503:
1681:
Planctomycetota can survive on high-molecular-weight polysaccharides as their only source of carbon, meaningthey must have the ability to incorporate complex carbon substrates into their cytoplasm. Three hypotheses have been put forth: First, the
Planctomycetota excrete an enzyme which, outside of the cell wall, degrades the complex substrates into smaller monosaccharides, which can more easily be transported through the different membranes. Second, the complex substrates become anchored to the outside of Planctomycetota, which are then able to slowly break down these substrates into oligosaccharides, which are able to be transported into the periplasm of Planctomycetota by specialized proteins. The third hypothesis involves the crateriform structures found on the outside of Planctomycetota cell walls. These structures have fibers lining their pits that may be able to absorb whole polysaccharides into the periplasm, where they would then be digested.
1511:
788:, regulatory systems, and protein interaction pathways. They are related to the functional organisation of the cell, which can be interpreted as an adaptation to a more complex lifestyle. The protein length is longer in the Gemmataceae than in most other bacteria and the genes have linkers. There is an overlap between the longest proteins in Planctomycetales and the shortest proteins in eukaryotes. In the terms of gene paralogy, protein length, and protein domain structures, prokaryotes and eukaryotes do not have sharp boundaries.
1702:. Planctomycetota are unique in that they have large invaginations of their cytoplasmic membrane, pulling away from the peptidoglycan cell wall and leaving room for the periplasm. Traditionally, the cytoplasmic membrane has been thought to be responsible for controlling the osmotic pressure of bacterial cells. Yet due to the folds in the cytoplasmic membrane, and the existence of large spaces of periplasm within Planctomycetota, their peptidoglycan acts as an osmotic barrier with the periplasm being isotonic to the cytosol.
48:
525:
1690:
28:
4392:
1465:
1734:. Up to 67% of dinitrogen gas production in the ocean can be attributed to anammox and about 50% of the nitrogen gas in the atmosphere is thought to be produced from anammox. Planctomycetota are the most dominant phylum of bacteria capable of performing anammox, thus the Planctomycetota capable of performing anammox play an important role in the global cycling of nitrogen.
1711:
1447:. Many Planctomycetota have the ability to breakdown extremely complex carbohydrates, making these nutrients available to other organisms. This ability to recycle carbon has been linked to specific C1 metabolism genes observed in many Planctomycetota and are suggested to play a significant role, but this area of research is still poorly understood.
1672:-like uptake system, which would be the first instance this function has been found outside of the eukaryotic domain. However, now that the existence of a rigid peptidoglycan cell wall has been confirmed, these vesicles to be able to pass through this cell wall seems unlikely. Additionally, deletion of one of these membrane coat proteins within
1776:
about
Planctomycetota secondary metabolites. This is unexpected, as the Planctomycetota have several key features as other known producers of bioactive molecules, such as the Myxobacteria. However, a number of ongoing studies serve as various first steps in including Planctomycetota in small-molecule drug development for humans.
593:
Planctomycetota is one of the only known phyla whose members lack FtsZ proteins. Bacteria in the
Chlamydiales, also a member of the PVC superphylum, also lack FtsZ. Although bacteria in the Planctomycetota lack FtsZ, two distinct modes of cell division have been observed. Most Planctomycetota divide
473:
Bacteria in the
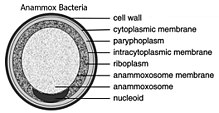
Planctomycetota that are anammox-capable form the order Brocadiales. The cells of anammox bacteria are often coccoid with a diameter of about 0.8 μm, and are suggested to contain three compartments, each surrounded by a membrane. The outer membrane encloses the cell and the protoplasm
1788:
The impacts of research on
Planctomycetota and their uses might be of global significance with regards to nutrient cycling processes and assist in furthering understanding for global marine biogeochemistry. However, with Planctomycetota's growing influences on metabolic processes involving water and
802:
Originally classified as a eukaryote due to morphology, the advent of genetic sequencing allowed researchers to agree that the
Planctomycetota belong to the domain Bacteria. Within that domain, Planctomycetota are classified as their own phylum, however, other researchers have argued they could also
520:
perform a lifestyle switch that is often associated with cell division. The sessile mother cell produces a free-swimming daughter cell. The daughter cell must then attach to a surface before starting the cycle over again. However, not all of the
Planctomycetota have a motile stage, and the lifestyle
506:
Representation of cell division modes in the last PVC common ancestor and in current the PVC superphylum: Outer membranes are represented by thick lines, and inner membranes are represented by thin lines. The peptidoglycan layer is displayed in dotted lines, and FtsZ proteins are displayed as a ring
464:
In marine environments, Planctomycetota are often suspended in the water column or present as biofilms on the surface of macroalgae, and are often exposed to harmful ultraviolet radiation. More highly pigmented species of the
Planctomycetota are more resistant to ultraviolet radiation, although this
1607:
Planctomycetota species are often associated with the surfaces of marine sponges. They interact with sponges either by attachment with a holdfast, or through a symbiotic relationship. A high diversity of
Planctomycetota is present as biofilms on sponges. The symbiotic relationship among sponges and
811:
transporters, and the
Verrucomicrobiota have also been found to have features common among eukaryotic cells. Thus, a common ancestor of this superphylum may have been the start of the eukaryotic lineage. While this is one possible explanation, because PVC is not the start of the bacterial tree, the
296:
1775:
Recently, interest has arisen in examining the Planctomycetota regarding their potential roles in biotechnology, mainly as a source of bioactive molecules, of interest mainly to the pharmaceutical industry. Bioactive compounds are mainly present as secondary metabolites, although little is known
1680:
uptake. In addition, with the use of cryoelectron tomography-based three-dimensional reconstruction of Planctomycetota has found that what were originally thought to be vesicles being held in the periplasm are actually just folds in the cytoplasmic membrane. Yet it has been demonstrated that the
1387:
communities in marine systems, and their vast distribution demonstrates their ability to inhabit many different environments. They can also adapt to both aerobic and anaerobic conditions. Many factors can affect their distribution, such as humidity, oxygen levels, and pH levels. Planctomycetota
1355:
Members of the Planctomycetota are found in a diverse range of environments, both geographically and ecologically, and occur in both aquatic and terrestrial habitats. In aquatic environments, they are found in both freshwater and marine systems. Planctomycetota were originally believed to exist
1457:, which are capable of breaking down sulfated heteropolysaccharides, which are produced by many groups of macroalgae. The breakdown of these sulfated heteropolysaccharides by Planctomycetota are then used as an energy source. Some Planctomycetota are suggested to be capable of breaking down
653:
A CSI has also been found to be shared by the entire PVC superphylum, including the Planctomycetota. Planctomycetota also contain an important conserved signature protein that has been characterized to play an important housekeeping function that is exclusive to members belonging to the PVC
1721:
is the process of oxidizing ammonium where nitrite acts as the electron acceptor. This process creates energy for the organism performing the reaction in the same way humans gain energy from oxidizing glucose. In a marine environment, this ultimately removes nitrogen from the water, as
405:
displayed varying interpretations of this suggested compartmentalization. The cytosol was suggested to be separated into compartments, both the paryphoplasm and pirellulosome, by an intracytoplasmic membrane. This interpretation has since been demonstrated to be incorrect. In fact, the
299:
Crateriform structures. (A) Crateriform structures from the outside of the cell (B–E) Micrographs of crateriform structures, perpendicular to the membrane outer membrane (OM), inner membrane (IM), cytoplasm (C), and periplasm (P). Scale bars, 50
1540:
were Planctomycetota. Almost 150 Planctomycetota species have been isolated from the biofilms of macroalgae, and these communities associated with macroalgae are mainly independent of changes in geographical distribution. This would suggest a
1374:
was used to detect Planctomycetota in various environments, and Planctomycetota are found in abundance in sphagnum bogs. Some Planctomycetota were found in the digestive systems of marine lifeforms, while others tend to live among eukaryotes.
695:
can be identified near the origin. The changes of genetic material is through internal chromosomal inversion, and not through lateral gene transfer. This creates a way of diversification in the Planctomycetota variants as multiple transposon
1626:
wetlands. Sphagnum wetlands store large amounts of carbon, contributing to the global carbon cycle. Planctomycetota play a considerable role in the degradation of sphagnum, accounting for roughly 15% of the bacterial community.
253:
Bacteria in the Planctomycetota are often small, spherical cells, but a large amount of morphological variation is seen. Members of the Planctomycetota also display distinct reproductive habits, with many species dividing by
590:, the protein present in eukaryotes, and is essential for septal formation during cell division. The lack of FtsZ proteins is often lethal. Peptidoglycan also play a considerable role in cell division by binary fission.
1697:
Almost all bacteria have a cytosol following the outer shape of their peptidoglycan cell wall. Eukaryotes are different in that they have their cytosol divided into multiple compartments to create organelles such as a
2139:
Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, et al. (December 2020). "Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay".
1762:
is unique within Planctomycetota. Sterol synthesis is suggested to be associated with regulation of membrane fluidity in Planctomycetota, and has been described as essential to the proper growth and reproduction of
1555:, the association of the Planctomycetota with kelp could indicate their significant role in coastal habitats. Planctomycetota also play an important role as components of detritus in the water column, also known as
1388:
diversity and abundance are strongly associated with relative humidity. The effects of oxygen levels demonstrate the energy needs of the individual. Many species of Planctomycetota are chemoheterotrophic, including
624:
of Planctomycetota, and is the only known species to divide by lateral budding at the middle of the cell. Lastly, members of the clade Saltatorellus are capable of switching between both binary fission and budding.
265:
Interest is growing in the Planctomycetota regarding biotechnology and human applications, mainly as a source of bioactive molecules. In addition, some Planctomycetota were recently described as human pathogens.
304:
The distinct morphological characteristics of bacteria in the Planctomycetota have been discussed extensively. The common morphology is often spherical cells roughly 2 μm in diameter, as observed in the species
1779:
Planctomycetota species are worthwhile considerations in challenging the current models for the origin of the nucleus, along with other aspects of origin and evolution of the eukaryotic endomembrane system.
245:
and were only later described as bacteria in 1972. Early examination of members of the Planctomycetota suggested a cell plan differing considerably from other bacteria, although they are now confirmed as
465:
is not yet well understood. It has since been shown that Planctomycetota synthesize C30 carotenoids from squalene and that this squalene route to C30 carotenoids is the most widespread in prokaryotes.
1551:
dominate the rocky coastlines of temperature regions, and provide habitat, shelter, and food for many organisms, including the Planctomycetota. Given the considerable role of kelp forests in coastal
502:
2332:
Kohn, Timo; Rast, Patrick; Kallscheuer, Nicolai; Wiegand, Sandra; Boedeker, Christian; Jetten, Mike S. M.; Jeske, Olga; Vollmers, John; Kaster, Anne-Kristin; Rohde, Manfred; Jogler, Mareike (2020).
1797:
Planctomycetota species were recently identified as being an opportunistic human pathogen, but a lack of culture media limits studies on the bacteria in the Planctomycetota as pathogens of humans.
2218:"Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories"
341:
display both large and small crateriform structures. Large crateriform structures often cover the cell surface, while small crateriform structures are often only at the end of the cell. Light
807:
Omnitrophica". Within this superphylum, its members have been found to be closely related through the creation of 16S rRNA trees. Both the Planctomycetota and Chlamydiota encode proteins for
499:
of roughly 6-14 days. In contrast, some other Planctomycetota have doubling times of around 30 days. Their high abundance in many ecosystems is surprising, given their slow growth rates.
642:(CSIs). These CSIs demarcate the group from neighboring phyla within the PVC group. An additional CSI has been found that is shared by all Planctomycetota species, with the exception of
309:. However, the diversity in cell shape often varies greatly in them. Ovoid and pear-shaped cells have been described in some species, and often occur in rosettes of three to 10 cells.
7298:
7164:
1871:
199:
and terrestrial habitats. They play a considerable role in global carbon and nitrogen cycles, with many species of this phylum capable of anaerobic ammonium oxidation, also known as
4133:
Izumi H, Sagulenko E, Webb RI, Fuerst JA (October 2013). "Isolation and diversity of planctomycetes from the sponge Niphates sp., seawater, and sediment of Moreton Bay, Australia".
601:
cells. However, bacterial budding observed in Planctomycetota is still poorly understood. Budding has been observed in both radial symmetric cells, such as bacteria in the species
337:
Unique appendages known as crateriform structures have been observed in species of Planctomycetota belonging to the class Planctomycetia. The outer surface of cells in the species
353:. The pili fibers in both these species were often associated with large crateriform structures; in contrast, the stalk fibers were associated with small crateriform structures.
1524:
Planctomycetota are often associated with marine surfaces high in nutrients. They occur as biofilms on algal surfaces in relatively high abundance. Macroalgae such as the kelps
773:, a subgroup within Planctomycetota, have low sequence similarity to eukaryotic proteins; however, they show highest sequence similarity to other Gemmataceae protein families.
1651:. The growth of many Planctomycetota is often supported by the essential nutrients provided by other bacteria within the community, and some Planctomycetota rely strongly on
638:
Planctomycetota are known for their unusual cellular characteristics, and their distinctness from all other bacteria is additionally supported by the shared presence of two
3427:"Molecular Signatures for the PVC Clade (Planctomycetes, Verrucomicrobia, Chlamydiae, and Lentisphaerae) of Bacteria Provide Insights into Their Evolutionary Relationships"
4437:
1356:
exclusively in aquatic environments, but they are now known to be also abundant in soils and hypersaline environments. They are widespread on five continents, including
6636:
1742:
The synthesis of sterols, often observed in eukaryotes and uncommon among bacteria, has been observed very rarely in Planctomycetota. The synthesis of sterols such as
803:
be categorized as part of a larger superphylum entitled PVC, which would encompass the phyla Verrucomicrobia, Chlamydiae and Lentisphaerae, and the candidate phylum "
241:. The phylum Planctomycetota is composed of the classes Planctomycetia and Phycisphaerae. First described in 1924, members of the Planctomycetota were identified as
7272:
2752:
Rivas-Marin, Elena; Stettner, Sean; Gottshall, Ekaterina Y.; Santana-Molina, Carlos; Helling, Mitch; Basile, Franco; Ward, Naomi L.; Devos, Damien P. (2019-07-02).
1894:
3255:"Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes"
410:, giving the appearance of compartmentalization within the cytosol. Planctomycetota therefore display the two compartments typical of Gram-negative bacteria, the
7177:
1898:
749:
sp. strain 1 incorporates 110 genes that contribute to encoding proteins that produce sulfatase enzymes. In comparison with a different species of prokaryotic,
6301:
5810:
5241:
4803:
4579:
3193:
Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, et al. (December 2020). "Three marine strains constitute the novel genus and species
474:
and the innermost membrane surrounds the anammoxosome, the central structure of anammox bacteria. The anammoxosome membrane is largely composed of unusual
377:
of glycans, present in all free-living bacteria, and its rigidity helps maintain integrity of the cell. Peptidoglycan synthesis is also essential during
2454:
Laterigemmans baculatus" gen. nov. sp. nov., the first representative of rod shaped planctomycetes with lateral budding in the family Pirellulaceae".
7246:
7151:
4002:"Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes"
1486:
Planctomycetota have often been observed in association with many organisms, including, macroalgae, microalgae, marine sponges, and plants such as
275:
has been identified specifically as comprising bacteria with unique characteristics among the Planctomycetota, such as their ability to synthesize
7285:
6342:
330:
Many Planctomycetota also have a holdfast, or stalk, which attaches the cell to a surface or substrate. Members of some species, though, such as
730:
gene that is expressed when the environment offers excessive ultraviolet radiation stress. Other stress responses include the decomposition of
4402:
3635:
Strous M, Pelletier E, Mangenot S (6 April 2006). "Deciphering the evolution and metabolism of an anammox bacterium from a community genome".
1608:
Planctomycetota contributes to the health of the sponge, and the sponge often provides suitable habitat and nutrients to the Planctomycetota.
4430:
4185:
812:
existence of eukaryotic traits and genes is more likely explained through lateral gene transfer, and not a more recent eukaryotic ancestor.
1583:
While macroalgae are well known substrates for Planctomycetota communities, their abundance has also been known to correlate with blooms of
7370:
3789:
819:
516:
Planctomycetota often perform a lifestyle switch between both a sessile stalked stage and a free-swimming stage. Members of the species
3481:"Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification"
4201:
Qian; et al. (2018). "Diversity and distribution of anammox bacteria in water column and sediments of the Eastern Indian Ocean".
1367:
753:
only 6 sulfatases occur and the genes that express these proteins are contained as two to five pairs, usually clustered in 22 groups.
2280:"The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance"
4396:
3129:
Kartal, Boran; de Almeida, Naomi M.; Maalcke, Wouter J.; Op den Camp, Huub J.M.; Jetten, Mike S.M.; Keltjens, Jan T. (2013-05-01).
4295:
Jing; et al. (2015). "A snapshot on spatial and vertical distribution of bacterial communities in the eastern Indian Ocean".
594:
by binary fission, mainly species of the class Phycisphaerae. In contrast, species of the class Planctomycetia divide by budding.
7337:
4475:
4423:
1428:
Planctomycetota have a significant impact on global biogeochemistry and climate, with their ability to mineralize and break down
491:
Planctomycetota species grow slowly, when compared to other bacteria, often forming rosette structures of 3-5 cells. The species
4470:
441:
Many Planctomycetota species display pink or orange coloring, suggested to result from the production of carotenoid pigments.
7290:
2398:
Lage OM, Bondoso J, Lobo-da-Cunha A (October 2013). "Insights into the ultrastructural morphology of novel Planctomycetes".
6309:
47:
4554:
4544:
4527:
3738:
1562:
As the climate continues to warm, the abundance of Planctomycetota associated with macroalgae might increase. The seaweed
612:
Considerable diversity has been observed in cell division among bacteria in the Planctomycetota. During cell division in
528:
Transmission electron micrographs of dividing cells displaying different cell division modes in the PVC superphylum. (a)
3717:
6335:
1510:
7182:
3880:
Buckley, Daniel H.; Huangyutitham, Varisa; Nelson, Tyrrell A.; Rumberger, Angelika; Thies, Janice E. (2006-07-01).
639:
1789:
air, it may also have a role in interchanges between oceans and atmosphere, potentially affecting climate change.
6479:
4959:
682:
688:
When comparing under a microscope, a defining characteristic for some Planctomycetota is that a single unlinked
373:
in their cell walls, and were instead suggested to have proteinaceous cell walls. Peptidoglycan is an essential
7365:
3821:
3794:
3767:
1575:
conditions, and the abundance of Planctomycetota increased substantially, as much as 10 times in some species.
826:
5826:
1620:
and displayed extremely high diversity. Planctomycetota have also been associated with lichen communities and
673:
sequenced. Extensive genome duplication takes up about 25% of the genome sequence. This may be a way for the
461:. Three different carotenoid pigments have been identified in two different strains of the Planctomycetota.
7375:
6564:
6278:
6122:
5918:
4559:
2632:
Rivas-Marin E, Peeters SH, Claret Fernández L, Jogler C, van Niftrik L, Wiegand S, Devos DP (January 2020).
780:. More than one thousand protein families were acquired by duplications and domain rearrangements. The new
315:
is a well studied species in the Planctomycetota with spherical cells. In contrast, bacteria in the species
5190:
1919:
Garrity GM, Holt JG. (2001). "The Road Map to the Manual". In Boone DR, Castenholz RW, Garrity GM. (eds.).
7119:
6328:
6103:
5310:
5290:
4774:
3857:
3848:
Fuerst, John (2004). "Planctomycetes: A phylum of emerging interest for microbial evolution and ecology".
366:
362:
247:
5742:
5643:
7324:
6792:
6442:
5636:
5617:
5444:
5364:
5143:
4522:
1536:
are suggested to be an important habitat for Planctomycetota. Roughly 70% of the bacterial community on
711:
361:
Early examination of the Planctomycetota suggested that their cell plan differed considerably from both
311:
271:
6204:
6030:
6006:
5977:
5864:
5788:
5672:
5650:
4891:
3011:"The squalene route to C30 carotenoid biosynthesis and the origins of carotenoid biosynthetic pathways"
2956:
Viana F, Lage OM, Oliveira R (October 2013). "High ultraviolet C resistance of marine Planctomycetes".
5910:
5754:
5376:
5339:
5317:
5226:
5214:
4335:"From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules"
3588:"Paralogization and New Protein Architectures in Planctomycetes Bacteria with Complex Cell Structures"
2279:
7342:
7233:
6457:
6173:
6110:
5992:
5886:
5202:
5131:
5108:
4725:
4688:
4636:
4249:
3893:
3644:
3533:
3266:
2855:
2769:
2645:
2084:
1526:
1515:
1077:
766:
5776:
5605:
5395:
5357:
3862:
7224:
6285:
6179:
5934:
5727:
5657:
5269:
5113:
4000:
Woebken D, Teeling H, Wecker P, Dumitriu A, Kostadinov I, Delong EF, et al. (September 2007).
3583:
2901:"Pink- and orange-pigmented planctomycetes produce saproxanthin-type carotenoids including a rare C
1636:
1143:
1069:
859:
785:
398:
123:
6266:
6192:
5624:
4333:
Jeske, Olga; Jogler, Mareike; Petersen, Jörn; Sikorski, Johannes; Jogler, Christian (2013-10-01).
2842:
Boedeker C, Schüler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, et al. (April 2017).
6787:
6469:
6228:
6018:
5715:
5407:
5257:
4539:
4517:
4505:
4480:
4370:
4312:
4218:
4158:
4031:
3882:"Diversity of Planctomycetes in Soil in Relation to Soil History and Environmental Heterogeneity"
3678:
3232:
3050:
2991:
2935:
2512:
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, et al. (January 2020).
2479:
2423:
2175:
1844:
1564:
1552:
797:
322:
Many Planctomycetota species display structures and appendages on the outer surface of the cell.
133:
42:
7329:
2634:"Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila"
2514:"Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology"
1668:
The existence of membrane coat proteins near the intracytoplasmic membrane could be used for an
685:
primer used often mismatches with the genes, creating difficulty when sequencing the genome.
609:, the daughter cells originate from the region opposite to the pole with the holdfast or stalk.
3077:
gen. nov., sp. nov., an Unusual Member of the Phylum Planctomycetes from the German Wadden Sea"
1616:
Planctomycetota were found to be highly abundant in lichen communities throughout northwestern
7311:
6579:
5965:
5526:
5432:
4549:
4512:
4500:
4490:
4362:
4354:
4277:
4181:
4150:
4112:
4072:
4023:
3979:
3971:
3927:
3909:
3670:
3617:
3609:
3559:
3502:
3458:
3404:
3364:
3292:
3224:
3170:
3162:
3108:
3042:
2983:
2927:
2899:
Kallscheuer N, Moreira C, Airs R, Llewellyn CA, Wiegand S, Jogler C, Lage OM (December 2019).
2881:
2805:
2787:
2731:
2671:
2599:
2543:
2471:
2415:
2373:
2355:
2307:
2299:
2249:
2167:
2112:
2038:
1836:
1648:
1401:
761:
Planctomycetota originate from within the Bacteria and these similarities between proteins in
731:
568:
259:
223:
4968:
406:
intracytoplasmic membrane is well known to be the cytoplasmic membrane which displays unique
7316:
6855:
6734:
6626:
6569:
6509:
6504:
6432:
6417:
6184:
5942:
5897:
5850:
5688:
4854:
4464:
4346:
4304:
4267:
4257:
4210:
4142:
4104:
4062:
4013:
3963:
3917:
3901:
3660:
3652:
3599:
3549:
3541:
3492:
3448:
3438:
3396:
3354:
3344:
3282:
3274:
3214:
3206:
3152:
3142:
3098:
3088:
3032:
3022:
2973:
2965:
2919:
2900:
2871:
2863:
2795:
2777:
2721:
2711:
2661:
2653:
2589:
2579:
2533:
2525:
2463:
2407:
2363:
2345:
2291:
2239:
2229:
2157:
2149:
2102:
2092:
2028:
1826:
1532:
1469:
1267:
1165:
992:
881:
762:
563:
458:
4108:
3387:
Bernander R, Ettema TJ (December 2010). "FtsZ-less cell division in archaea and bacteria".
1750:. Lanosterol is common in eukaryotes and two other groups of bacteria, both methylotrophic
7277:
7028:
6933:
6895:
6850:
6782:
6499:
6484:
6377:
6235:
6139:
5952:
5947:
5701:
5481:
5296:
5009:
4980:
4930:
4765:
4660:
4628:
3009:
Santana-Molina C, Henriques V, Hornero-Méndez D, Devos DP, Rivas-Marin E (December 2022).
1644:
1180:
896:
219:
69:
2334:"The Microbiome of Posidonia oceanica Seagrass Leaves Can Be Dominated by Planctomycetes"
2071:
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, et al. (July 2003).
6320:
4253:
3897:
3648:
3537:
3270:
2859:
2773:
2649:
2088:
567:. The dominant form of reproduction observed in almost all bacteria is cell division by
524:
7088:
7033:
7023:
6865:
6679:
6631:
6544:
6519:
6514:
6494:
6489:
6392:
6382:
6215:
6154:
6145:
6042:
5998:
5806:
5769:
5683:
5575:
5516:
5019:
4996:
4699:
4612:
4599:
4594:
4495:
4415:
4272:
4237:
3952:"The planctomycetes: Emerging models for microbial ecology, evolution and cell biology"
3922:
3881:
3554:
3521:
3453:
3426:
3359:
3320:
3287:
3254:
3103:
3072:
3037:
3010:
2876:
2843:
2800:
2753:
2726:
2699:
2666:
2633:
2594:
2567:
2538:
2513:
2368:
2333:
2244:
2217:
1751:
1596:
1569:
1210:
926:
558:
231:
145:
102:
2107:
2072:
1689:
393:
Planctomycetota were once thought to display distinct compartmentalization within the
27:
7359:
6812:
6797:
6759:
6684:
6620:
6606:
6554:
6549:
6463:
6452:
6447:
6427:
6422:
6397:
6221:
5875:
5594:
5534:
5511:
5450:
5237:
5120:
5098:
5067:
5041:
4942:
4756:
4647:
4623:
4604:
4575:
3236:
3054:
2939:
2483:
2179:
1848:
1727:
1677:
1588:
1223:
1129:
961:
846:
689:
496:
378:
370:
97:
4374:
4316:
4222:
4162:
4035:
2995:
2427:
7008:
7003:
6978:
6953:
6943:
6870:
6739:
6613:
6574:
6559:
6534:
6529:
6474:
6407:
6081:
6071:
5795:
5587:
5559:
5468:
5388:
5324:
5179:
5154:
5077:
4673:
4655:
4051:"Community composition of the Planctomycetota associated with different macroalgae"
3682:
1755:
1710:
1699:
1640:
1444:
1436:
1282:
1007:
572:
426:
407:
4407:
1464:
7209:
7142:
3951:
2716:
2700:"Three-dimensional reconstruction of bacteria with a complex endomembrane system"
2295:
7259:
7218:
7063:
7038:
6983:
6958:
6880:
6875:
6840:
6830:
6524:
6437:
6053:
5957:
5564:
5491:
4214:
4178:
Cultivation, Detection, andEcophysiology of AnaerobicAmmonium-Oxidizing Bacteria
3967:
2467:
1669:
1556:
1548:
1458:
1397:
1099:
777:
770:
715:
616:, a tubular structure is connected from the bud to the mother cell. The species
454:
227:
4391:
3210:
3071:
Kohn T, Heuer A, Jogler M, Vollmers J, Boedeker C, Bunk B, et al. (2016).
2782:
2698:
Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP (2013-05-21).
2657:
2153:
2077:
Proceedings of the National Academy of Sciences of the United States of America
369:
bacteria. Until recently, bacteria in the Planctomycetota were thought to lack
7169:
7096:
7058:
6988:
6968:
6963:
6948:
6918:
6890:
6845:
6835:
6807:
6802:
6744:
6719:
6412:
6387:
6166:
6058:
5837:
5569:
5496:
4833:
4799:
4679:
4446:
4403:
Beyond Prokaryotes and Eukaryotes : Planctomycetes and Cell Organization.
4350:
4308:
4146:
4095:
Fuerst JA (October 2005). "Intracellular compartmentation in planctomycetes".
3400:
2969:
2529:
2411:
2234:
1743:
1731:
1635:
Planctomycetota display associations with other bacterial communities, mainly
1584:
1475:
1440:
1384:
1357:
1245:
946:
808:
727:
667:
442:
342:
109:
7303:
4358:
3975:
3913:
3816:
3613:
3582:
Mahajan, Mayank; Yee, Benjamin; Hägglund, Emil; Guy, Lionel; Fuerst, John A;
3443:
3349:
3166:
3093:
2791:
2584:
2359:
2350:
2303:
2073:"Complete genome sequence of the marine planctomycete Pirellula sp. strain 1"
1863:
700:
in these regions have reverse orientation that transfers to rearrangements.
521:
switch observed in many species may not be common among all Planctomycetota.
7013:
6993:
6885:
6764:
6699:
6689:
6674:
6669:
6650:
6402:
5549:
4815:
4334:
4067:
4050:
3604:
3587:
3497:
3480:
3147:
3130:
3027:
2923:
2097:
2033:
2016:
1652:
1599:
provide nutrients for Planctomycetota, which could explain the association.
1542:
1499:
1495:
1491:
1451:
1361:
1091:
742:
666:
has been estimated to be over 7 million bases, making it one of the largest
475:
415:
411:
323:
242:
113:
4366:
4281:
4154:
4116:
4076:
4027:
3931:
3674:
3621:
3563:
3506:
3462:
3408:
3368:
3296:
3228:
3174:
3112:
3046:
2987:
2931:
2885:
2809:
2735:
2675:
2603:
2547:
2475:
2419:
2377:
2311:
2253:
2171:
2116:
2042:
1840:
1831:
1814:
707:, and have genes that are required for heterotactic acid fermentation. The
4018:
4001:
3983:
3696:
7203:
7136:
7068:
7048:
7043:
7018:
6998:
6973:
6938:
6928:
6923:
6754:
6729:
6709:
6369:
4534:
4450:
3905:
3762:
3545:
3278:
2450:
Kumar D, Kumar G, Jagadeeshwari U, Sasikala C, Ramana CV (April 2021). ""
1622:
1429:
781:
678:
674:
192:
59:
7156:
3656:
3219:
2867:
2162:
295:
7073:
7053:
6749:
6724:
6714:
6704:
6660:
6597:
4821:
4262:
3157:
2754:"Essentiality of sterol synthesis genes in the planctomycete bacterium
1718:
1617:
1413:
1409:
704:
597:
The mechanisms involved in budding have been described extensively for
587:
580:
394:
374:
255:
208:
204:
200:
7264:
3665:
2978:
703:
Some Planctomycetota thrive in regions containing highly concentrated
6913:
6905:
6774:
1592:
1487:
1454:
1405:
708:
692:
670:
450:
430:
276:
212:
188:
7113:
7251:
6822:
6694:
1931:) (2nd ed.). New York, NY: Springer-Verlag. pp. 119–166.
1815:"Valid publication of the names of forty-two phyla of prokaryotes"
735:
681:, allowing for redundancy if a part of the genome is damaged. The
621:
605:, and axially symmetric cells. During cell division in members of
598:
586:
FtsZ proteins are suggested to be similar in structure to that of
446:
326:, common in most bacteria, have also been observed in the species
196:
557:
The current understanding of bacterial cell division is based on
258:, in contrast to all other free-living bacteria, which divide by
6860:
6352:
776:
There is massive emergence of novel protein families within the
718:
radiation protection response, and is associated with the genes
697:
576:
7117:
6324:
4419:
1443:, the primary source of carbon used by Planctomycetota is from
714:
plays a key role in this process. The genetic process also has
579:. In contrast, many bacteria in the Planctomycetota divide by
7238:
3321:"Evolutionary Cell Biology of Division Mode in the Bacterial
433:, which is suggested to be associated with sterol synthesis.
203:. Many Planctomycetota occur in relatively high abundance as
1417:
3520:
Lagkouvardos I, Jehl MA, Rattei T, Horn M (January 2014).
2844:"Determining the bacterial cell biology of Planctomycetes"
1416:. Planctomycetota can also inhabit regions with ranges in
3131:"How to make a living from anaerobic ammonium oxidation"
4238:"Anammox Planctomycetes have a peptidoglycan cell wall"
2568:"Planctomycetes as Novel Source of Bioactive Molecules"
1872:
List of Prokaryotic names with Standing in Nomenclature
650:
forms a deep branch within the Planctomycetota phylum.
385:
were found to have peptidoglycan in their cell walls.
4049:
Bondoso J, Balagué V, Gasol JM, Lage OM (June 2014).
2286:. Environmental biotechnology/Energy biotechnology.
7193:
7126:
7086:
6904:
6821:
6773:
6658:
6649:
6596:
6368:
6258:
6132:
5933:
5818:
5805:
5548:
5480:
5422:
5349:
5249:
5236:
5172:
5091:
5060:
5053:
4958:
4798:
4746:
4587:
4574:
4203:
International Biodeterioration & Biodegradation
3850:
World Federation for Culture Collections Newsletter
1494:. They have also been observed inhabiting deep-sea
495:is suggested to be relatively fast growing, with a
345:demonstrated fibers of both stalk and pili type in
3391:. Growth and development: eukaryotes/prokaryotes.
4180:. San Diego CA: Academic Press. pp. 89–108.
207:, often associating with other organisms such as
6637:Archaeal Richmond Mine acidophilic nanoorganisms
2222:Frontiers in Cellular and Infection Microbiology
1435:Planctomycetota play a considerable role in the
3474:
3472:
3253:Jogler C, Glöckner FO, Kolter R (August 2011).
2015:Wiegand S, Jogler M, Jogler C (November 2018).
2278:Wagner, Michael; Horn, Matthias (2006-06-01).
1498:, where they are dominant organisms living on
6336:
4431:
1899:National Center for Biotechnology Information
1559:, given their ability to attach to surfaces.
575:of both peptidoglycans and proteins known as
8:
3420:
3418:
2216:Kaboré OD, Godreuil S, Drancourt M (2020).
1439:. As both obligate and facultative aerobic
1383:Planctomycetota account for roughly 11% of
7114:
6655:
6593:
6365:
6361:
6343:
6329:
6321:
5815:
5246:
5057:
4584:
4438:
4424:
4416:
3522:"Signature protein of the PVC superphylum"
3319:Rivas-Marín E, Canosa I, Devos DP (2016).
1927:and the deeply branching and phototrophic
1921:Bergey's Manual of Systematic Bacteriology
1520:a species of kelp in the phylum Ochrophyta
1424:Ecological impacts and global carbon cycle
26:
17:
4271:
4261:
4066:
4017:
3921:
3861:
3664:
3603:
3553:
3496:
3452:
3442:
3425:Gupta RS, Bhandari V, Naushad HS (2012).
3358:
3348:
3286:
3218:
3197:conspicua in the phylum Planctomycetes".
3156:
3146:
3102:
3092:
3036:
3026:
2977:
2875:
2799:
2781:
2725:
2715:
2665:
2593:
2583:
2537:
2367:
2349:
2243:
2233:
2161:
2106:
2096:
2032:
1830:
1709:
1688:
1509:
1463:
1379:Environmental influences on distribution
814:
523:
501:
294:
250:, but with many unique characteristics.
1805:
1758:. The synthesis of sterols observed in
469:Unique characteristics of anammox cells
4328:
4326:
4128:
4126:
4109:10.1146/annurev.micro.59.030804.121258
4090:
4088:
4086:
3995:
3993:
3945:
3943:
3941:
3886:Applied and Environmental Microbiology
3875:
3873:
3843:
3841:
3839:
3577:
3575:
3573:
3526:Applied and Environmental Microbiology
3382:
3380:
3378:
3314:
3312:
3310:
3308:
3306:
3259:Applied and Environmental Microbiology
3248:
3246:
3124:
3122:
3066:
3064:
2951:
2949:
2747:
2745:
2561:
2559:
2557:
2327:
2325:
2323:
2321:
1706:Anaerobic ammonium oxidation (anammox)
825:120 single copy marker proteins based
3188:
3186:
3184:
2837:
2835:
2833:
2831:
2829:
2827:
2825:
2823:
2821:
2819:
2693:
2691:
2689:
2687:
2685:
2627:
2625:
2623:
2621:
2619:
2617:
2615:
2613:
2566:Graça AP, Calisto R, Lage OM (2016).
2507:
2505:
2503:
2501:
2499:
2497:
2495:
2493:
2445:
2443:
2441:
2439:
2437:
2393:
2391:
2389:
2387:
2273:
2271:
2269:
2267:
2265:
2263:
2211:
2209:
2134:
2132:
2130:
2128:
2126:
2010:
2008:
2006:
2004:
2002:
2000:
1998:
1996:
1994:
1992:
1990:
1988:
1986:
1984:
1982:
1980:
1978:
1976:
1974:
1972:
1970:
1968:
1966:
1964:
1962:
1960:
1958:
7:
2207:
2205:
2203:
2201:
2199:
2197:
2195:
2193:
2191:
2189:
2066:
2064:
2062:
2060:
2058:
2056:
2054:
2052:
1956:
1954:
1952:
1950:
1948:
1946:
1944:
1942:
1940:
1938:
1771:Biotechnology and human applications
1612:Lichen communities and sphagnum bogs
218:Planctomycetota are included in the
2456:Systematic and Applied Microbiology
1693:Labeled diagram of an anammox cell.
1655:relationships with other bacteria.
1260:
1238:
1216:
1158:
1136:
1124:
1064:
1057:
1050:
985:
939:
932:
874:
852:
842:
835:
2912:Environmental Microbiology Reports
1450:Planctomycetota also display many
741:Many Planctomycetota also express
14:
1714:Diagram of the anammox mechanisms
381:. Recently, those in the species
4390:
2284:Current Opinion in Biotechnology
2017:"On the maverick Planctomycetes"
1482:Association with other organisms
1474:a species of kelp in the phylum
421:The excess membrane observed in
46:
4236:Teeseling; et al. (2015).
3718:"LTP_all tree in newick format"
3592:Molecular Biology and Evolution
3389:Current Opinion in Microbiology
1432:particles in the water column.
1:
4097:Annual Review of Microbiology
769:. Gained protein families in
482:Life history and reproduction
3950:Fuerst, J. A. (1995-07-01).
2717:10.1371/journal.pbio.1001565
2296:10.1016/j.copbio.2006.05.005
1793:Planctomycetota as pathogens
1579:Microalgae and diatom blooms
1400:anaerobic species occupying
646:This supports the idea that
457:bacteria to protect against
429:of the cell relative to its
4215:10.1016/j.ibiod.2018.05.015
3968:10.1099/13500872-141-7-1493
3739:"LTP_12_2021 Release Notes"
2468:10.1016/j.syapm.2021.126188
1813:Oren A, Garrity GM (2021).
1631:Other bacterial communities
1587:such as diatoms. Blooms of
1568:was incubated under higher
1404:regions, can use elemental
7392:
7371:Environmental microbiology
3211:10.1007/s10482-019-01375-4
2783:10.1038/s41467-019-10983-7
2658:10.1038/s41598-019-56978-8
2154:10.1007/s10482-019-01367-4
1351:Distribution and abundance
795:
640:conserved signature indels
282:Phylum of aquatic bacteria
234:, Kiritimatiellaeota, and
6592:
6364:
6360:
6355:phyla/divisions by domain
6295:
5832:"Acidulodesulfobacteriia"
4458:
4351:10.1007/s10482-013-0007-1
4309:10.1007/s13131-016-0871-4
4147:10.1007/s10482-013-0003-5
4055:FEMS Microbiology Ecology
3485:FEMS Microbiology Reviews
3431:Frontiers in Microbiology
3401:10.1016/j.mib.2010.10.005
3337:Frontiers in Microbiology
3135:FEMS Microbiology Reviews
3081:Frontiers in Microbiology
2970:10.1007/s10482-013-0027-x
2572:Frontiers in Microbiology
2530:10.1038/s41564-019-0588-1
2412:10.1007/s10482-013-9969-2
2338:Frontiers in Microbiology
2235:10.3389/fcimb.2020.519301
2021:FEMS Microbiology Reviews
1819:Int J Syst Evol Microbiol
1730:and is released into the
1420:levels from 4.2 to 11.6.
1280:
1265:
1258:
1243:
1236:
1221:
1214:
1178:
1163:
1156:
1141:
1134:
1122:
1096:
1074:
1062:
1055:
1005:
990:
983:
959:
944:
937:
930:
894:
879:
872:
857:
850:
840:
824:
817:
683:polymerase chain reaction
644:Kuenenia stuttgartiensis.
389:Internal cell composition
291:Cell shape and appendages
148:1987) Cavalier-Smith 2002
139:
132:
94:
89:
43:Scientific classification
41:
34:
25:
20:
6089:"Thermodesulfovibrionia"
5827:Acidulodesulfobacteriota
4842:"Thermosediminibacteria"
4780:"Marinamargulisbacteria"
4297:Acta Oceanologica Sinica
3822:Genome Taxonomy Database
3795:Genome Taxonomy Database
3768:Genome Taxonomy Database
3444:10.3389/fmicb.2012.00327
3350:10.3389/fmicb.2016.01964
3094:10.3389/fmicb.2016.02079
3075:Fuerstia marisgermanicae
3015:Proc Natl Acad Sci U S A
2585:10.3389/fmicb.2016.01241
2351:10.3389/fmicb.2020.01458
1901:(NCBI) taxonomy database
614:Fuerstia marisgermanicae
286:Structure and morphology
6565:Thermodesulfobacteriota
6123:Thermodesulfobacteriota
5924:"Thermosulfidibacteria"
4783:"Riflemargulisbacteria"
4339:Antonie van Leeuwenhoek
4135:Antonie van Leeuwenhoek
4068:10.1111/1574-6941.12258
3763:"GTDB release 08-RS214"
3199:Antonie van Leeuwenhoek
3148:10.1111/1574-6976.12014
3028:10.1073/pnas.2210081119
2958:Antonie van Leeuwenhoek
2924:10.1111/1758-2229.12796
2400:Antonie van Leeuwenhoek
2142:Antonie van Leeuwenhoek
2098:10.1073/pnas.1431443100
765:and eukaryotes reflect
751:Pseudomonas aeruginosa,
720:recA, lexA, uvrA, uvrB,
658:General characteristics
629:Genetic characteristics
542:Phycisphaera mikurensis
317:Planctopirus limnophila
307:Aquisphaera giovannonii
6279:Salinosulfoleibacteria
6097:"Lambdaproteobacteria"
5919:Thermosulfidibacterota
5196:"Macinerneyibacteriia"
4839:"Thermoanaerobacteria"
4766:Vampirovibrionophyceae
4176:Kartal, Boran (2011).
3790:"bac120_r214.sp_label"
3479:Gupta RS (July 2016).
1832:10.1099/ijsem.0.005056
1726:gas cannot be used by
1715:
1694:
1521:
1478:
664:Rhodopirellula baltica
549:
508:
301:
248:Gram-negative bacteria
191:of widely distributed
7325:Paleobiology Database
6793:Neocallimastigomycota
5191:Macinerneyibacteriota
4242:Nature Communications
4019:10.1038/ismej.2007.63
3605:10.1093/molbev/msz287
3498:10.1093/femsre/fuw011
2848:Nature Communications
2762:Nature Communications
2756:Gemmata obscuriglobus
2034:10.1093/femsre/fuy029
1862:Euzéby JP, Parte AC.
1746:has been observed in
1713:
1692:
1676:found no decrease in
1513:
1467:
745:genes. The genome of
712:lactate dehydrogenase
571:, which involves the
548:. Scale bars, 0.5 μm.
546:Chlamydia trachomatis
538:Lentisphaera araneosa
534:Chthoniobacter flavus
530:Gemmata obscuriglobus
527:
505:
357:Cell wall composition
312:Gemmata obscuriglobus
298:
272:Gemmata obscuriglobus
164:Garrity and Holt 2001
84:Garrity and Holt 2021
36:Brocadia anammoxidans
6458:Coprothermobacterota
6174:Polarisedimenticolia
6162:"Guanabaribacteriia"
6104:Schekmaniibacteriota
5845:"Desulfurobacteriia"
5760:"Handelsmanbacteria"
5748:"Krumholzibacteriia"
5311:Firestoneibacteriota
5291:Desantisiibacteriota
5109:Coprothermobacterota
4989:"Absconditabacteria"
4906:"Desulfitobacteriia"
4775:Margulisiibacteriota
3906:10.1128/AEM.00149-06
3546:10.1128/AEM.02655-13
3279:10.1128/AEM.05132-11
1893:Sayers; et al.
1553:primary productivity
1527:Laminaria hyperborea
1516:Laminaria hyperborea
767:convergent evolution
634:Molecular signatures
397:. Three-dimensional
195:, occurring in both
152:"Planctomycetaeota"
6308:Alternative views:
6180:Thermoanaerobaculia
5983:"Anaeroferrophilia"
5935:Deltaproteobacteria
5907:Desulfobacterota G
5770:Neomarinimicrobiota
5743:Krumholzibacteriota
5644:Edwardsiibacteriota
5370:"Hinthialibacteria"
5149:"Thermodesulfobiia"
5114:Coprothermobacteria
4926:"Halanaerobiaeota"
4874:"Thermaerobacteria"
4761:"Sericytochromatia"
4254:2015NatCo...6.6878V
3898:2006ApEnM..72.4522B
3657:10.1038/nature04647
3649:2006Natur.440..790S
3538:2014ApEnM..80..440L
3271:2011ApEnM..77.5826J
3021:(52): e2210081119.
2868:10.1038/ncomms14853
2860:2017NatCo...814853B
2774:2019NatCo..10.2916R
2650:2020NatSR..10...66R
2518:Nature Microbiology
2089:2003PNAS..100.8298G
1923:. Vol. 1 (The
1637:Alphaproteobacteria
1437:global carbon cycle
1394:Thermostilla marina
1144:Sedimentisphaerales
860:Sedimentisphaerales
786:signal transduction
757:Molecular evolution
726:, in addition to a
662:The genome size of
399:electron tomography
90:Classes and orders
6788:Blastocladiomycota
6036:"Methylomirabilia"
5986:"Deferrimicrobiia"
5870:"Calescibacteriia"
5696:"Raymondbacteriia"
5678:"Fermentibacteria"
5637:Delongiibacteriota
5618:Coatesiibacteriota
5445:Lindowiibacteriota
5365:Hinthialibacterota
5144:Thermodesulfobiota
4992:"Andersenbacteria"
4918:"Syntrophomonadia"
4909:"Desulfotomaculia"
4900:"Carboxydothermia"
4731:"Eudoremicrobiia"
4716:"Heboniibacteriia"
4263:10.1038/ncomms7878
3584:Andersson, Siv G E
2638:Scientific Reports
1716:
1695:
1685:Osmotic regulation
1565:Caulerpa taxifolia
1522:
1479:
798:Bacterial taxonomy
648:K. stuttgartiensis
618:Kolteria novifilia
550:
509:
401:reconstruction of
332:Isosphaera pallida
319:have ovoid cells.
302:
168:"Planctomycetota"
142:"Planctobacteria"
7353:
7352:
7312:Open Tree of Life
7120:Taxon identifiers
7108:
7107:
7104:
7103:
7082:
7081:
6645:
6644:
6588:
6587:
6580:Verrucomicrobiota
6318:
6317:
6254:
6253:
6250:
6249:
6205:Leptospirillaeota
6159:"Fischerbacteria"
6031:Methylomirabilota
6007:Desulfuromonadota
5978:Deferrimicrobiota
5902:"Deferribacteria"
5865:Calescibacteriota
5855:"Campylobacteria"
5789:Tianyaibacteriota
5763:"Latescibacteria"
5673:Fermentibacterota
5651:Eiseniibacteriota
5553:(Sphingobacteria)
5527:Verrucomicrobiota
5485:(Planctobacteria)
5382:"Hydrogenedentia"
5284:"Tritonobacteria"
5168:
5167:
5034:"Saccharimonadia"
5015:"Howlettbacteria"
4954:
4953:
4938:"Selenobacteria"
4903:"Dehalobacteriia"
4897:"Carboxydocellia"
4892:Desulfotomaculota
4882:"Hydrogenisporia"
4871:"Symbiobacteriia"
4860:"Proteinivoracia"
4704:"Abditibacteriia"
4617:"Geothermincolia"
4187:978-0-12-381294-0
3643:(7085): 790–794.
3205:(12): 1797–1809.
2148:(12): 1751–1766.
1649:Verrucomicrobiota
1441:chemoheterotrophs
1412:and respire with
1402:hydrothermal vent
1343:
1342:
1338:
1337:
1329:
1328:
1320:
1319:
1311:
1310:
1302:
1301:
1293:
1292:
1200:
1199:
1191:
1190:
1111:
1110:
1045:
1044:
1036:
1035:
1027:
1026:
1018:
1017:
972:
971:
916:
915:
907:
906:
732:hydrogen peroxide
620:forms a distinct
507:of gray circles.
334:lack a holdfast.
224:Verrucomicrobiota
181:
180:
175:
165:
162:"Planctomycetes"
159:
149:
85:
7383:
7346:
7345:
7333:
7332:
7320:
7319:
7307:
7306:
7294:
7293:
7281:
7280:
7268:
7267:
7255:
7254:
7242:
7241:
7229:
7228:
7227:
7214:
7213:
7212:
7186:
7185:
7173:
7172:
7160:
7159:
7147:
7146:
7145:
7115:
6856:Anthocerotophyta
6735:Hemimastigophora
6656:
6627:Nitrososphaerota
6594:
6570:Thermomicrobiota
6510:Kiritimatiellota
6505:Ignavibacteriota
6470:Deferribacterota
6433:Campylobacterota
6418:Bdellovibrionota
6366:
6362:
6345:
6338:
6331:
6322:
6210:"Leptospirillia"
6185:Vicinamibacteria
6116:"Entotheonellia"
6012:Desulfuromonadia
5943:Bdellovibrionota
5911:Syntrophorhabdia
5898:Deferribacterota
5851:Campylobacterota
5816:
5755:Latescibacterota
5689:Chitinivibrionia
5666:"Tariuqbacteria"
5539:Verrucomicrobiia
5377:Hydrogenedentota
5340:Ratteibacteriota
5333:"Velamenicoccia"
5318:Goldiibacteriota
5281:"Erginobacteria"
5275:"Ancaeobacteria"
5247:
5227:Walliibacteriota
5215:Rifleibacteriota
5208:"Muiribacteriia"
5137:"Lithacetigenia"
5058:
5028:"Microgenomatia"
5005:"Doudnabacteria"
4850:"Dethiobacteria"
4809:
4789:"Termititenacia"
4753:Cyanobacteriota
4737:"Eremiobacteria"
4668:"Limnocylindria"
4642:"Bipolaricaulia"
4585:
4487:
4467:
4440:
4433:
4426:
4417:
4395:Data related to
4394:
4379:
4378:
4330:
4321:
4320:
4292:
4286:
4285:
4275:
4265:
4233:
4227:
4226:
4198:
4192:
4191:
4173:
4167:
4166:
4130:
4121:
4120:
4092:
4081:
4080:
4070:
4046:
4040:
4039:
4021:
4006:The ISME Journal
3997:
3988:
3987:
3962:(7): 1493–1506.
3947:
3936:
3935:
3925:
3892:(7): 4522–4531.
3877:
3868:
3867:
3865:
3845:
3834:
3833:
3831:
3829:
3813:
3807:
3806:
3804:
3802:
3786:
3780:
3779:
3777:
3775:
3759:
3753:
3752:
3750:
3748:
3743:
3735:
3729:
3728:
3726:
3724:
3714:
3708:
3707:
3705:
3703:
3693:
3687:
3686:
3668:
3632:
3626:
3625:
3607:
3598:(4): 1020–1040.
3579:
3568:
3567:
3557:
3517:
3511:
3510:
3500:
3476:
3467:
3466:
3456:
3446:
3422:
3413:
3412:
3384:
3373:
3372:
3362:
3352:
3316:
3301:
3300:
3290:
3250:
3241:
3240:
3222:
3190:
3179:
3178:
3160:
3150:
3126:
3117:
3116:
3106:
3096:
3068:
3059:
3058:
3040:
3030:
3006:
3000:
2999:
2981:
2953:
2944:
2943:
2909:
2896:
2890:
2889:
2879:
2839:
2814:
2813:
2803:
2785:
2749:
2740:
2739:
2729:
2719:
2695:
2680:
2679:
2669:
2629:
2608:
2607:
2597:
2587:
2563:
2552:
2551:
2541:
2509:
2488:
2487:
2447:
2432:
2431:
2395:
2382:
2381:
2371:
2353:
2329:
2316:
2315:
2275:
2258:
2257:
2247:
2237:
2213:
2184:
2183:
2165:
2136:
2121:
2120:
2110:
2100:
2083:(14): 8298–303.
2068:
2047:
2046:
2036:
2012:
1933:
1932:
1916:
1910:
1909:
1907:
1906:
1895:"Planctomycetes"
1890:
1884:
1883:
1881:
1879:
1859:
1853:
1852:
1834:
1810:
1765:G. obscuriglobus
1760:G. obscuriglobus
1748:G. obscuriglobus
1738:Sterol synthesis
1538:Ecklonia radiata
1533:Ecklonia radiata
1470:Ecklonia radiata
1390:G. obscuriglobus
1268:Planctomycetales
1261:
1239:
1217:
1166:Tepidisphaerales
1159:
1137:
1125:
1065:
1058:
1051:
993:Planctomycetales
986:
940:
933:
882:Tepidisphaerales
875:
853:
843:
836:
815:
763:Planctomycetales
564:Escherichia coli
459:oxidative stress
445:are produced by
423:G. obscuriglobus
403:G. obscuriglobus
383:G. obscuriglobus
351:G. obscuriglobus
169:
163:
153:
143:
83:
51:
50:
30:
21:Planctomycetota
18:
7391:
7390:
7386:
7385:
7384:
7382:
7381:
7380:
7366:Planctomycetota
7356:
7355:
7354:
7349:
7341:
7336:
7328:
7323:
7315:
7310:
7302:
7297:
7289:
7284:
7276:
7271:
7263:
7258:
7250:
7245:
7237:
7232:
7223:
7222:
7217:
7208:
7207:
7202:
7189:
7181:
7176:
7170:planctomycetota
7168:
7163:
7155:
7150:
7141:
7140:
7135:
7128:Planctomycetota
7122:
7111:
7109:
7100:
7078:
7029:Gnathostomulida
6934:Xenacoelomorpha
6900:
6896:Flowering plant
6851:Marchantiophyta
6817:
6783:Chytridiomycota
6769:
6641:
6584:
6540:Planctomycetota
6500:Gemmatimonadota
6485:Elusimicrobiota
6378:Acidobacteriota
6356:
6349:
6319:
6314:
6302:Bergey's Manual
6291:
6246:
6241:"Pseudomonadia"
6151:"Aminicenantia"
6140:Acidobacteriota
6128:
6111:Tectimicrobiota
5993:Deferrisomatota
5953:Bdellovibrionia
5948:Bacteriovoracia
5929:
5887:Dadaibacteriota
5809:
5801:
5796:Zixiibacteriota
5782:"Marinisomatia"
5736:"Stahlbacteria"
5709:"Glassbacteria"
5702:Gemmatimonadota
5611:"Cloacimonadia"
5576:Ignavibacteriia
5552:
5544:
5531:Kiritimatiellia
5521:"Uabimicrobiia"
5504:Planctomycetota
5484:
5476:
5425:bacteriobiontes
5424:
5418:
5401:"Saltatorellae"
5345:
5297:Elusimicrobiota
5240:
5232:
5220:"Ozemibacteria"
5203:Muiribacteriota
5164:
5132:Lithacetigenota
5121:Dictyoglomerota
5087:
5049:
5037:"Torokbacteria"
5031:"Paceibacteria"
5025:"Kazanbacteria"
5010:Gracilibacteria
5002:"Dojkabacteria"
4981:Patescibacteria
4974:"Elulimicrobia"
4950:
4868:"Sulfobacillia"
4807:
4806:
4802:
4794:
4786:"Saganbacteria"
4747:Cyanoprokaryota
4742:
4726:Eremiobacterota
4694:"Dormibacteria"
4689:Dormiibacterota
4665:Ktedonobacteria
4661:Dehalococcoidia
4637:Bipolaricaulota
4629:Thermoleophilia
4620:"Humimicrobiia"
4578:
4570:
4569:
4485:
4463:
4454:
4444:
4397:Planctomycetota
4387:
4382:
4332:
4331:
4324:
4294:
4293:
4289:
4235:
4234:
4230:
4200:
4199:
4195:
4188:
4175:
4174:
4170:
4132:
4131:
4124:
4094:
4093:
4084:
4048:
4047:
4043:
3999:
3998:
3991:
3949:
3948:
3939:
3879:
3878:
3871:
3863:10.1.1.538.2883
3847:
3846:
3837:
3827:
3825:
3817:"Taxon History"
3815:
3814:
3810:
3800:
3798:
3788:
3787:
3783:
3773:
3771:
3761:
3760:
3756:
3746:
3744:
3741:
3737:
3736:
3732:
3722:
3720:
3716:
3715:
3711:
3701:
3699:
3695:
3694:
3690:
3634:
3633:
3629:
3581:
3580:
3571:
3519:
3518:
3514:
3478:
3477:
3470:
3424:
3423:
3416:
3386:
3385:
3376:
3327:Verrucomicrobia
3318:
3317:
3304:
3252:
3251:
3244:
3192:
3191:
3182:
3128:
3127:
3120:
3070:
3069:
3062:
3008:
3007:
3003:
2955:
2954:
2947:
2907:
2904:
2898:
2897:
2893:
2841:
2840:
2817:
2751:
2750:
2743:
2710:(5): e1001565.
2697:
2696:
2683:
2631:
2630:
2611:
2565:
2564:
2555:
2511:
2510:
2491:
2449:
2448:
2435:
2397:
2396:
2385:
2331:
2330:
2319:
2277:
2276:
2261:
2215:
2214:
2187:
2138:
2137:
2124:
2070:
2069:
2050:
2014:
2013:
1936:
1918:
1917:
1913:
1904:
1902:
1892:
1891:
1887:
1877:
1875:
1861:
1860:
1856:
1812:
1811:
1807:
1803:
1795:
1786:
1773:
1740:
1725:
1708:
1687:
1666:
1661:
1645:Gemmatimonadota
1633:
1614:
1605:
1597:dinoflagellates
1581:
1573:
1508:
1484:
1426:
1381:
1353:
1348:
1339:
1330:
1321:
1312:
1303:
1294:
1201:
1192:
1181:Phycisphaerales
1112:
1078:Uabimicrobiales
1046:
1037:
1028:
1019:
973:
917:
908:
897:Phycisphaerales
818:16S rRNA based
800:
794:
759:
660:
636:
631:
559:model organisms
555:
514:
489:
484:
476:ladderane-based
471:
439:
391:
359:
293:
288:
283:
220:PVC superphylum
185:Planctomycetota
82:
80:Planctomycetota
70:PVC superphylum
45:
12:
11:
5:
7389:
7387:
7379:
7378:
7376:Bacteria phyla
7373:
7368:
7358:
7357:
7351:
7350:
7348:
7347:
7334:
7321:
7308:
7304:planctomycetes
7295:
7282:
7269:
7256:
7243:
7230:
7225:Planctomycetes
7215:
7199:
7197:
7195:Planctomycetes
7191:
7190:
7188:
7187:
7174:
7161:
7148:
7132:
7130:
7124:
7123:
7118:
7106:
7105:
7102:
7101:
7094:
7092:
7089:Incertae sedis
7084:
7083:
7080:
7079:
7077:
7076:
7071:
7066:
7061:
7056:
7051:
7046:
7041:
7036:
7034:Micrognathozoa
7031:
7026:
7024:Acanthocephala
7021:
7016:
7011:
7006:
7001:
6996:
6991:
6986:
6981:
6976:
6971:
6966:
6961:
6956:
6951:
6946:
6941:
6936:
6931:
6926:
6921:
6916:
6910:
6908:
6902:
6901:
6899:
6898:
6893:
6888:
6883:
6878:
6873:
6868:
6866:Lycopodiophyta
6863:
6858:
6853:
6848:
6843:
6838:
6833:
6827:
6825:
6819:
6818:
6816:
6815:
6810:
6805:
6800:
6795:
6790:
6785:
6779:
6777:
6771:
6770:
6768:
6767:
6762:
6757:
6752:
6747:
6742:
6737:
6732:
6727:
6722:
6717:
6712:
6707:
6702:
6697:
6692:
6687:
6682:
6680:Ancyromonadida
6677:
6672:
6666:
6664:
6653:
6647:
6646:
6643:
6642:
6640:
6639:
6634:
6632:Thermoproteota
6629:
6624:
6617:
6610:
6602:
6600:
6590:
6589:
6586:
6585:
6583:
6582:
6577:
6572:
6567:
6562:
6557:
6552:
6547:
6545:Pseudomonadota
6542:
6537:
6532:
6527:
6522:
6520:Mycoplasmatota
6517:
6515:Lentisphaerota
6512:
6507:
6502:
6497:
6495:Fusobacteriota
6492:
6490:Fibrobacterota
6487:
6482:
6477:
6472:
6467:
6460:
6455:
6450:
6445:
6440:
6435:
6430:
6425:
6420:
6415:
6410:
6405:
6400:
6395:
6393:Armatimonadota
6390:
6385:
6383:Actinomycetota
6380:
6374:
6372:
6358:
6357:
6350:
6348:
6347:
6340:
6333:
6325:
6316:
6315:
6313:
6312:
6306:
6296:
6293:
6292:
6290:
6289:
6282:
6275:
6274:
6273:
6262:
6260:
6256:
6255:
6252:
6251:
6248:
6247:
6245:
6244:
6243:
6242:
6239:
6232:
6225:
6216:Pseudomonadota
6213:
6212:
6211:
6201:
6200:
6199:
6189:
6188:
6187:
6182:
6177:
6170:
6163:
6160:
6157:
6155:Blastocatellia
6152:
6149:
6146:Acidobacteriia
6136:
6134:
6130:
6129:
6127:
6126:
6119:
6118:
6117:
6107:
6100:
6099:
6098:
6092:
6091:
6090:
6087:
6079:
6078:
6077:
6069:
6068:
6067:
6064:
6063:"Bradymonadia"
6061:
6051:
6050:
6049:
6048:"Moduliflexia"
6039:
6038:
6037:
6027:
6026:
6025:
6015:
6014:
6013:
6003:
6002:
6001:
5999:Deferrisomatia
5989:
5988:
5987:
5984:
5974:
5973:
5972:
5962:
5961:
5960:
5955:
5950:
5939:
5937:
5931:
5930:
5928:
5927:
5926:
5925:
5915:
5914:
5913:
5905:
5904:
5903:
5895:
5894:
5893:
5892:"Dadabacteria"
5883:
5882:
5881:
5873:
5872:
5871:
5861:
5860:
5859:
5856:
5848:
5847:
5846:
5843:
5835:
5834:
5833:
5822:
5820:
5813:
5807:Proteobacteria
5803:
5802:
5800:
5799:
5792:
5785:
5784:
5783:
5777:Marinisomatota
5773:
5766:
5765:
5764:
5761:
5751:
5750:
5749:
5739:
5738:
5737:
5734:
5733:"Hydrothermia"
5724:
5723:
5722:
5712:
5711:
5710:
5707:
5706:Gemmatimonadia
5699:
5698:
5697:
5694:
5691:
5684:Fibrobacterota
5681:
5680:
5679:
5669:
5668:
5667:
5664:
5663:"Electryoneia"
5654:
5647:
5640:
5633:
5632:
5631:
5630:"Cosmopolitia"
5621:
5614:
5613:
5612:
5606:Cloacimonadota
5602:
5601:
5600:
5592:
5591:
5590:
5585:
5582:
5581:"Kapabacteria"
5579:
5572:
5567:
5556:
5554:
5546:
5545:
5543:
5542:
5541:
5540:
5537:
5532:
5524:
5523:
5522:
5519:
5517:Planctomycetia
5514:
5509:
5501:
5500:
5499:
5488:
5486:
5478:
5477:
5475:
5474:
5473:
5472:
5465:
5462:
5459:
5458:"Brachyspirae"
5456:
5455:"Brevinematia"
5448:
5441:
5440:
5439:
5428:
5426:
5420:
5419:
5417:
5416:
5415:
5414:
5404:
5403:
5402:
5396:Saltatorellota
5392:
5389:Poribacteriota
5385:
5384:
5383:
5373:
5372:
5371:
5361:
5358:Abyssubacteria
5353:
5351:
5347:
5346:
5344:
5343:
5336:
5335:
5334:
5331:
5321:
5314:
5307:
5306:
5305:
5302:
5301:Elusimicrobiia
5294:
5287:
5286:
5285:
5282:
5279:
5278:"Auribacteria"
5276:
5266:
5265:
5264:
5253:
5251:
5244:
5234:
5233:
5231:
5230:
5223:
5222:
5221:
5211:
5210:
5209:
5199:
5198:
5197:
5187:
5186:
5185:
5180:Fusobacteriota
5176:
5174:
5170:
5169:
5166:
5165:
5163:
5162:
5161:
5160:
5152:
5151:
5150:
5140:
5139:
5138:
5128:
5127:
5126:
5125:Dictyoglomeria
5118:
5117:
5116:
5106:
5105:
5104:
5095:
5093:
5089:
5088:
5086:
5085:
5084:
5083:
5075:
5074:
5073:
5064:
5062:
5055:
5051:
5050:
5048:
5047:
5046:
5045:
5038:
5035:
5032:
5029:
5026:
5023:
5020:Katanobacteria
5016:
5013:
5006:
5003:
5000:
4997:Berkelbacteria
4993:
4990:
4987:
4977:
4976:
4975:
4964:
4962:
4956:
4955:
4952:
4951:
4949:
4948:
4947:
4946:
4936:
4935:
4934:
4924:
4923:
4922:
4921:"Thermincolia"
4919:
4916:
4913:
4910:
4907:
4904:
4901:
4898:
4888:
4887:
4886:
4883:
4877:
4876:
4875:
4872:
4869:
4863:
4862:
4861:
4858:
4855:Natranaerobiia
4851:
4845:
4844:
4843:
4840:
4837:
4827:
4826:
4825:
4812:
4810:
4796:
4795:
4793:
4792:
4791:
4790:
4787:
4784:
4781:
4771:
4770:
4769:
4762:
4759:
4750:
4748:
4744:
4743:
4741:
4740:
4739:
4738:
4735:
4729:
4722:
4721:
4720:
4717:
4714:
4713:Fimbriimonadia
4711:
4710:Chthonomonadia
4708:
4705:
4700:Armatimonadota
4697:
4696:
4695:
4685:
4684:
4683:
4671:
4670:
4669:
4666:
4663:
4658:
4653:
4645:
4644:
4643:
4633:
4632:
4631:
4626:
4621:
4618:
4615:
4613:Coriobacteriia
4610:
4609:"Aquicultoria"
4607:
4602:
4600:Acidimicrobiia
4595:Actinomycetota
4591:
4589:
4582:
4572:
4571:
4568:
4567:
4566:
4565:
4564:
4563:
4560:Mesomycetozoea
4557:
4552:
4542:
4532:
4531:
4530:
4525:
4520:
4515:
4510:
4509:
4508:
4496:Diaphoretickes
4493:
4488:
4483:
4478:
4473:
4468:
4460:
4459:
4456:
4455:
4453:classification
4445:
4443:
4442:
4435:
4428:
4420:
4414:
4413:
4411:at Microbewiki
4405:
4400:
4399:at Wikispecies
4386:
4385:External links
4383:
4381:
4380:
4345:(4): 551–567.
4322:
4287:
4228:
4193:
4186:
4168:
4122:
4103:(1): 299–328.
4082:
4041:
3989:
3937:
3869:
3835:
3808:
3781:
3754:
3730:
3709:
3688:
3627:
3586:(2019-12-11).
3569:
3512:
3468:
3414:
3374:
3323:Planctomycetes
3302:
3265:(16): 5826–9.
3242:
3180:
3141:(3): 428–461.
3118:
3060:
3001:
2945:
2918:(6): 741–748.
2902:
2891:
2815:
2741:
2681:
2609:
2553:
2524:(1): 126–140.
2489:
2433:
2383:
2317:
2290:(3): 241–249.
2259:
2185:
2122:
2048:
2027:(6): 739–760.
1934:
1911:
1885:
1866:Planctomycetes
1854:
1804:
1802:
1799:
1794:
1791:
1785:
1784:Climate change
1782:
1772:
1769:
1752:Pseudomonadota
1739:
1736:
1723:
1707:
1704:
1686:
1683:
1665:
1662:
1660:
1657:
1632:
1629:
1613:
1610:
1604:
1603:Marine sponges
1601:
1580:
1577:
1571:
1545:relationship.
1507:
1504:
1483:
1480:
1425:
1422:
1380:
1377:
1352:
1349:
1347:
1344:
1341:
1340:
1336:
1335:
1332:
1331:
1327:
1326:
1323:
1322:
1318:
1317:
1314:
1313:
1309:
1308:
1305:
1304:
1300:
1299:
1296:
1295:
1291:
1290:
1287:
1286:
1279:
1276:
1275:
1272:
1271:
1264:
1259:
1257:
1254:
1253:
1250:
1249:
1242:
1237:
1235:
1232:
1231:
1228:
1227:
1220:
1215:
1213:
1211:Planctomycetia
1207:
1206:
1203:
1202:
1198:
1197:
1194:
1193:
1189:
1188:
1185:
1184:
1177:
1174:
1173:
1170:
1169:
1162:
1157:
1155:
1152:
1151:
1148:
1147:
1140:
1135:
1133:
1123:
1121:
1118:
1117:
1114:
1113:
1109:
1108:
1105:
1104:
1095:
1087:
1086:
1083:
1082:
1073:
1063:
1061:
1056:
1054:
1049:
1047:
1043:
1042:
1039:
1038:
1034:
1033:
1030:
1029:
1025:
1024:
1021:
1020:
1016:
1015:
1012:
1011:
1004:
1001:
1000:
997:
996:
989:
984:
982:
979:
978:
975:
974:
970:
969:
966:
965:
958:
955:
954:
951:
950:
943:
938:
936:
931:
929:
927:Planctomycetia
923:
922:
919:
918:
914:
913:
910:
909:
905:
904:
901:
900:
893:
890:
889:
886:
885:
878:
873:
871:
868:
867:
864:
863:
856:
851:
849:
841:
839:
834:
831:
830:
823:
793:
790:
758:
755:
659:
656:
635:
632:
630:
627:
569:binary fission
554:
551:
513:
510:
488:
485:
483:
480:
470:
467:
453:, and by some
438:
435:
390:
387:
371:peptidoglycans
358:
355:
328:P. limnophila.
292:
289:
287:
284:
281:
260:binary fission
232:Lentisphaerota
179:
178:
177:
176:
166:
160:
150:
146:Cavalier-Smith
137:
136:
130:
129:
128:
127:
117:
105:
103:Planctomycetia
100:
92:
91:
87:
86:
77:
73:
72:
67:
63:
62:
57:
53:
52:
39:
38:
32:
31:
23:
22:
13:
10:
9:
6:
4:
3:
2:
7388:
7377:
7374:
7372:
7369:
7367:
7364:
7363:
7361:
7344:
7339:
7335:
7331:
7326:
7322:
7318:
7313:
7309:
7305:
7300:
7296:
7292:
7287:
7283:
7279:
7274:
7270:
7266:
7261:
7257:
7253:
7248:
7244:
7240:
7235:
7231:
7226:
7220:
7216:
7211:
7205:
7201:
7200:
7198:
7196:
7192:
7184:
7179:
7175:
7171:
7166:
7162:
7158:
7153:
7149:
7144:
7138:
7134:
7133:
7131:
7129:
7125:
7121:
7116:
7112:
7099:
7098:
7093:
7091:
7090:
7085:
7075:
7072:
7070:
7067:
7065:
7062:
7060:
7057:
7055:
7052:
7050:
7047:
7045:
7042:
7040:
7037:
7035:
7032:
7030:
7027:
7025:
7022:
7020:
7017:
7015:
7012:
7010:
7007:
7005:
7002:
7000:
6997:
6995:
6992:
6990:
6987:
6985:
6982:
6980:
6977:
6975:
6972:
6970:
6967:
6965:
6962:
6960:
6957:
6955:
6952:
6950:
6947:
6945:
6942:
6940:
6937:
6935:
6932:
6930:
6927:
6925:
6922:
6920:
6917:
6915:
6912:
6911:
6909:
6907:
6903:
6897:
6894:
6892:
6889:
6887:
6884:
6882:
6879:
6877:
6874:
6872:
6869:
6867:
6864:
6862:
6859:
6857:
6854:
6852:
6849:
6847:
6844:
6842:
6839:
6837:
6834:
6832:
6829:
6828:
6826:
6824:
6820:
6814:
6813:Basidiomycota
6811:
6809:
6806:
6804:
6801:
6799:
6798:Glomeromycota
6796:
6794:
6791:
6789:
6786:
6784:
6781:
6780:
6778:
6776:
6772:
6766:
6763:
6761:
6760:Stramenopiles
6758:
6756:
6753:
6751:
6748:
6746:
6743:
6741:
6738:
6736:
6733:
6731:
6728:
6726:
6723:
6721:
6718:
6716:
6713:
6711:
6708:
6706:
6703:
6701:
6698:
6696:
6693:
6691:
6688:
6686:
6685:Apusomonadida
6683:
6681:
6678:
6676:
6673:
6671:
6668:
6667:
6665:
6662:
6657:
6654:
6652:
6648:
6638:
6635:
6633:
6630:
6628:
6625:
6622:
6621:Nanoarchaeota
6618:
6615:
6611:
6608:
6607:Euryarchaeota
6604:
6603:
6601:
6599:
6595:
6591:
6581:
6578:
6576:
6573:
6571:
6568:
6566:
6563:
6561:
6558:
6556:
6555:Spirochaetota
6553:
6551:
6550:Rhodothermota
6548:
6546:
6543:
6541:
6538:
6536:
6533:
6531:
6528:
6526:
6523:
6521:
6518:
6516:
6513:
6511:
6508:
6506:
6503:
6501:
6498:
6496:
6493:
6491:
6488:
6486:
6483:
6481:
6480:Dictyoglomota
6478:
6476:
6473:
6471:
6468:
6465:
6464:Cyanobacteria
6461:
6459:
6456:
6454:
6453:Chrysiogenota
6451:
6449:
6448:Chloroflexota
6446:
6444:
6441:
6439:
6436:
6434:
6431:
6429:
6428:Calditrichota
6426:
6424:
6423:Caldisericota
6421:
6419:
6416:
6414:
6411:
6409:
6406:
6404:
6401:
6399:
6398:Atribacterota
6396:
6394:
6391:
6389:
6386:
6384:
6381:
6379:
6376:
6375:
6373:
6371:
6367:
6363:
6359:
6354:
6346:
6341:
6339:
6334:
6332:
6327:
6326:
6323:
6311:
6307:
6304:
6303:
6298:
6297:
6294:
6287:
6286:Teskebacteria
6283:
6280:
6276:
6272:"Qinglongiia"
6271:
6270:
6268:
6264:
6263:
6261:
6257:
6240:
6237:
6236:Magnetococcia
6233:
6230:
6229:Mariprofundia
6226:
6223:
6222:Caulobacteria
6219:
6218:
6217:
6214:
6209:
6208:
6206:
6202:
6198:"Canglongiia"
6197:
6196:
6194:
6190:
6186:
6183:
6181:
6178:
6175:
6171:
6168:
6164:
6161:
6158:
6156:
6153:
6150:
6147:
6143:
6142:
6141:
6138:
6137:
6135:
6131:
6124:
6120:
6115:
6114:
6112:
6108:
6105:
6101:
6096:
6095:
6093:
6088:
6085:
6084:
6083:
6080:
6075:
6074:
6073:
6070:
6065:
6062:
6060:
6057:
6056:
6055:
6052:
6047:
6046:
6044:
6043:Moduliflexota
6040:
6035:
6034:
6032:
6028:
6023:
6022:
6020:
6016:
6011:
6010:
6008:
6004:
6000:
5997:
5996:
5994:
5990:
5985:
5982:
5981:
5979:
5975:
5970:
5969:
5967:
5963:
5959:
5956:
5954:
5951:
5949:
5946:
5945:
5944:
5941:
5940:
5938:
5936:
5932:
5923:
5922:
5920:
5916:
5912:
5909:
5908:
5906:
5901:
5900:
5899:
5896:
5891:
5890:
5888:
5884:
5879:
5878:
5877:
5876:Chrysiogenota
5874:
5869:
5868:
5866:
5862:
5858:Desulfurellia
5857:
5854:
5853:
5852:
5849:
5844:
5841:
5840:
5839:
5836:
5831:
5830:
5828:
5824:
5823:
5821:
5817:
5814:
5812:
5808:
5804:
5797:
5793:
5790:
5786:
5781:
5780:
5778:
5774:
5771:
5767:
5762:
5759:
5758:
5756:
5752:
5747:
5746:
5744:
5740:
5735:
5732:
5731:
5729:
5728:Hydrothermota
5725:
5720:
5719:
5717:
5713:
5708:
5705:
5704:
5703:
5700:
5695:
5693:Fibrobacteria
5692:
5690:
5687:
5686:
5685:
5682:
5677:
5676:
5674:
5670:
5665:
5662:
5661:
5659:
5658:Electryoneota
5655:
5652:
5648:
5645:
5641:
5638:
5634:
5629:
5628:
5626:
5622:
5619:
5615:
5610:
5609:
5607:
5603:
5598:
5597:
5596:
5595:Calditrichota
5593:
5589:
5586:
5583:
5580:
5577:
5573:
5571:
5568:
5566:
5563:
5562:
5561:
5558:
5557:
5555:
5551:
5547:
5538:
5536:
5535:Lentisphaeria
5533:
5530:
5529:
5528:
5525:
5520:
5518:
5515:
5513:
5512:Phycisphaeria
5510:
5507:
5506:
5505:
5502:
5498:
5495:
5494:
5493:
5490:
5489:
5487:
5483:
5479:
5470:
5466:
5464:"Leptospiria"
5463:
5461:"Exilispiria"
5460:
5457:
5454:
5453:
5452:
5451:Spirochaetota
5449:
5446:
5442:
5437:
5436:
5434:
5430:
5429:
5427:
5421:
5412:
5411:
5409:
5405:
5400:
5399:
5397:
5393:
5390:
5386:
5381:
5380:
5378:
5374:
5369:
5368:
5366:
5362:
5359:
5355:
5354:
5352:
5348:
5341:
5337:
5332:
5330:"Omnitrophia"
5329:
5328:
5326:
5322:
5319:
5315:
5312:
5308:
5304:Endomicrobiia
5303:
5300:
5299:
5298:
5295:
5292:
5288:
5283:
5280:
5277:
5274:
5273:
5271:
5270:Auribacterota
5267:
5262:
5261:
5259:
5255:
5254:
5252:
5248:
5245:
5243:
5239:
5238:Hydrobacteria
5235:
5228:
5224:
5219:
5218:
5216:
5212:
5207:
5206:
5204:
5200:
5195:
5194:
5192:
5188:
5184:Fusobacteriia
5183:
5182:
5181:
5178:
5177:
5175:
5173:Fusobacterida
5171:
5159:"Thermotogia"
5158:
5157:
5156:
5153:
5148:
5147:
5145:
5141:
5136:
5135:
5133:
5129:
5124:
5123:
5122:
5119:
5115:
5112:
5111:
5110:
5107:
5102:
5101:
5100:
5099:Caldisericota
5097:
5096:
5094:
5090:
5081:
5080:
5079:
5076:
5071:
5070:
5069:
5068:Atribacterota
5066:
5065:
5063:
5061:Synergistetes
5059:
5056:
5052:
5043:
5042:Wirthbacteria
5039:
5036:
5033:
5030:
5027:
5024:
5021:
5017:
5014:
5011:
5007:
5004:
5001:
4998:
4994:
4991:
4988:
4985:
4984:
4982:
4978:
4973:
4972:
4970:
4966:
4965:
4963:
4961:
4957:
4944:
4943:Selenomonadia
4940:
4939:
4937:
4932:
4931:Halanaerobiia
4928:
4927:
4925:
4920:
4917:
4915:"Peptococcia"
4914:
4911:
4908:
4905:
4902:
4899:
4896:
4895:
4893:
4889:
4884:
4881:
4880:
4878:
4873:
4870:
4867:
4866:
4864:
4859:
4856:
4852:
4849:
4848:
4846:
4841:
4838:
4835:
4831:
4830:
4828:
4823:
4819:
4818:
4817:
4814:
4813:
4811:
4805:
4801:
4797:
4788:
4785:
4782:
4779:
4778:
4776:
4772:
4767:
4763:
4760:
4758:
4757:Cyanobacteria
4755:
4754:
4752:
4751:
4749:
4745:
4736:
4733:
4732:
4730:
4727:
4723:
4718:
4715:
4712:
4709:
4707:Armatimonadia
4706:
4703:
4702:
4701:
4698:
4693:
4692:
4690:
4686:
4681:
4677:
4676:
4675:
4672:
4667:
4664:
4662:
4659:
4657:
4654:
4652:"Caldilineia"
4651:
4650:
4649:
4648:Chloroflexota
4646:
4641:
4640:
4638:
4634:
4630:
4627:
4625:
4624:Rubrobacteria
4622:
4619:
4616:
4614:
4611:
4608:
4606:
4605:Actinomycetia
4603:
4601:
4598:
4597:
4596:
4593:
4592:
4590:
4586:
4583:
4581:
4580:BV1, BV3, BV5
4577:
4576:Terrabacteria
4573:
4561:
4558:
4556:
4553:
4551:
4548:
4547:
4546:
4543:
4541:
4538:
4537:
4536:
4533:
4529:
4526:
4524:
4523:Stramenopiles
4521:
4519:
4516:
4514:
4511:
4507:
4504:
4503:
4502:
4499:
4498:
4497:
4494:
4492:
4489:
4486:(major groups
4484:
4482:
4479:
4477:
4474:
4472:
4469:
4466:
4462:
4461:
4457:
4452:
4448:
4441:
4436:
4434:
4429:
4427:
4422:
4421:
4418:
4412:
4410:
4406:
4404:
4401:
4398:
4393:
4389:
4388:
4384:
4376:
4372:
4368:
4364:
4360:
4356:
4352:
4348:
4344:
4340:
4336:
4329:
4327:
4323:
4318:
4314:
4310:
4306:
4302:
4298:
4291:
4288:
4283:
4279:
4274:
4269:
4264:
4259:
4255:
4251:
4247:
4243:
4239:
4232:
4229:
4224:
4220:
4216:
4212:
4208:
4204:
4197:
4194:
4189:
4183:
4179:
4172:
4169:
4164:
4160:
4156:
4152:
4148:
4144:
4141:(4): 533–46.
4140:
4136:
4129:
4127:
4123:
4118:
4114:
4110:
4106:
4102:
4098:
4091:
4089:
4087:
4083:
4078:
4074:
4069:
4064:
4061:(3): 445–56.
4060:
4056:
4052:
4045:
4042:
4037:
4033:
4029:
4025:
4020:
4015:
4012:(5): 419–35.
4011:
4007:
4003:
3996:
3994:
3990:
3985:
3981:
3977:
3973:
3969:
3965:
3961:
3957:
3953:
3946:
3944:
3942:
3938:
3933:
3929:
3924:
3919:
3915:
3911:
3907:
3903:
3899:
3895:
3891:
3887:
3883:
3876:
3874:
3870:
3864:
3859:
3855:
3851:
3844:
3842:
3840:
3836:
3824:
3823:
3818:
3812:
3809:
3797:
3796:
3791:
3785:
3782:
3770:
3769:
3764:
3758:
3755:
3740:
3734:
3731:
3719:
3713:
3710:
3698:
3692:
3689:
3684:
3680:
3676:
3672:
3667:
3662:
3658:
3654:
3650:
3646:
3642:
3638:
3631:
3628:
3623:
3619:
3615:
3611:
3606:
3601:
3597:
3593:
3589:
3585:
3578:
3576:
3574:
3570:
3565:
3561:
3556:
3551:
3547:
3543:
3539:
3535:
3531:
3527:
3523:
3516:
3513:
3508:
3504:
3499:
3494:
3491:(4): 520–53.
3490:
3486:
3482:
3475:
3473:
3469:
3464:
3460:
3455:
3450:
3445:
3440:
3436:
3432:
3428:
3421:
3419:
3415:
3410:
3406:
3402:
3398:
3395:(6): 747–52.
3394:
3390:
3383:
3381:
3379:
3375:
3370:
3366:
3361:
3356:
3351:
3346:
3342:
3338:
3334:
3332:
3328:
3324:
3315:
3313:
3311:
3309:
3307:
3303:
3298:
3294:
3289:
3284:
3280:
3276:
3272:
3268:
3264:
3260:
3256:
3249:
3247:
3243:
3238:
3234:
3230:
3226:
3221:
3216:
3212:
3208:
3204:
3200:
3196:
3189:
3187:
3185:
3181:
3176:
3172:
3168:
3164:
3159:
3154:
3149:
3144:
3140:
3136:
3132:
3125:
3123:
3119:
3114:
3110:
3105:
3100:
3095:
3090:
3086:
3082:
3078:
3076:
3067:
3065:
3061:
3056:
3052:
3048:
3044:
3039:
3034:
3029:
3024:
3020:
3016:
3012:
3005:
3002:
2997:
2993:
2989:
2985:
2980:
2975:
2971:
2967:
2964:(4): 585–95.
2963:
2959:
2952:
2950:
2946:
2941:
2937:
2933:
2929:
2925:
2921:
2917:
2913:
2906:
2895:
2892:
2887:
2883:
2878:
2873:
2869:
2865:
2861:
2857:
2853:
2849:
2845:
2838:
2836:
2834:
2832:
2830:
2828:
2826:
2824:
2822:
2820:
2816:
2811:
2807:
2802:
2797:
2793:
2789:
2784:
2779:
2775:
2771:
2767:
2763:
2759:
2757:
2748:
2746:
2742:
2737:
2733:
2728:
2723:
2718:
2713:
2709:
2705:
2701:
2694:
2692:
2690:
2688:
2686:
2682:
2677:
2673:
2668:
2663:
2659:
2655:
2651:
2647:
2643:
2639:
2635:
2628:
2626:
2624:
2622:
2620:
2618:
2616:
2614:
2610:
2605:
2601:
2596:
2591:
2586:
2581:
2577:
2573:
2569:
2562:
2560:
2558:
2554:
2549:
2545:
2540:
2535:
2531:
2527:
2523:
2519:
2515:
2508:
2506:
2504:
2502:
2500:
2498:
2496:
2494:
2490:
2485:
2481:
2477:
2473:
2469:
2465:
2462:(2): 126188.
2461:
2457:
2453:
2446:
2444:
2442:
2440:
2438:
2434:
2429:
2425:
2421:
2417:
2413:
2409:
2406:(4): 467–76.
2405:
2401:
2394:
2392:
2390:
2388:
2384:
2379:
2375:
2370:
2365:
2361:
2357:
2352:
2347:
2343:
2339:
2335:
2328:
2326:
2324:
2322:
2318:
2313:
2309:
2305:
2301:
2297:
2293:
2289:
2285:
2281:
2274:
2272:
2270:
2268:
2266:
2264:
2260:
2255:
2251:
2246:
2241:
2236:
2231:
2227:
2223:
2219:
2212:
2210:
2208:
2206:
2204:
2202:
2200:
2198:
2196:
2194:
2192:
2190:
2186:
2181:
2177:
2173:
2169:
2164:
2159:
2155:
2151:
2147:
2143:
2135:
2133:
2131:
2129:
2127:
2123:
2118:
2114:
2109:
2104:
2099:
2094:
2090:
2086:
2082:
2078:
2074:
2067:
2065:
2063:
2061:
2059:
2057:
2055:
2053:
2049:
2044:
2040:
2035:
2030:
2026:
2022:
2018:
2011:
2009:
2007:
2005:
2003:
2001:
1999:
1997:
1995:
1993:
1991:
1989:
1987:
1985:
1983:
1981:
1979:
1977:
1975:
1973:
1971:
1969:
1967:
1965:
1963:
1961:
1959:
1957:
1955:
1953:
1951:
1949:
1947:
1945:
1943:
1941:
1939:
1935:
1930:
1926:
1922:
1915:
1912:
1900:
1896:
1889:
1886:
1873:
1869:
1867:
1858:
1855:
1850:
1846:
1842:
1838:
1833:
1828:
1824:
1820:
1816:
1809:
1806:
1800:
1798:
1792:
1790:
1783:
1781:
1777:
1770:
1768:
1766:
1761:
1757:
1753:
1749:
1745:
1737:
1735:
1733:
1729:
1728:phytoplankton
1720:
1712:
1705:
1703:
1701:
1691:
1684:
1682:
1679:
1678:macromolecule
1675:
1674:P. limnophila
1671:
1663:
1658:
1656:
1654:
1650:
1646:
1642:
1638:
1630:
1628:
1625:
1624:
1619:
1611:
1609:
1602:
1600:
1598:
1594:
1590:
1589:cyanobacteria
1586:
1578:
1576:
1574:
1567:
1566:
1560:
1558:
1554:
1550:
1546:
1544:
1539:
1535:
1534:
1529:
1528:
1519:
1517:
1512:
1505:
1503:
1501:
1497:
1493:
1489:
1481:
1477:
1473:
1471:
1466:
1462:
1460:
1456:
1453:
1448:
1446:
1445:carbohydrates
1442:
1438:
1433:
1431:
1423:
1421:
1419:
1415:
1411:
1407:
1403:
1399:
1395:
1391:
1386:
1378:
1376:
1373:
1372:hybridization
1371:
1368:Fluorescence
1365:
1363:
1359:
1350:
1345:
1334:
1333:
1325:
1324:
1316:
1315:
1307:
1306:
1298:
1297:
1289:
1288:
1285:
1284:
1278:
1277:
1274:
1273:
1270:
1269:
1263:
1262:
1256:
1255:
1252:
1251:
1248:
1247:
1241:
1240:
1234:
1233:
1230:
1229:
1226:
1225:
1224:Isosphaerales
1219:
1218:
1212:
1209:
1208:
1205:
1204:
1196:
1195:
1187:
1186:
1183:
1182:
1176:
1175:
1172:
1171:
1168:
1167:
1161:
1160:
1154:
1153:
1150:
1149:
1146:
1145:
1139:
1138:
1131:
1130:Phycisphaeria
1127:
1126:
1120:
1119:
1116:
1115:
1107:
1106:
1103:
1101:
1093:
1089:
1088:
1085:
1084:
1081:
1079:
1071:
1070:Uabimicrobiia
1067:
1066:
1060:
1059:
1053:
1052:
1048:
1041:
1040:
1032:
1031:
1023:
1022:
1014:
1013:
1010:
1009:
1003:
1002:
999:
998:
995:
994:
988:
987:
981:
980:
977:
976:
968:
967:
964:
963:
962:Isosphaerales
957:
956:
953:
952:
949:
948:
942:
941:
935:
934:
928:
925:
924:
921:
920:
912:
911:
903:
902:
899:
898:
892:
891:
888:
887:
884:
883:
877:
876:
870:
869:
866:
865:
862:
861:
855:
854:
848:
847:Phycisphaeria
845:
844:
838:
837:
833:
832:
828:
821:
816:
813:
810:
806:
799:
791:
789:
787:
783:
779:
774:
772:
768:
764:
756:
754:
752:
748:
744:
739:
737:
733:
729:
725:
721:
717:
713:
710:
706:
701:
699:
694:
691:
686:
684:
680:
676:
672:
669:
665:
657:
655:
654:superphylum.
651:
649:
645:
641:
633:
628:
626:
623:
619:
615:
610:
608:
607:P. limnophila
604:
603:P. limnophila
600:
595:
591:
589:
584:
582:
578:
574:
570:
566:
565:
560:
552:
547:
543:
539:
535:
531:
526:
522:
519:
518:P. limnophila
511:
504:
500:
498:
497:doubling time
494:
493:P. limnophila
486:
481:
479:
477:
468:
466:
462:
460:
456:
455:heterotrophic
452:
448:
444:
436:
434:
432:
428:
424:
419:
417:
413:
409:
408:invaginations
404:
400:
396:
388:
386:
384:
380:
379:cell division
376:
372:
368:
367:Gram-negative
364:
363:Gram-positive
356:
354:
352:
348:
347:P. limnophila
344:
340:
339:P. limnophila
335:
333:
329:
325:
320:
318:
314:
313:
308:
297:
290:
285:
280:
278:
274:
273:
267:
263:
261:
257:
251:
249:
244:
240:
237:
233:
229:
225:
221:
216:
214:
210:
206:
202:
198:
194:
190:
186:
173:
167:
161:
157:
151:
147:
141:
140:
138:
135:
131:
125:
124:Uabimicrobiia
122:
118:
115:
112:
111:
106:
104:
101:
99:
98:Phycisphaerae
96:
95:
93:
88:
81:
78:
75:
74:
71:
68:
66:Superphylum:
65:
64:
61:
58:
55:
54:
49:
44:
40:
37:
33:
29:
24:
19:
16:
7194:
7127:
7110:
7095:
7087:
7009:Orthonectida
7004:Gastrotricha
6979:Nematomorpha
6954:Chaetognatha
6944:Hemichordate
6871:Pteridophyta
6740:Malawimonada
6614:Korarchaeota
6575:Thermotogota
6560:Synergistota
6539:
6535:Nitrospirota
6530:Nitrospinota
6475:Deinococcota
6408:Bacteroidota
6300:
6267:Qinglongiota
6193:Canglongiota
6082:Nitrospirota
6072:Nitrospinota
6024:"Lernaellia"
5880:Chrysiogenia
5721:"Heilongiia"
5625:Cosmopoliota
5599:Calditrichia
5588:Rhodothermia
5584:"Kryptoniia"
5560:Bacteroidota
5503:
5469:Spirochaetia
5413:"Sumerlaeia"
5325:Omnitrophota
5263:"Aerophobia"
5155:Thermotogota
5103:Caldisericia
5078:Synergistota
5072:Atribacteria
5054:Thermotogida
4885:Limnochordia
4879:Bacillota G
4865:Bacillota E
4847:Bacillota D
4829:Bacillota A
4719:"Zipacnadia"
4674:Deinococcota
4656:Chloroflexia
4545:Opisthokonta
4408:
4342:
4338:
4303:(6): 85–93.
4300:
4296:
4290:
4245:
4241:
4231:
4206:
4202:
4196:
4177:
4171:
4138:
4134:
4100:
4096:
4058:
4054:
4044:
4009:
4005:
3959:
3956:Microbiology
3955:
3889:
3885:
3853:
3849:
3826:. Retrieved
3820:
3811:
3799:. Retrieved
3793:
3784:
3772:. Retrieved
3766:
3757:
3745:. Retrieved
3733:
3721:. Retrieved
3712:
3700:. Retrieved
3691:
3640:
3636:
3630:
3595:
3591:
3532:(2): 440–5.
3529:
3525:
3515:
3488:
3484:
3434:
3430:
3392:
3388:
3340:
3336:
3333:Superphylum"
3330:
3326:
3322:
3262:
3258:
3220:10033/623180
3202:
3198:
3195:Crateriforma
3194:
3138:
3134:
3084:
3080:
3074:
3018:
3014:
3004:
2961:
2957:
2915:
2911:
2894:
2854:(1): 14853.
2851:
2847:
2765:
2761:
2755:
2707:
2704:PLOS Biology
2703:
2641:
2637:
2575:
2571:
2521:
2517:
2459:
2455:
2451:
2403:
2399:
2341:
2337:
2287:
2283:
2225:
2221:
2163:10033/623182
2145:
2141:
2080:
2076:
2024:
2020:
1928:
1924:
1920:
1914:
1903:. Retrieved
1888:
1876:. Retrieved
1865:
1857:
1825:(10): 5056.
1822:
1818:
1808:
1796:
1787:
1778:
1774:
1764:
1759:
1756:myxobacteria
1747:
1741:
1717:
1696:
1673:
1667:
1641:Bacteroidota
1634:
1621:
1615:
1606:
1582:
1563:
1561:
1549:Kelp forests
1547:
1537:
1531:
1525:
1523:
1514:
1485:
1468:
1449:
1434:
1427:
1408:to generate
1398:thermophilic
1393:
1389:
1382:
1369:
1366:
1354:
1283:Pirellulales
1281:
1266:
1244:
1222:
1179:
1164:
1142:
1097:
1075:
1008:Pirellulales
1006:
991:
960:
945:
895:
880:
858:
804:
801:
784:function in
775:
760:
750:
746:
740:
723:
719:
702:
687:
677:to adapt to
663:
661:
652:
647:
643:
637:
617:
613:
611:
606:
602:
596:
592:
585:
562:
556:
553:Reproduction
545:
541:
537:
533:
529:
517:
515:
492:
490:
472:
463:
440:
427:surface area
425:triples the
422:
420:
402:
392:
382:
360:
350:
346:
338:
336:
331:
327:
321:
316:
310:
306:
303:
270:
269:The species
268:
264:
252:
239:Omnitrophica
238:
235:
217:
184:
182:
171:
155:
120:
108:
79:
35:
15:
7260:iNaturalist
7219:Wikispecies
7064:Brachiopoda
7039:Cycliophora
6984:Onychophora
6959:Kinorhyncha
6881:Ginkgophyta
6876:Cycadophyta
6841:Chlorophyta
6831:Glaucophyta
6525:Myxococcota
6443:Chlorobiota
6438:Chlamydiota
6310:Wikispecies
6305:(2001–2012)
6086:Nitrospiria
6076:Nitrospinia
6054:Myxococcota
6019:Lernaellota
5958:Oligoflexia
5716:Heilongiota
5565:Bacteroidia
5508:"Brocadiia"
5492:Chlamydiota
5423:Spirochaeto
5408:Sumerlaeota
5258:Aerophobota
5092:Thermocalda
5082:Synergistia
4912:"Moorellia"
4834:Clostridiia
4680:Deinococcia
4447:Prokaryotes
3747:23 February
3723:23 February
3702:23 February
3158:2066/103425
2905:carotenoid"
2768:(1): 2916.
1670:endocytosis
1664:Endocytosis
1557:marine snow
1459:carrageenan
1385:prokaryotic
1100:Brocadiales
778:Gemmataceae
771:Gemmataceae
716:ultraviolet
668:prokaryotic
443:Carotenoids
228:Chlamydiota
222:along with
211:and marine
7360:Categories
7097:Parakaryon
7059:Entoprocta
6994:Arthropoda
6989:Tardigrada
6969:Priapulida
6964:Loricifera
6949:Echinoderm
6919:Ctenophora
6891:Gnetophyta
6846:Charophyta
6836:Rhodophyta
6808:Ascomycota
6803:Zygomycota
6745:Metamonada
6720:Euglenozoa
6705:Ciliophora
6413:Balneolota
6388:Aquificota
6167:Holophagia
6066:Polyangiia
6059:Myxococcia
5838:Aquificota
5819:Aquificida
5570:Chlorobiia
5497:Chlamydiia
5438:"Babeliae"
4800:Firmicutes
4734:"Xenobiia"
3666:2066/35981
3331:Chlamydiae
2979:1822/50752
2452:Candidatus
2228:: 519301.
1905:2016-03-20
1878:August 26,
1801:References
1744:lanosterol
1732:atmosphere
1659:Physiology
1585:microalgae
1506:Macroalgae
1500:tube worms
1496:cold seeps
1492:bryophytes
1476:Ochrophyta
1358:Antarctica
1246:Gemmatales
947:Gemmatales
809:nucleotide
805:Candidatus
796:See also:
728:photolyase
343:microscopy
243:eukaryotes
236:Candidatus
209:macroalgae
7210:Q18674594
7143:Q18674593
7014:Dicyemida
6886:Pinophyta
6765:Telonemia
6700:Cryptista
6690:Breviatea
6675:Amoebozoa
6670:Alveolata
6651:Eukaryote
6403:Bacillota
5971:"Binatia"
5842:Aquificia
5550:FCB group
5482:PVC group
4960:CPR group
4816:Bacillota
4540:Amoebozoa
4518:Alveolata
4506:Cryptista
4481:Eukaryota
4359:1572-9699
4209:: 52–62.
3976:1350-0872
3914:0099-2240
3858:CiteSeerX
3697:"The LTP"
3614:0737-4038
3237:209516837
3167:0168-6445
3055:254909480
2940:204244301
2792:2041-1723
2644:(1): 66.
2484:232091472
2360:1664-302X
2304:0958-1669
2180:208641991
1849:239887308
1653:symbiotic
1543:symbiotic
1452:sulfatase
1362:Australia
1092:Brocadiia
829:08-RS214
822:_12_2021
792:Phylogeny
747:Pirellula
743:sulfatase
736:oxidation
679:mutations
573:synthesis
512:Lifecycle
416:periplasm
412:cytoplasm
114:Brocadiae
7204:Wikidata
7157:10815135
7137:Wikidata
7074:Annelida
7069:Mollusca
7049:Phoronid
7044:Nemertea
7019:Rotifera
6999:Flatworm
6974:Nematoda
6939:Chordate
6929:Cnidaria
6924:Placozoa
6755:Rhizaria
6730:Haptista
6710:Cercozoa
6370:Bacteria
6299:Source:
5966:Binatota
5433:Babelota
5242:BV2, BV4
4822:Bacillia
4535:Amorphea
4513:Rhizaria
4501:Hacrobia
4491:Excavata
4476:Bacteria
4451:Bacteria
4375:18752686
4367:23982431
4317:89295982
4282:25962786
4248:: 6878.
4223:90491277
4163:12315225
4155:23959164
4117:15910279
4077:24266389
4036:23859244
4028:18043661
3932:16820439
3675:16598256
3622:31808939
3564:24185849
3507:27279642
3463:23060863
3409:21050804
3369:28018303
3343:: 1964.
3297:21724885
3229:31894495
3175:23210799
3113:28066393
3087:: 2079.
3047:36534808
2996:13153498
2988:24052365
2932:31600855
2886:28393831
2810:31266954
2736:23700385
2676:31919386
2604:27570520
2578:: 1241.
2548:31740763
2476:33647766
2428:17003623
2420:23857394
2378:32754127
2344:: 1458.
2312:16704931
2254:33330115
2172:31802338
2117:12835416
2043:30052954
1929:Bacteria
1841:34694987
1623:Sphagnum
1430:detritus
782:paralogs
675:organism
561:such as
544:and (e)
478:lipids.
437:Pigments
324:Flagella
205:biofilms
193:bacteria
170:Whitman
134:Synonyms
76:Phylum:
60:Bacteria
56:Domain:
7054:Bryozoa
6750:Provora
6725:Jakobea
6715:Discoba
6661:Protist
6598:Archaea
6351:Extant
6094:SAR324
5350:Clade 2
5250:Clade 1
4969:Elulota
4550:Animals
4471:Archaea
4409:Gemmata
4273:4432595
4250:Bibcode
3984:7551018
3923:1489350
3894:Bibcode
3683:4402553
3645:Bibcode
3555:3911108
3534:Bibcode
3454:3444138
3437:: 327.
3360:5147048
3288:3165242
3267:Bibcode
3104:5177795
3038:9907078
2877:5394234
2856:Bibcode
2801:6606645
2770:Bibcode
2727:3660258
2667:6952346
2646:Bibcode
2595:4982196
2539:7286433
2369:7366357
2245:7734314
2085:Bibcode
1925:Archaea
1719:Anammox
1700:nucleus
1618:Siberia
1593:diatoms
1488:lichens
1455:enzymes
1414:nitrate
1410:sulfide
1370:in situ
1346:Ecology
705:nitrate
671:genomes
588:tubulin
581:budding
395:cytosol
375:polymer
277:sterols
256:budding
213:sponges
201:anammox
197:aquatic
7343:565455
7330:325165
7317:553035
7291:956119
7265:151858
7183:203682
6914:Sponge
6906:Animal
6259:others
6133:others
4808:Low GC
4588:others
4528:Plants
4465:Domain
4373:
4365:
4357:
4315:
4280:
4270:
4221:
4184:
4161:
4153:
4115:
4075:
4034:
4026:
3982:
3974:
3930:
3920:
3912:
3860:
3828:10 May
3801:10 May
3774:10 May
3681:
3673:
3637:Nature
3620:
3612:
3562:
3552:
3505:
3461:
3451:
3407:
3367:
3357:
3295:
3285:
3235:
3227:
3173:
3165:
3111:
3101:
3053:
3045:
3035:
2994:
2986:
2938:
2930:
2884:
2874:
2808:
2798:
2790:
2734:
2724:
2674:
2664:
2602:
2592:
2546:
2536:
2482:
2474:
2426:
2418:
2376:
2366:
2358:
2310:
2302:
2252:
2242:
2178:
2170:
2115:
2108:166223
2105:
2041:
1874:(LPSN)
1847:
1839:
1647:, and
1595:, and
1406:sulfur
709:enzyme
693:operon
487:Growth
447:plants
431:volume
189:phylum
187:are a
174:. 2018
158:. 2015
7338:WoRMS
7273:IRMNG
6823:Plant
6775:Fungi
6695:CRuMs
4555:Fungi
4371:S2CID
4313:S2CID
4219:S2CID
4159:S2CID
4032:S2CID
3742:(PDF)
3679:S2CID
3233:S2CID
3051:S2CID
2992:S2CID
2936:S2CID
2908:(PDF)
2480:S2CID
2424:S2CID
2176:S2CID
1845:S2CID
698:genes
622:clade
599:yeast
451:fungi
172:et al
156:et al
154:Oren
7299:LPSN
7286:ITIS
7247:GBIF
7239:7716
7178:NCBI
7165:LPSN
7152:GBIF
6861:Moss
6353:life
4986:ABY1
4363:PMID
4355:ISSN
4278:PMID
4182:ISBN
4151:PMID
4113:PMID
4073:PMID
4024:PMID
3980:PMID
3972:ISSN
3928:PMID
3910:ISSN
3830:2023
3803:2023
3776:2023
3749:2021
3725:2021
3704:2021
3671:PMID
3618:PMID
3610:ISSN
3560:PMID
3503:PMID
3459:PMID
3405:PMID
3365:PMID
3293:PMID
3225:PMID
3171:PMID
3163:ISSN
3109:PMID
3043:PMID
2984:PMID
2928:PMID
2882:PMID
2806:PMID
2788:ISSN
2732:PMID
2672:PMID
2600:PMID
2544:PMID
2472:PMID
2416:PMID
2374:PMID
2356:ISSN
2308:PMID
2300:ISSN
2250:PMID
2168:PMID
2113:PMID
2039:PMID
1880:2021
1837:PMID
1754:and
1530:and
1490:and
1396:, a
1360:and
827:GTDB
734:and
724:uvrC
690:rRNA
577:FtsZ
540:(d)
536:(c)
532:(b)
449:and
414:and
365:and
349:and
183:The
7278:188
7234:EoL
5811:BV2
4804:BV3
4347:doi
4343:104
4305:doi
4268:PMC
4258:doi
4211:doi
4207:133
4143:doi
4139:104
4105:doi
4063:doi
4014:doi
3964:doi
3960:141
3918:PMC
3902:doi
3661:hdl
3653:doi
3641:440
3600:doi
3550:PMC
3542:doi
3493:doi
3449:PMC
3439:doi
3397:doi
3355:PMC
3345:doi
3283:PMC
3275:doi
3215:hdl
3207:doi
3203:113
3153:hdl
3143:doi
3099:PMC
3089:doi
3033:PMC
3023:doi
3019:119
2974:hdl
2966:doi
2962:104
2920:doi
2872:PMC
2864:doi
2796:PMC
2778:doi
2722:PMC
2712:doi
2662:PMC
2654:doi
2590:PMC
2580:doi
2534:PMC
2526:doi
2464:doi
2408:doi
2404:104
2364:PMC
2346:doi
2292:doi
2240:PMC
2230:doi
2158:hdl
2150:doi
2146:113
2103:PMC
2093:doi
2081:100
2029:doi
1827:doi
820:LTP
738:.
722:and
121:Ca.
110:Ca.
7362::
7340::
7327::
7314::
7301::
7288::
7275::
7262::
7252:90
7249::
7236::
7221::
7206::
7180::
7167::
7154::
7139::
6269:"
6207:"
6195:"
6113:"
6045:"
6033:"
6021:"
6009:"
5995:"
5980:"
5968:"
5921:"
5889:"
5867:"
5829:"
5779:"
5757:"
5745:"
5730:"
5718:"
5675:"
5660:"
5627:"
5608:"
5435:"
5410:"
5398:"
5379:"
5367:"
5327:"
5272:"
5260:"
5217:"
5205:"
5193:"
5146:"
5134:"
4983:"
4971:"
4894:"
4777:"
4691:"
4639:"
4449::
4369:.
4361:.
4353:.
4341:.
4337:.
4325:^
4311:.
4301:35
4299:.
4276:.
4266:.
4256:.
4244:.
4240:.
4217:.
4205:.
4157:.
4149:.
4137:.
4125:^
4111:.
4101:59
4099:.
4085:^
4071:.
4059:88
4057:.
4053:.
4030:.
4022:.
4008:.
4004:.
3992:^
3978:.
3970:.
3958:.
3954:.
3940:^
3926:.
3916:.
3908:.
3900:.
3890:72
3888:.
3884:.
3872:^
3856:.
3854:38
3852:.
3838:^
3819:.
3792:.
3765:.
3677:.
3669:.
3659:.
3651:.
3639:.
3616:.
3608:.
3596:37
3594:.
3590:.
3572:^
3558:.
3548:.
3540:.
3530:80
3528:.
3524:.
3501:.
3489:40
3487:.
3483:.
3471:^
3457:.
3447:.
3433:.
3429:.
3417:^
3403:.
3393:13
3377:^
3363:.
3353:.
3339:.
3335:.
3305:^
3291:.
3281:.
3273:.
3263:77
3261:.
3257:.
3245:^
3231:.
3223:.
3213:.
3201:.
3183:^
3169:.
3161:.
3151:.
3139:37
3137:.
3133:.
3121:^
3107:.
3097:.
3083:.
3079:.
3063:^
3049:.
3041:.
3031:.
3017:.
3013:.
2990:.
2982:.
2972:.
2960:.
2948:^
2934:.
2926:.
2916:11
2914:.
2910:.
2903:45
2880:.
2870:.
2862:.
2850:.
2846:.
2818:^
2804:.
2794:.
2786:.
2776:.
2766:10
2764:.
2760:.
2744:^
2730:.
2720:.
2708:11
2706:.
2702:.
2684:^
2670:.
2660:.
2652:.
2642:10
2640:.
2636:.
2612:^
2598:.
2588:.
2574:.
2570:.
2556:^
2542:.
2532:.
2520:.
2516:.
2492:^
2478:.
2470:.
2460:44
2458:.
2436:^
2422:.
2414:.
2402:.
2386:^
2372:.
2362:.
2354:.
2342:11
2340:.
2336:.
2320:^
2306:.
2298:.
2288:17
2282:.
2262:^
2248:.
2238:.
2226:10
2224:.
2220:.
2188:^
2174:.
2166:.
2156:.
2144:.
2125:^
2111:.
2101:.
2091:.
2079:.
2075:.
2051:^
2037:.
2025:42
2023:.
2019:.
1937:^
1897:.
1870:.
1868:""
1864:""
1843:.
1835:.
1823:71
1821:.
1817:.
1767:.
1643:,
1639:,
1591:,
1570:CO
1502:.
1461:.
1418:pH
1392:.
1364:.
1132:"
1102:"
1094:"
1080:"
1072:"
583:.
418:.
300:nm
262:.
230:,
226:,
215:.
6663:"
6659:"
6623:"
6619:"
6616:"
6612:"
6609:"
6605:"
6466:"
6462:"
6344:e
6337:t
6330:v
6288:"
6284:"
6281:"
6277:"
6265:"
6238:"
6234:"
6231:"
6227:"
6224:"
6220:"
6203:"
6191:"
6176:"
6172:"
6169:"
6165:"
6148:"
6144:"
6125:"
6121:"
6109:"
6106:"
6102:"
6041:"
6029:"
6017:"
6005:"
5991:"
5976:"
5964:"
5917:"
5885:"
5863:"
5825:"
5798:"
5794:"
5791:"
5787:"
5775:"
5772:"
5768:"
5753:"
5741:"
5726:"
5714:"
5671:"
5656:"
5653:"
5649:"
5646:"
5642:"
5639:"
5635:"
5623:"
5620:"
5616:"
5604:"
5578:"
5574:"
5471:"
5467:"
5447:"
5443:"
5431:"
5406:"
5394:"
5391:"
5387:"
5375:"
5363:"
5360:"
5356:"
5342:"
5338:"
5323:"
5320:"
5316:"
5313:"
5309:"
5293:"
5289:"
5268:"
5256:"
5229:"
5225:"
5213:"
5201:"
5189:"
5142:"
5130:"
5044:"
5040:"
5022:"
5018:"
5012:"
5008:"
4999:"
4995:"
4979:"
4967:"
4945:"
4941:"
4933:"
4929:"
4890:"
4857:"
4853:"
4836:"
4832:"
4824:"
4820:"
4773:"
4768:"
4764:"
4728:"
4724:"
4687:"
4682:"
4678:"
4635:"
4562:)
4439:e
4432:t
4425:v
4377:.
4349::
4319:.
4307::
4284:.
4260::
4252::
4246:6
4225:.
4213::
4190:.
4165:.
4145::
4119:.
4107::
4079:.
4065::
4038:.
4016::
4010:1
3986:.
3966::
3934:.
3904::
3896::
3866:.
3832:.
3805:.
3778:.
3751:.
3727:.
3706:.
3685:.
3663::
3655::
3647::
3624:.
3602::
3566:.
3544::
3536::
3509:.
3495::
3465:.
3441::
3435:3
3411:.
3399::
3371:.
3347::
3341:7
3329:-
3325:-
3299:.
3277::
3269::
3239:.
3217::
3209::
3177:.
3155::
3145::
3115:.
3091::
3085:7
3073:"
3057:.
3025::
2998:.
2976::
2968::
2942:.
2922::
2888:.
2866::
2858::
2852:8
2812:.
2780::
2772::
2758:"
2738:.
2714::
2678:.
2656::
2648::
2606:.
2582::
2576:7
2550:.
2528::
2522:5
2486:.
2466::
2430:.
2410::
2380:.
2348::
2314:.
2294::
2256:.
2232::
2182:.
2160::
2152::
2119:.
2095::
2087::
2045:.
2031::
1908:.
1882:.
1851:.
1829::
1724:2
1722:N
1572:2
1518:,
1472:,
1128:"
1098:"
1090:"
1076:"
1068:"
279:.
144:(
126:"
119:"
116:"
107:"
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.