:
1394:
1393:
1389:
1361:
1360:
1356:
1320:
1319:
1310:
1270:
1269:
1262:
1214:
1213:
1209:
1171:
1170:
1166:
1122:
1121:
1117:
1103:
1102:
1098:
1077:(7): 1493–506.
1066:
1061:
1060:
1056:
1002:
1001:
997:
939:
938:
931:
898:(5): e1001565.
885:
884:
873:
844:Current Biology
837:
836:
825:
815:
813:
805:
804:
800:
762:
761:
746:
713:(1): e1000281.
700:
699:
692:
688:
638:
603:
598:
568:dinoflagellates
552:
501:
496:
463:generation time
434:
414:outer membranes
383:
335:Planctomycetota
308:
307:
306:
305:
304:
297:
289:
288:
281:
270:
262:
243:
238:
209:Planctomycetota
178:
175:
169:
156:
142:
139:
137:
136:
122:
108:
94:
80:
77:Planctomycetota
66:
17:
12:
11:
5:
2178:
2176:
2168:
2167:
2162:
2152:
2151:
2145:
2144:
2142:
2141:
2128:
2115:
2102:
2089:
2076:
2063:
2050:
2037:
2024:
2008:
2006:
2000:
1999:
1994:
1988:
1987:
1972:
1971:External links
1969:
1967:
1966:
1939:(8): 534–535.
1923:
1872:
1823:
1761:
1702:
1673:(5): 1439–45.
1653:
1602:
1553:
1493:
1436:
1387:
1374:(3): 289–292.
1354:
1308:
1260:
1207:
1164:
1115:
1096:
1054:
1015:(18): 8184–8.
995:
929:
871:
823:
798:
771:(3): 261–268.
744:
689:
687:
684:
637:
634:
624:analysis, the
602:
599:
597:
594:
560:liquid crystal
551:
548:
500:
497:
495:
492:
453:reproduces by
433:
430:
382:
379:
298:
291:
290:
282:
275:
274:
273:
272:
271:
269:
266:
261:
258:
242:
239:
237:
234:
180:
179:
176:
165:
164:
158:
157:
150:
148:
144:
143:
138:
130:
128:
124:
123:
116:
114:
110:
109:
102:
100:
96:
95:
91:Planctomycetia
88:
86:
82:
81:
74:
72:
68:
67:
62:
60:
56:
55:
49:
48:
37:
36:
28:
27:
15:
13:
10:
9:
6:
4:
3:
2:
2177:
2166:
2163:
2161:
2158:
2157:
2155:
2138:
2133:
2129:
2125:
2120:
2116:
2112:
2107:
2103:
2099:
2094:
2090:
2086:
2081:
2077:
2073:
2068:
2064:
2060:
2055:
2051:
2047:
2042:
2038:
2034:
2029:
2025:
2020:
2014:
2010:
2009:
2007:
2005:
2001:
1997:
1992:
1986:
1984:
1980:
1975:
1974:
1970:
1962:
1958:
1954:
1950:
1946:
1942:
1938:
1934:
1927:
1924:
1919:
1915:
1910:
1905:
1900:
1895:
1891:
1887:
1883:
1876:
1873:
1868:
1864:
1859:
1854:
1850:
1846:
1842:
1838:
1834:
1827:
1824:
1819:
1815:
1810:
1805:
1800:
1795:
1791:
1787:
1783:
1779:
1775:
1768:
1766:
1762:
1757:
1753:
1748:
1743:
1738:
1733:
1729:
1725:
1721:
1717:
1713:
1706:
1703:
1698:
1694:
1689:
1684:
1680:
1676:
1672:
1668:
1664:
1657:
1654:
1649:
1645:
1640:
1635:
1630:
1625:
1621:
1617:
1613:
1606:
1603:
1598:
1594:
1589:
1584:
1580:
1576:
1572:
1568:
1564:
1557:
1554:
1549:
1545:
1540:
1535:
1530:
1525:
1521:
1517:
1513:
1511:
1502:
1500:
1498:
1494:
1489:
1485:
1480:
1475:
1471:
1467:
1463:
1459:
1455:
1451:
1447:
1440:
1437:
1432:
1428:
1423:
1418:
1414:
1410:
1406:
1402:
1398:
1391:
1388:
1382:
1377:
1373:
1369:
1365:
1358:
1355:
1350:
1346:
1341:
1336:
1332:
1328:
1324:
1317:
1315:
1313:
1309:
1304:
1300:
1295:
1290:
1286:
1282:
1278:
1274:
1267:
1265:
1261:
1256:
1252:
1248:
1244:
1239:
1234:
1230:
1226:
1222:
1218:
1211:
1208:
1203:
1199:
1195:
1191:
1187:
1183:
1180:(4): 451–66.
1179:
1175:
1168:
1165:
1160:
1156:
1151:
1146:
1142:
1138:
1135:(11): 810–7.
1134:
1130:
1126:
1119:
1116:
1111:
1107:
1100:
1097:
1092:
1088:
1084:
1080:
1076:
1072:
1065:
1058:
1055:
1050:
1046:
1041:
1036:
1031:
1026:
1022:
1018:
1014:
1010:
1006:
999:
996:
991:
987:
982:
977:
972:
967:
963:
959:
956:(3): e91344.
955:
951:
947:
945:
936:
934:
930:
925:
921:
916:
911:
906:
901:
897:
893:
889:
882:
880:
878:
876:
872:
867:
863:
858:
853:
849:
845:
841:
834:
832:
830:
828:
824:
812:
808:
802:
799:
794:
790:
786:
782:
778:
774:
770:
766:
759:
757:
755:
753:
751:
749:
745:
740:
736:
731:
726:
721:
716:
712:
708:
704:
697:
695:
691:
685:
683:
681:
677:
674:
670:
666:
662:
658:
654:
650:
646:
642:
635:
633:
631:
627:
623:
619:
615:
611:
607:
600:
595:
593:
591:
587:
583:
582:Transcription
579:
577:
574:tolerance of
573:
569:
565:
561:
557:
549:
547:
545:
541:
537:
534:. A study of
533:
529:
526:. It has 67%
525:
521:
517:
513:
509:
506:
498:
493:
491:
489:
485:
479:
477:
472:
468:
464:
460:
456:
452:
443:
438:
431:
429:
427:
426:bioinformatic
423:
419:
415:
411:
407:
403:
400:
396:
392:
391:peptidoglycan
388:
380:
378:
375:
371:
366:
364:
360:
356:
352:
348:
347:Gram-negative
344:
340:
336:
332:
328:
325:
321:
317:
316:cell membrane
313:
302:
295:
286:
279:
267:
265:
259:
257:
255:
251:
247:
240:
235:
233:
232:
228:
224:
220:
216:
214:
210:
207:
203:
200:
199:heterotrophic
196:
192:
191:Gram-negative
188:
187:
174:
172:
166:
163:
162:Binomial name
159:
155:
154:
149:
146:
145:
135:
134:
129:
126:
125:
121:
120:
115:
112:
111:
107:
106:
101:
98:
97:
93:
92:
87:
84:
83:
79:
78:
73:
70:
69:
65:
61:
58:
57:
54:
50:
46:
42:
38:
34:
29:
26:
22:
19:
2003:
1982:
1978:
1936:
1932:
1926:
1889:
1885:
1875:
1840:
1836:
1826:
1781:
1777:
1719:
1715:
1705:
1670:
1666:
1656:
1619:
1615:
1605:
1570:
1566:
1556:
1519:
1515:
1509:
1453:
1449:
1439:
1404:
1400:
1390:
1371:
1367:
1357:
1330:
1326:
1294:10261/129431
1279:(1): 14–20.
1276:
1272:
1238:10261/129395
1223:(2): 271–4.
1220:
1216:
1210:
1177:
1173:
1167:
1132:
1128:
1118:
1109:
1105:
1099:
1074:
1071:Microbiology
1070:
1057:
1012:
1008:
998:
953:
949:
943:
895:
892:PLOS Biology
891:
847:
843:
814:. Retrieved
801:
768:
764:
710:
707:PLOS Biology
706:
672:
656:
640:
639:
625:
622:phylogenetic
605:
604:
580:
575:
555:
553:
539:
504:
502:
480:
467:cell culture
450:
448:
441:
432:Reproduction
417:
409:
386:
384:
373:
367:
350:
327:cell nucleus
311:
309:
300:
284:
263:
245:
244:
218:
217:
212:
185:
184:
183:
170:
168:
152:
151:
132:
131:
117:
103:
89:
75:
44:
24:
18:
1843:: 364–381.
1573:: 870–886.
645:endocytosis
636:Endocytosis
586:translation
422:biochemical
363:prokaryotes
343:Chlamydiota
331:superphylum
256:per cell.
231:prokaryotes
119:Gemmataceae
2154:Categories
816:30 October
686:References
655:, or that
647:. Active,
614:lanosterol
596:Physiology
528:GC content
488:nanometers
428:analysis.
359:eukaryotes
324:eukaryotic
223:morphology
105:Gemmatales
2019:Q21349510
1129:BioEssays
669:clathrins
572:radiation
564:chromatin
550:Nucleoids
520:megabases
484:cytoplasm
476:lag phase
395:cell wall
355:evolution
147:Species:
2085:10798519
2013:Wikidata
1961:12013205
1953:20665955
1918:20566852
1867:20333205
1818:14660793
1756:25024214
1697:19074379
1648:22993511
1597:21048002
1548:19144151
1522:(1): 4.
1488:25964217
1456:: 7116.
1431:26483522
1349:24144586
1303:24286661
1255:16557669
1247:24292377
1202:14283647
1194:23912444
1159:21858844
1112:(9): 44.
1049:11607213
990:24632833
950:PLOS ONE
924:23700385
866:24028944
793:24707205
739:20087413
676:proteome
665:homology
399:archaeal
370:tomogram
254:flagella
202:bacteria
113:Family:
71:Phylum:
64:Bacteria
59:Domain:
2072:3228999
2059:1001488
2028:BacDive
1909:2919973
1858:2817430
1786:Bibcode
1747:4121771
1724:Bibcode
1688:2648202
1639:3440768
1622:: 326.
1588:3000692
1539:2656463
1479:4432640
1458:Bibcode
1422:4751799
1150:3795523
1091:7551018
1017:Bibcode
981:3954628
958:Bibcode
915:3660258
785:6486770
730:2799638
680:sterols
628:sterol
618:parkeol
610:sterols
471:sessile
459:fission
455:budding
402:S-layer
204:of the
195:aerobic
133:Gemmata
127:Genus:
99:Order:
85:Class:
2137:686207
2098:961865
1981:at Bac
1959:
1951:
1916:
1906:
1865:
1855:
1816:
1809:307571
1806:
1754:
1744:
1695:
1685:
1646:
1636:
1595:
1585:
1546:
1536:
1486:
1476:
1429:
1419:
1347:
1301:
1253:
1245:
1200:
1192:
1157:
1147:
1089:
1047:
1037:
988:
978:
922:
912:
864:
791:
783:
737:
727:
536:indels
516:genome
508:genome
341:, and
227:genome
206:phylum
2080:IRMNG
2046:3FK9R
2033:11936
1957:S2CID
1251:S2CID
1198:S2CID
1067:(PDF)
1040:52471
789:S2CID
590:genes
524:genes
393:(PG)
2119:NCBI
2106:LPSN
2093:ITIS
2067:GBIF
1983:Dive
1949:PMID
1914:PMID
1863:PMID
1814:PMID
1752:PMID
1693:PMID
1644:PMID
1593:PMID
1544:PMID
1484:PMID
1427:PMID
1345:PMID
1331:1843
1299:PMID
1243:PMID
1190:PMID
1155:PMID
1087:PMID
1045:PMID
986:PMID
920:PMID
862:PMID
818:2015
781:PMID
735:PMID
616:and
584:and
532:FtsZ
503:The
445:1μm.
424:and
361:and
2124:114
2054:EoL
2041:CoL
1941:doi
1904:PMC
1894:doi
1890:107
1853:PMC
1845:doi
1804:PMC
1794:doi
1782:100
1742:PMC
1732:doi
1720:111
1683:PMC
1675:doi
1671:191
1634:PMC
1624:doi
1583:PMC
1575:doi
1534:PMC
1524:doi
1474:PMC
1466:doi
1417:PMC
1409:doi
1405:198
1376:doi
1335:doi
1289:hdl
1281:doi
1233:hdl
1225:doi
1221:105
1182:doi
1178:104
1145:PMC
1137:doi
1079:doi
1075:141
1035:PMC
1025:doi
976:PMC
966:doi
910:PMC
900:doi
852:doi
773:doi
725:PMC
715:doi
667:to
649:ATP
588:of
408:in
320:DNA
2156::
2134::
2121::
2108::
2095::
2082::
2069::
2056::
2043::
2030::
2015::
1955:.
1947:.
1935:.
1912:.
1902:.
1888:.
1884:.
1861:.
1851:.
1839:.
1835:.
1812:.
1802:.
1792:.
1780:.
1776:.
1764:^
1750:.
1740:.
1730:.
1718:.
1714:.
1691:.
1681:.
1669:.
1665:.
1642:.
1632:.
1618:.
1614:.
1591:.
1581:.
1569:.
1565:.
1542:.
1532:.
1520:10
1518:.
1514:.
1496:^
1482:.
1472:.
1464:.
1452:.
1448:.
1425:.
1415:.
1403:.
1399:.
1372:37
1370:.
1366:.
1343:.
1329:.
1325:.
1311:^
1297:.
1287:.
1277:22
1275:.
1263:^
1249:.
1241:.
1231:.
1219:.
1196:.
1188:.
1176:.
1153:.
1143:.
1133:33
1131:.
1127:.
1108:.
1085:.
1073:.
1069:.
1043:.
1033:.
1023:.
1013:88
1011:.
1007:.
984:.
974:.
964:.
952:.
948:.
932:^
918:.
908:.
896:11
894:.
890:.
874:^
860:.
848:23
846:.
842:.
826:^
809:.
787:.
779:.
769:50
767:.
747:^
733:.
723:.
709:.
705:.
693:^
578:.
546:.
337:,
250:μm
211:.
197:,
193:,
1963:.
1943::
1937:8
1920:.
1896::
1869:.
1847::
1841:1
1820:.
1796::
1788::
1758:.
1734::
1726::
1699:.
1677::
1650:.
1626::
1620:3
1599:.
1577::
1571:2
1550:.
1526::
1490:.
1468::
1460::
1454:6
1433:.
1411::
1384:.
1378::
1351:.
1337::
1305:.
1291::
1283::
1257:.
1235::
1227::
1204:.
1184::
1161:.
1139::
1110:3
1093:.
1081::
1051:.
1027::
1019::
992:.
968::
960::
954:9
926:.
902::
868:.
854::
820:.
795:.
775::
741:.
717::
711:8
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.