1125:(SFVT) approach for HLA genetic analysis that categorizes HLA proteins into biologically relevant smaller sequence features (SFs), and their variant types (VTs). Sequence features are combinations of amino acid sites defined based on structural information (e.g., beta-sheet 1), functional information (e.g., peptide antigen-binding), and polymorphism. These sequence features can be overlapping and continuous or discontinuous in the linear sequence. Variant types for each sequence feature are defined based upon all known polymorphisms in the HLA locus being described. SFVT categorization of HLA is applied in genetic association analysis so that the effects and roles of the epitopes shared by several HLA alleles can be identified. Sequence features and their variant types have been described for all classical HLA proteins; the international repository of HLA SFVTs will be maintained at IMGT/HLA database. A tool to convert HLA alleles into their component SFVTs can be found on the Immunology Database and Analysis Portal (ImmPort) website.
1650:
receptors and receptor isoforms. Over the years, serotyping antibodies became more refined as techniques for increasing sensitivity improved and new serotyping antibodies continue to appear. One of the goals of serotype analysis is to fill gaps in the analysis. It is possible to predict based on 'square root','maximum-likelihood' method, or analysis of familial haplotypes to account for adequately typed alleles. These studies using serotyping techniques frequently revealed, in particular for non-European or north East Asian populations many null or blank serotypes. This was particularly problematic for the Cw locus until recently, and almost half of the Cw serotypes went untyped in the 1991 survey of the human population.
395:. T-cells have receptors that are similar to B-cell receptors, and each T-cell recognizes only a few MHC class II-peptide combinations. Once a T-cell recognizes a peptide within an MHC class II molecule, it can stimulate B-cells that also recognize the same molecule in their B-cell receptors. Thus, T-cells help B-cells make antibodies to the same foreign antigens. Each HLA can bind many peptides, and each person has 3 HLA types and can have 4 isoforms of DP, 4 isoforms of DQ and 4 Isoforms of DR (2 of DRB1, and 2 of DRB3, DRB4, or DRB5) for a total of 12 isoforms. In such heterozygotes, it is difficult for disease-related proteins to escape detection.
849:
could not completely distinguish alleles and so stopped at this level. The third through fourth digits specify a nonsynonymous allele. Digits five through six denote any synonymous mutations within the coding frame of the gene. The seventh and eighth digits distinguish mutations outside the coding region. Letters such as L, N, Q, or S may follow an allele's designation to specify an expression level or other non-genomic data known about it. Thus, a completely described allele may be up to 9 digits long, not including the HLA-prefix and locus notation.
1663:
designated as Dw types. Serotyped DR1 has cellularly defined as either of Dw1 or of Dw20 and so on for other serotyped DRs. Table shows associated cellular specificities for DR alleles. However, cellular typing has inconsistency in the reaction between cellular-type individuals, sometimes resulting differently from predicted. Together with difficulty of cellular assay in generating and maintaining cellular typing reagents, cellular assay is being replaced by DNA-based typing method.
1846:
receptors are capable of binding an almost endless variation of peptides of 9 amino acids or longer in length, protecting interbreeding subpopulations from nascent or epidemic diseases. Individuals in a population frequently have different haplotypes, and this results in many combinations, even in small groups. This diversity enhances the survival of such groups, and thwarts evolution of epitopes in pathogens, which would otherwise be able to be shielded from the immune system.
864:
42:
1842:
have fewer alleles, combinations of A1:B1 can produce a theoretical potential of 7,755 DQ and 5,270 DP αβ heterodimers, respectively. While nowhere near this number of isoforms exist in the human population, each individual can carry 4 variable DQ and DP isoforms, increasing the potential number of antigens that these receptors can present to the immune system.
1138:
been detected at least five times in unrelated individuals via the application of a sequence-based typing (SBT) method, or at least three times via a SBT method and in a specific haplotype in unrelated individuals. Rare alleles are defined as those that have been reported one to four times, and very rare alleles as those reported only once.
194:), all of which are the HLA Class1 group, present peptides from inside the cell. For example, if the cell is infected by a virus, the HLA system brings fragments of the virus to the surface of the cell so that the cell can be destroyed by the immune system. These peptides are produced from digested proteins that are broken down in the
1833:
the world where
Western Europeans have migrated. The "A3-B7-DR2-DQ1" is more widely spread, from Eastern Asia to Iberia. The Super-B8 haplotype is associated with a number of diet-associated autoimmune diseases. There are 100,000s of extended haplotypes, but only a few show a visible and nodal character in the human population.
408:. Because of the importance of HLA in transplantation, the HLA loci are some of the most frequently typed by serology and PCR. It has been shown that high resolution HLA typing (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1 and HLA-DPB1) may be relevant in transplantation to identify a full match, even when the donor is related.
91:
287:. They may protect against cancers or fail to protect (if down-regulated by an infection). HLA may also be related to people's perception of the odor of other people, and may be involved in mate selection, as at least one study found a lower-than-expected rate of HLA similarity between spouses in an isolated community.
679:
enteropathy-associated T-cell lymphoma cases. More often, however, HLA molecules play a protective role, recognizing increases in antigens that are not tolerated because of low levels in the normal state. Abnormal cells might be targeted for apoptosis, which is thought to mediate many cancers before diagnosis.
885:
variant of the nucleotide (DNA) sequence at a locus, such that each allele differs from all other alleles in at least one (single nucleotide polymorphism, SNP) position. Most of these changes result in a change in the amino acid sequences that result in slight to major functional differences in the protein.
1845:
Studies of the variable positions of DP, DR, and DQ reveal that peptide antigen contact residues on class II molecules are most frequently the site of variation in the protein primary structure. Therefore, through a combination of intense allelic variation and/or subunit pairing, the class II peptide
1841:
Studies of humans and animals imply a heterozygous selection mechanism operating on these loci as an explanation for this variability. One proposed mechanism is sexual selection in which females are able to detect males with different HLA relative to their own type. While the DQ and DP encoding loci
1752:
For many populations, such as the
Japanese or European populations, so many patients have been typed that new alleles are relatively rare, and thus SSP-PCR is more than adequate for allele resolution. Haplotypes can be obtained by typing family members in areas of the world where SSP-PCR is unable to
1653:
There are several types of serotypes. A broad antigen serotype is a crude measure of identity of cells. For example, HLA A9 serotype recognizes cells of A23- and A24-bearing individuals. It may also recognize cells that A23 and A24 miss because of small variations. A23 and A24 are split antigens, but
892:
However, the gene frequencies of the most common alleles (>5%) of HLA-A, -B, -C and HLA-DPA1, -DPB1, -DQA1, -DQB1, and -DRB1 from South
America have been reported from the typing and sequencing carried out in genetic diversity studies and cases and controls. In addition, information on the allele
871:
MHC loci are some of the most genetically variable coding loci in mammals, and the human HLA loci are no exceptions. Despite the fact that the human population went through a constriction several times during its history that was capable of fixing many loci, the HLA loci appear to have survived such
1692:
Gene typing is different from gene sequencing and serotyping. With this strategy, PCR primers specific to a variant region of DNA are used (called sequence-specific primers). If a product of the right size is found, the assumption is that the HLA allele has been identified. New gene sequences often
1679:
In the end, a workshop, based on sequence, decides which new allele goes into which serogroup either by sequence or by reactivity. Once the sequence is verified, it is assigned a number. For example, a new allele of B44 may get a serotype (i.e. B44) and allele ID i.e. B*44:65, as it is the 65th B44
1662:
A representative cellular assay is the mixed lymphocyte culture (MLC) and used to determine the HLA class II types. The cellular assay is more sensitive in detecting HLA differences than serotyping. This is because minor differences unrecognized by alloantisera can stimulate T cells. This typing is
848:
Modern HLA alleles are typically noted with a variety of levels of detail. Most designations begin with HLA- and the locus name, then * and some (even) number of digits specifying the allele. The first two digits specify a group of alleles, also known as supertypes. Older typing methodologies often
1832:
These haplotypes can be used to trace migrations in the human population because they are often much like a fingerprint of an event that has occurred in evolution. The Super-B8 haplotype is enriched in the
Western Irish, declines along gradients away from that region, and is found only in areas of
1134:
variation in the rate at which of individual HLA alleles are detected, attempts have been made to categorize alleles at each expressed HLA locus in terms of their prevalence. The result is a catalog of common and well-documented (CWD) HLA alleles, and a catalogue of rare and very rare HLA alleles.
1133:
Although the number of individual HLA alleles that have been identified is large, approximately 40% of these alleles appear to be unique, having only been identified in single individuals. Roughly a third of alleles have been reported more than three times in unrelated individuals. Because of this
1649:
In order to create a typing reagent, blood from animals or humans would be taken, the blood cells allowed to separate from the serum, and the serum diluted to its optimal sensitivity and used to type cells from other individuals or animals. Thus, serotyping became a way of crudely identifying HLA
1137:
Common HLA alleles are defined as having been observed with a frequency of at least 0.001 in reference populations of at least 1500 individuals. Well-documented HLA alleles were originally defined as having been reported at least three times in unrelated individuals, and are now defined as having
888:
There are issues that limit this variation. Certain alleles like DQA1*05:01 and DQA1*05:05 encode proteins with identically processed products. Other alleles like DQB1*0201 and DQB1*0202 produce proteins that are functionally similar. For class II (DR, DP and DQ), amino acid variants within the
884:
Six loci have over 100 alleles that have been detected in the human population. Of these, the most variable are HLA B and HLA DRB1. As of 2012, the number of alleles that have been determined are listed in the table below. To interpret this table, it is necessary to consider that an allele is a
290:
Aside from the genes encoding the six major antigen-presenting proteins, many other genes, many involved in immune function, are located on the HLA complex. Diversity of HLAs in the human population is one aspect of disease defense, and, as a result, the chance of two unrelated individuals with
1675:
Broad antigen types are still useful, such as typing very diverse populations with many unidentified HLA alleles (Africa, Arabia, Southeastern Iran and
Pakistan, India). Africa, Southern Iran, and Arabia show the difficulty in typing areas that were settled earlier. Allelic diversity makes it
1671:
Minor reactions to subregions that show similarity to other types can be observed to the gene products of alleles of a serotype group. The sequence of the antigens determines the antibody reactivities, and so having a good sequencing capability (or sequence-based typing) obviates the need for
1120:
The large extent of variability in HLA genes poses significant challenges in investigating the role of HLA genetic variations in diseases. Disease association studies typically treat each HLA allele as a single complete unit, which does not illuminate the parts of the molecule associated with
1861:
Donor-specific HLA antibodies have been found to be associated with graft failure in renal, heart, lung, and liver transplantation. These donor-specific HLA antibodies can exist pretransplant as consequence of sensitization to prior transplants or through pregnancies, but also occur de novo
387:
The image off to the side shows a piece of a poisonous bacterial protein (SEI peptide) bound within the binding cleft portion of the HLA-DR1 molecule. In the illustration far below, a different view, one can see an entire DQ with a bound peptide in a similar cleft, as viewed from the side.
678:
Some HLA-mediated diseases are directly involved in the promotion of cancer. Gluten-sensitive enteropathy is associated with increased prevalence of enteropathy-associated T-cell lymphoma, and DR3-DQ2 homozygotes are within the highest risk group, with close to 80% of gluten-sensitive
1854:
HLA antibodies are typically not naturally occurring, and with few exceptions are formed as a result of an immunologic challenge to a foreign material containing non-self HLAs via blood transfusion, pregnancy (paternally inherited antigens), or organ or tissue transplant.
1618:
or, shortened, B27). A parallel system that allowed more refined definition of alleles was developed. In this system, an "HLA" is used in conjunction with a letter, *, and a four-or-more-digit number (e.g., HLA-B*08:01, A*68:01, A*24:02:01N N=Null) to designate a specific
403:
Any cell displaying some other HLA type is "non-self" and is seen as an invader by the body's immune system, resulting in the rejection of the tissue bearing those cells. This is particularly important in the case of transplanted tissue, because it could lead to
876:
coefficient for these loci. In addition, some HLA loci are among the fastest-evolving coding regions in the human genome. One mechanism of diversification has been noted in the study of
Amazonian tribes of South America that appear to have undergone intense
872:
a constriction with a great deal of variation. Of the 9 loci mentioned above, most retained a dozen or more allele-groups for each locus, far more preserved variation than the vast majority of human loci. This is consistent with a heterozygous or
661:
HLA typing has led to some improvement and acceleration in the diagnosis of celiac disease and type 1 diabetes; however, for DQ2 typing to be useful, it requires either high-resolution B1*typing (resolving *02:01 from *02:02), DQA1*typing, or DR
670:, it is the only effective means of discriminating between first-degree relatives that are at risk from those that are not at risk, prior to the appearance of sometimes-irreversible symptoms such as allergies and secondary autoimmune disease.
214:-positive or cytotoxic T-cells) that destroy cells. Some new work has proposed that antigens longer than 10 amino acids, 11-14 amino acids, can be presented on MHC I, eliciting a cytotoxic T-cell response. MHC class I proteins associate with
3190:
Gonzalez-Galarza FF, Mack SJ, Hollenbach J, Fernandez-Vina M, Setterholm M, Kempenich J, et al. (February 2013). "16(th) IHIW: extending the number of resources and bioinformatics analysis for the investigation of HLA rare alleles".
1753:
recognize alleles and typing requires the sequencing of new alleles. Areas of the world where SSP-PCR or serotyping may be inadequate include
Central Africa, Eastern Africa, parts of southern Africa, Arabia, S. Iran, Pakistan, and India.
1613:
There are two parallel systems of nomenclature that are applied to HLA. The first, and oldest, system is based on serological (antibody based) recognition. In this system, antigens were eventually assigned letters and numbers (e.g.,
1898:
2482:
Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, et al. (June 2004). "HLA-DQ relative risks for coeliac disease in
European populations: a study of the European Genetics Cluster on Coeliac Disease".
3241:
Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, et al. (May 2007). "Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics".
3644:
Senev A, Coemans M, Lerut E, Van Sandt V, Kerkhofs J, Daniëls L, Driessche MV, Compernolle V, Sprangers B, Van Loon E, Callemeyn J, Claas F, Tambur AR, Verbeke G, Kuypers D, Emonds MP, Naesens M (September 2020).
893:
frequencies of HLA-I and HLA-II genes for the
European population has been compiled. In both cases the distribution of allele frequencies reveals a regional variation related with the history of the populations.
3443:
Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H (November 2004). "Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of
Pakistan".
760:
There are three major and two minor MHC class II proteins encoded by the HLA. The genes of the class II combine to form heterodimeric (αβ) protein receptors that are typically expressed on the surface of
470:
2880:
Nunes JM, Buhler S, Roessli D, Sanchez-Mazas A (May 2014). "The HLA-net GENE pipeline for effective HLA data analysis and its application to 145 population samples from Europe and neighbouring areas".
3408:
Valluri V, Valluei V, Mustafa M, Santhosh A, Middleton D, Alvares M, et al. (August 2005). "Frequencies of HLA-A, HLA-B, HLA-DR, and HLA-DQ phenotypes in the United Arab Emirates population".
318:
DR protein (DRA:DRB1*0101 gene products) with bound Staphylococcal enterotoxin ligand (subunit I-C), view is top down showing all DR amino acid residues within 5 Angstroms of the SEI peptide.
3601:
Oshima M, Deitiker P, Ashizawa T, Atassi MZ (May 2002). "Vaccination with a MHC class II peptide attenuates cellular and humoral responses against tAChR and suppresses clinical EAMG".
1693:
result in an increasing appearance of ambiguity. Because gene typing is based on SSP-PCR, it is possible that new variants, in particular in the class I and DRB1 loci, may be missed.
881:
between variable alleles and loci within each HLA gene class. Less frequently, longer-range productive recombinations through HLA genes have been noted producing chimeric genes.
836:
The other MHC class II proteins, DM and DO, are used in the internal processing of antigens, loading the antigenic peptides generated from pathogens onto the HLA molecules of
3138:
Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy JM, Marsh SG, Aubrey M, et al. (December 2009). "A bioinformatics approach to ascertaining the rarity of HLA alleles".
3647:"Eplet Mismatch Load and De Novo Occurrence of Donor-Specific Anti-HLA Antibodies, Rejection, and Graft Failure after Kidney Transplantation: An Observational Cohort Study"
1999:"The Role of Major Histocompatibility Complex in Organ Transplantation- Donor Specific Anti-Major Histocompatibility Complex Antibodies Analysis Goes to the Next Stage"
853:
168:
376:
Through a similar process, proteins (both native and foreign, such as the proteins of viruses) produced inside most cells are displayed on HLAs (to be specific,
311:
uses the HLAs to differentiate self cells and non-self cells. Any cell displaying that person's HLA type belongs to that person and is therefore not an invader.
269:. Predicting which (fragments of) antigens will be presented to the immune system by a certain HLA type is difficult, but the technology involved is improving.
1696:
For example, SSP-PCR within the clinical situation is often used for identifying HLA phenotypes. An example of an extended phenotype for a person might be:
295:
is extremely low. HLA genes have historically been identified as a result of the ability to successfully transplant organs between HLA-similar individuals.
1680:
allele discovered. Marsh et al. (2005) can be considered a code book for HLA serotypes and genotypes, and a new book biannually with monthly updates in
2048:
Matsumura M, Fremont DH, Peterson PA, Wilson IA (August 1992). "Emerging principles for the recognition of peptide antigens by MHC class I molecules".
2917:"A new HLA map of Europe: Regional genetic variation and its implication for peopling history, disease-association studies and tissue transplantation"
391:
When bound, peptides are presented to T-cells. T-cells require presentation via MHC molecules to recognize foreign antigens—a requirement known as
1877:
1672:
serological reactions. Therefore, different serotype reactions may indicate the need to sequence a person's HLA to determine a new gene sequence.
1676:
necessary to use broad antigen typing followed by gene sequencing because there is an increased risk of misidentifying by serotyping techniques.
3863:
315:
3750:
2761:
2466:
2432:
1862:
post-transplantation. There is a clear link between the risk of HLA antibody sensitisation and the donor-recipient HLA (molecular) mismatch.
3479:
Shankarkumar U, Prasanavar D, Ghosh K, Mohanty D (May 2003). "HLA A*02 allele frequencies and B haplotype associations in Western Indians".
3373:
Bodmer JG, Marsh SG, Albert ED, Bodmer WF, Dupont B, Erlich HA, et al. (May 1992). "Nomenclature for factors of the HLA system, 1991".
3115:
4129:
2128:"Empirical Evaluation of the Use of Computational HLA Binding as an Early Filter to the Mass Spectrometry-Based Epitope Discovery Workflow"
2177:
Galbraith W, Wagner MC, Chao J, Abaza M, Ernst LA, Nederlof MA, et al. (1991). "Imaging cytometry by multiparameter fluorescence".
753:
666:. Current serotyping can resolve, in one step, DQ8. HLA typing in autoimmunity is being increasingly used as a tool in diagnosis. In
3782:
3765:
3360:
1881:
4063:
3872:
123:
1146:
While the current CWD and rare or very rare designations were developed using different datasets and different versions of the
2126:
Bouzid R, de Beijer MT, Luijten RJ, Bezstarosti K, Kessler AL, Bruno MJ, Peppelenbosch MP, Demmers JA, Buschow SI (May 2021).
1761:
An HLA haplotype is a series of HLA "genes" (loci-alleles) by chromosome, one passed from the mother and one from the father.
756:
Illustration of an HLA-DQ molecule (magenta and blue) with a bound ligand (yellow) floating on the plasma membrane of the cell
4139:
4124:
2831:"Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country"
2730:
1635:(or rarely, D locus). Every two years, a nomenclature is put forth to aid researchers in interpreting serotypes to alleles.
4134:
1122:
3514:
Apanius V, Penn D, Slev PR, Ruff LR, Potts WK (1997). "The nature of selection on the major histocompatibility complex".
639:
543:
497:
1858:
Antibodies against disease-associated HLA haplotypes have been proposed as a treatment for severe autoimmune diseases.
3026:
2667:"Gamete-level immunogenetic incompatibility in humans-towards deeper understanding of fertilization and infertility?"
622:
and other diseases. People with certain HLA antigens are more likely to develop certain autoimmune diseases, such as
249:) present antigens from outside of the cell to T-lymphocytes. These particular antigens stimulate multiplication of
4093:
3856:
2364:"The Case for High Resolution Extended 6-Loci HLA Typing for Identifying Related Donors in the Indian Subcontinent"
4083:
3792:
2091:
Burrows SR, Rossjohn J, McCluskey J (January 2006). "Have we cut ourselves too short in mapping CTL epitopes?".
3825:
762:
583:
568:
533:
4114:
3796:
3101:
837:
769:
647:
366:
346:
4119:
4088:
1764:
The phenotype exampled above is one of the more common in Ireland and is the result of two common genetic
627:
455:
435:
307:
encoded by HLAs are those on the outer part of body cells that are (in effect) unique to that person. The
158:
901:
Number of variant alleles at class I loci according to the IMGT-HLA database, last updated October 2018:
485:
3849:
1147:
219:
2777:
Parham P, Ohta T (April 1996). "Population biology of antigen presentation by MHC class I molecules".
2618:"Post-copulatory genetic matchmaking: HLA-dependent effects of cervical mucus on human sperm function"
3559:
3550:
Wedekind C, Seebeck T, Bettens F, Paepke AJ (June 1995). "MHC-dependent mate preferences in humans".
2786:
2057:
742:
631:
558:
512:
405:
284:
215:
150:
146:
3821:
1884:
may be used to give rise to a sibling with matching HLA, although there are ethical considerations.
3050:"Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis"
873:
692:
651:
619:
523:
373:, which then produce a variety of effects and cell-to-cell interactions to eliminate the pathogen.
77:
2616:
Jokiniemi A, Magris M, Ritari J, Kuusipalo L, Lundgren T, Partanen J, Kekäläinen J (August 2020).
3626:
3583:
3216:
3119:
2810:
2704:
2292:
445:
130:
2569:"Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups"
1654:
antibodies specific to either are typically used more often than antibodies to broad antigens.
4068:
3761:
3746:
3717:
3676:
3618:
3575:
3531:
3496:
3461:
3425:
3390:
3356:
3323:
3259:
3208:
3155:
3079:
2992:
2938:
2897:
2862:
2802:
2757:
2696:
2647:
2598:
2549:
2500:
2462:
2428:
2385:
2344:
2284:
2243:
2194:
2159:
2108:
2073:
2030:
1953:
1796:
889:
receptor's peptide-binding cleft tend to produce molecules with different binding capability.
643:
320:
277:
266:
2452:
3707:
3666:
3658:
3610:
3567:
3523:
3488:
3453:
3417:
3382:
3313:
3305:
3251:
3200:
3147:
3069:
3061:
2982:
2974:
2928:
2889:
2852:
2842:
2794:
2686:
2678:
2637:
2629:
2588:
2580:
2539:
2531:
2492:
2375:
2334:
2326:
2274:
2263:"Serotyping for homotransplantation. XVII. Preliminary studies of HL-A subunits and alleles"
2233:
2225:
2186:
2149:
2139:
2100:
2065:
2020:
2010:
1943:
1935:
1749:
In general, this is identical to the extended serotype: A1,A3,B7,B8,DR3,DR15(2), DQ2,DQ6(1)
1624:
381:
292:
172:
115:
3841:
3048:
Karp DR, Marthandan N, Marsh SG, Ahn C, Arnett FC, Deluca DS, et al. (February 2010).
2961:
Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. (April 2010).
4030:
878:
635:
623:
602:
392:
283:
HLAs have other roles. They are important in disease defense. They are the major cause of
134:
2665:
Jokiniemi A, Kuusipalo L, Ritari J, Koskela S, Partanen J, Kekäläinen J (November 2020).
1971:
1150:, the approximate fraction of alleles at each HLA locus in each category is shown below.
3563:
3292:
Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, et al. (April 2013).
2790:
2319:
Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences
2218:
Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences
2061:
17:
3671:
3646:
3318:
3293:
3074:
3049:
2987:
2962:
2857:
2830:
2691:
2642:
2617:
2593:
2568:
2544:
2519:
2339:
2314:
2238:
2213:
2154:
2127:
2025:
1998:
1948:
1923:
1871:
667:
342:
138:
3492:
2315:"Immunological considerations for embryonic and induced pluripotent stem cell banking"
314:
4108:
3742:
3527:
3457:
3421:
3386:
3220:
3151:
2978:
2708:
2496:
2279:
2262:
703:
MHC class I proteins form a functional receptor on most nucleated cells of the body.
423:
370:
308:
273:
250:
207:
119:
3587:
2814:
2296:
388:
Disease-related peptides fit into these "slots" much like a hand fits into a glove.
3924:
3630:
1924:"The HLA system: genetics, immunology, clinical testing, and clinical implications"
1632:
830:
4 β-chains (only 3 possible per person), encoded by HLA-DRB1, DRB3, DRB4, DRB5 loci
362:
350:
226:
111:
2722:
691:
with respect to certain genetic characteristics. This has led to a field known as
206:
in length. Foreign antigens presented by MHC class I attract T-lymphocytes called
3255:
82:
70:
4078:
3881:
3836:
2798:
2362:
Agarwal RK, Kumari A, Sedai A, Parmar L, Dhanya R, Faulkner L (September 2017).
1628:
688:
377:
265:
to produce antibodies to that specific antigen. Self-antigens are suppressed by
179:
2380:
2363:
3614:
2682:
2666:
2535:
752:
663:
655:
203:
195:
3712:
3695:
2847:
2518:
Kurkó J, Besenyei T, Laki J, Glant TT, Mikecz K, Szekanecz Z (October 2013).
2104:
1939:
3352:
2567:
Mignot E, Lin L, Rogers W, Honda Y, Qiu X, Lin X, et al. (March 2001).
2144:
2069:
1997:
Nakamura T, Shirouzu T, Nakata K, Yoshimura N, Ushigome H (September 2019).
1899:
List of human leukocyte antigen alleles associated with cutaneous conditions
863:
142:
3721:
3680:
3662:
3622:
3571:
3500:
3465:
3429:
3327:
3263:
3212:
3159:
3083:
3018:
2996:
2942:
2901:
2866:
2700:
2651:
2633:
2602:
2553:
2504:
2389:
2348:
2330:
2247:
2229:
2190:
2163:
2112:
2034:
1957:
3579:
3535:
3394:
3294:"Common and well-documented HLA alleles: 2012 update to the CWD catalogue"
2806:
2288:
2198:
2077:
983:
903:
4073:
4040:
4035:
4025:
4008:
4003:
3998:
3993:
3988:
3976:
3971:
3696:"Preimplantation diagnosis for Fanconi anemia combined with HLA matching"
3065:
2015:
1829:(By serotyping A3-Cw7-B7-DR15-DQ6 or the older version "A3-B7-DR2-DQ1")
1644:
1536:
1502:
1468:
1434:
1404:
1374:
1344:
1310:
814:
807:
793:
380:) on the cell surface. Infected cells can be recognized and destroyed by
354:
338:
258:
65:
3694:
Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A (June 2001).
41:
4020:
3959:
3954:
3942:
3937:
3831:
1615:
597:
593:
553:
507:
492:
480:
465:
430:
358:
304:
199:
163:
154:
3309:
3204:
2933:
2916:
2893:
324:
4015:
3983:
3966:
3949:
3932:
2457:
Salmon J, Wallace DJ, Dubois EL, Kirou KA, Hahn B, Lehman TA (2007).
1620:
981:
Number of variant alleles at class II loci (DM, DO, DP, DQ, and DR):
822:
801:
784:
777:
773:
262:
246:
242:
238:
234:
230:
3812:
HistoCheck HLA matching tool for organ and stem cell transplantation
3832:
dbMHC Home, NCBI's database of the Major Histocompatibility Complex
3816:
2829:
Requena D, Médico A, Chacón RD, Ramírez M, Marín-Sánchez O (2020).
2584:
749:
binds with major and minor gene subunits to produce a heterodimer.
3914:
3909:
3904:
3899:
3894:
3889:
1276:
1242:
1208:
862:
751:
738:
734:
730:
723:
718:
713:
313:
191:
187:
183:
89:
3837:
Rare Alleles Project at the AlleleFrequencies Net Database (AFND)
3778:
3343:
Hurley CK (1997). "DNA-based typing of HLA for transplantation."
3097:
2214:"Mammalian social odours: attraction and individual recognition"
1893:
1802:
which is called ' 'super B8' ' or ' 'ancestral haplotype' ' and
706:
There are three major and three minor MHC class I genes in HLA.
107:
3845:
2261:
Singal DP, Mickey MR, Mittal KK, Terasaki PI (November 1968).
254:
211:
90:
618:
HLA types are inherited, and some of them are connected with
3811:
2520:"Genetics of rheumatoid arthritis - a comprehensive review"
122:. The HLA system is also known as the human version of the
3807:
American Society for Histocompatibility and Immunogenetics
3787:
218:, which, unlike the HLA proteins, is encoded by a gene on
161:. The proteins encoded by certain genes are also known as
3802:
British Society for Histocompatibility and Immunogenetics
988:
908:
3801:
3806:
349:(APCs) engulf the pathogen through a process called
4051:
3923:
3880:
3116:"Immunology Database and Analysis Portal (ImmPort)"
2461:. Philadelphia: Lippincott Williams & Wilkins.
76:
64:
56:
51:
34:
3817:Allele Frequencies at Variable Immune related loci
2963:"Nomenclature for factors of the HLA system, 2010"
1109:DRB3, DRB4, DRB5 have variable presence in humans
357:from the pathogen are digested into small pieces (
198:. In general, these particular peptides are small
3019:"Statistics < IMGT/HLA < IPD < EMBL-EBI"
2423:Mitchell RS, Kumar V, Abbas AK, Fausto N (2007).
361:) and loaded on to HLA antigens (to be specific,
3347:Leffell MS, Donnenberg AD, Rose NR, eds. (1997)
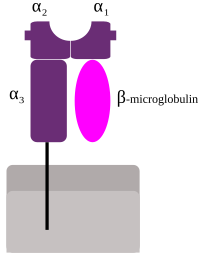
2313:Taylor CJ, Bolton EM, Bradley JA (August 2011).
1142:Table of HLA alleles in each prevalence category
2308:
2306:
2451:Values are given for Caucasians, according to
1917:
1915:
1913:
854:History and naming of human leukocyte antigens
3857:
1121:disease. Karp D. R. et al. describes a novel
8:
3756:Zsolt Harsanyi and Richard Hutton, Richard,
3133:
3131:
3129:
2915:Sanchez-Mazas A, Buhler S, Nunes JM (2013).
2524:Clinical Reviews in Allergy & Immunology
257:-positive T cells), which in turn stimulate
153:, which means that they have many different
3185:
3183:
3181:
3179:
3177:
3175:
3173:
3171:
3169:
2417:
2415:
2413:
2411:
2409:
2407:
2405:
2403:
2401:
2399:
2368:Biology of Blood and Marrow Transplantation
2003:International Journal of Molecular Sciences
3864:
3850:
3842:
1627:. HLA loci can be further classified into
768:Major MHC class II proteins only occur on
3824:at the U.S. National Library of Medicine
3758:Genetic Prophecy: Beyond the Double Helix
3711:
3670:
3339:
3337:
3317:
3073:
2986:
2932:
2856:
2846:
2690:
2641:
2592:
2543:
2447:
2445:
2379:
2338:
2278:
2237:
2212:Brennan PA, Kendrick KM (December 2006).
2153:
2143:
2024:
2014:
1947:
2956:
2954:
2952:
1152:
1129:Common, well-documented and rare alleles
410:
129:Mutations in HLA genes may be linked to
3287:
3285:
3283:
3281:
3279:
3277:
3275:
3273:
3236:
3234:
3232:
3230:
3193:International Journal of Immunogenetics
3104:from the original on 24 September 2006.
3012:
3010:
3008:
3006:
1909:
1878:hematopoietic stem cell transplantation
46:Schematic representation of MHC class I
3029:from the original on 20 September 2012
2754:Evolution and the Spiral of Technology
141:. The HLA gene complex resides on a 3
31:
7:
2723:"HLA Nomenclature @ hla.alleles.org"
1116:Sequence feature variant type (SFVT)
2573:American Journal of Human Genetics
867:Codominant expression of HLA genes
365:). They are then displayed by the
118:responsible for regulation of the
25:
3783:European Bioinformatics Institute
1882:preimplantation genetic diagnosis
789:α-chain encoded by HLA-DPA1 locus
687:There is evidence for non-random
157:, allowing them to fine-tune the
4064:Minor histocompatibility antigen
3873:Major histocompatibility complex
3552:Proceedings. Biological Sciences
3528:10.1615/critrevimmunol.v17.i2.40
3458:10.1111/j.1399-0039.2004.00302.x
3422:10.1111/j.1399-0039.2005.00441.x
3152:10.1111/j.1399-0039.2009.01361.x
2979:10.1111/j.1399-0039.2010.01466.x
2622:Proceedings. Biological Sciences
2497:10.1111/j.0001-2815.2004.00237.x
2280:10.1097/00007890-196811000-00005
827:α-chain encoded by HLA-DRA locus
642:(systemic lupus erythematosus),
124:major histocompatibility complex
40:
2733:from the original on 2 May 2018
1972:"Human leukocyte antigen (HLA)"
984:
904:
291:identical HLA molecules on all
3516:Critical Reviews in Immunology
1866:HLA matching for sick siblings
149:at 21.3. HLA genes are highly
1:
3493:10.1016/S0198-8859(03)00032-6
3349:Handbook of Human Immunology.
1123:sequence feature variant type
126:(MHC) found in many animals.
3387:10.1016/0198-8859(92)90079-3
3256:10.1016/j.humimm.2007.01.014
544:Systemic lupus erythematosus
498:Systemic lupus erythematosus
420:Diseases with increased risk
412:HLA and autoimmune diseases
4130:Genes on human chromosome 6
2799:10.1126/science.272.5258.67
2459:Dubois' lupus erythematosus
1876:In some diseases requiring
27:Genes on human chromosome 6
4156:
4094:Cluster of differentiation
3779:IMGT/HLA Sequence Database
2427:. Philadelphia: Saunders.
2381:10.1016/j.bbmt.2017.05.030
1869:
1642:
851:
341:enters the body, specific
285:organ transplant rejection
94:HLA region of Chromosome 6
4084:Human blood group systems
3760:, London: Granada, 1982 (
3615:10.1080/08916930290022270
2683:10.1038/s41437-020-0350-8
2536:10.1007/s12016-012-8346-7
1922:Choo SY (February 2007).
1837:Role of allelic variation
1609:Serotype and allele names
1108:
913:
897:Tables of variant alleles
552:
506:
471:21-hydroxylase deficiency
429:
276:encode components of the
39:
3826:Medical Subject Headings
3822:Human+leukocyte+antigens
3713:10.1001/jama.285.24.3130
3351:pp. 521–55, Boca Raton:
3054:Human Molecular Genetics
2848:10.3389/fimmu.2020.02008
2105:10.1016/j.it.2005.11.001
1940:10.3349/ymj.2007.48.1.11
1022:
1005:
1002:
999:
996:
993:
916:
770:antigen-presenting cells
763:antigen-presenting cells
584:Diabetes mellitus type 1
569:Diabetes mellitus type 1
534:Diabetes mellitus type 1
367:antigen-presenting cells
347:antigen-presenting cells
18:Human Leukocyte Antigens
4089:Cell adhesion molecules
4059:Human leukocyte antigen
3797:The Anthony Nolan Trust
2835:Frontiers in Immunology
2756:. Trafford Publishing.
2425:Robbins Basic Pathology
2145:10.3390/cancers13102307
2070:10.1126/science.1323878
838:antigen-presenting cell
648:inclusion body myositis
167:, as a result of their
114:in humans which encode
106:) system or complex of
100:human leukocyte antigen
35:Human leukocyte antigen
3739:The Compatibility Gene
3663:10.1681/ASN.2020010019
3572:10.1098/rspb.1995.0087
2634:10.1098/rspb.2020.1682
2331:10.1098/rstb.2011.0030
2230:10.1098/rstb.2006.1931
2191:10.1002/cyto.990120702
1928:Yonsei Medical Journal
868:
757:
628:ankylosing spondylitis
456:Acute anterior uveitis
436:Ankylosing spondylitis
333:In infectious diseases
329:
272:HLAs corresponding to
225:HLAs corresponding to
178:HLAs corresponding to
159:adaptive immune system
95:
4140:Blood antigen systems
4125:Human MHC haplogroups
3793:HLA Informatics Group
1643:Further information:
866:
852:Further information:
755:
317:
210:(also referred to as
116:cell-surface proteins
93:
4135:Transfusion medicine
2093:Trends in Immunology
2016:10.3390/ijms20184544
632:rheumatoid arthritis
620:autoimmune disorders
559:Rheumatoid arthritis
513:Autoimmune hepatitis
406:transplant rejection
3564:1995RSPSB.260..245W
3098:"IMGT/HLA Database"
2791:1996Sci...272...67P
2752:Shennan DH (2006).
2455:(right column) in:
2062:1992Sci...257..927M
1604:Examining HLA types
1171:No. well-documented
985:
905:
874:balancing selection
813:β-chain encoded by
806:α-chain encoded by
792:β-chain encoded by
693:genetic matchmaking
413:
371:CD4+ helper T cells
147:chromosome 6, p-arm
131:autoimmune diseases
3066:10.1093/hmg/ddp521
3017:EBI Web Services.
2628:(1933): 20201682.
869:
758:
709:Major MHC class I
446:Reactive arthritis
411:
399:In graft rejection
330:
267:regulatory T cells
169:historic discovery
96:
4102:
4101:
4069:Blood transfusion
3751:978-0-241-95675-5
3737:Daniel M. Davis,
3310:10.1111/tan.12093
3205:10.1111/iji.12030
2934:10.1159/000360855
2894:10.1111/tan.12356
2763:978-1-55212-518-2
2468:978-0-7817-9394-0
2434:978-1-4160-2973-1
2325:(1575): 2312–22.
2224:(1476): 2061–78.
1797:A1-Cw7-B8-DR3-DQ2
1601:
1600:
1176:% well-documented
1148:IMGT/HLA Database
1113:
1112:
979:
978:
683:In mate selection
644:myasthenia gravis
611:
610:
278:complement system
173:organ transplants
88:
87:
16:(Redirected from
4147:
3866:
3859:
3852:
3843:
3726:
3725:
3715:
3691:
3685:
3684:
3674:
3651:J Am Soc Nephrol
3641:
3635:
3634:
3598:
3592:
3591:
3547:
3541:
3539:
3511:
3505:
3504:
3481:Human Immunology
3476:
3470:
3469:
3440:
3434:
3433:
3405:
3399:
3398:
3375:Human Immunology
3370:
3364:
3341:
3332:
3331:
3321:
3289:
3268:
3267:
3244:Human Immunology
3238:
3225:
3224:
3187:
3164:
3163:
3135:
3124:
3123:
3122:on 26 July 2011.
3118:. Archived from
3112:
3106:
3105:
3094:
3088:
3087:
3077:
3045:
3039:
3038:
3036:
3034:
3014:
3001:
3000:
2990:
2958:
2947:
2946:
2936:
2912:
2906:
2905:
2877:
2871:
2870:
2860:
2850:
2826:
2820:
2818:
2774:
2768:
2767:
2749:
2743:
2742:
2740:
2738:
2719:
2713:
2712:
2694:
2662:
2656:
2655:
2645:
2613:
2607:
2606:
2596:
2564:
2558:
2557:
2547:
2515:
2509:
2508:
2479:
2473:
2472:
2449:
2440:
2438:
2419:
2394:
2393:
2383:
2374:(9): 1592–1596.
2359:
2353:
2352:
2342:
2310:
2301:
2300:
2282:
2258:
2252:
2251:
2241:
2209:
2203:
2202:
2174:
2168:
2167:
2157:
2147:
2123:
2117:
2116:
2088:
2082:
2081:
2056:(5072): 927–34.
2045:
2039:
2038:
2028:
2018:
1994:
1988:
1987:
1985:
1983:
1968:
1962:
1961:
1951:
1919:
1828:
1824:
1820:
1816:
1812:
1808:
1794:
1790:
1786:
1782:
1778:
1774:
1746:
1742:
1738:
1734:
1730:
1726:
1722:
1718:
1714:
1710:
1706:
1702:
1153:
1006:Theor. possible
986:
906:
729:Minor genes are
652:Sjögren syndrome
524:Sjögren syndrome
486:Behçet's Disease
414:
327:
216:β2-microglobulin
202:, of about 8-10
44:
32:
21:
4155:
4154:
4150:
4149:
4148:
4146:
4145:
4144:
4105:
4104:
4103:
4098:
4047:
3919:
3876:
3870:
3788:hla.alleles.org
3775:
3734:
3729:
3693:
3692:
3688:
3657:(9): 2193–204.
3643:
3642:
3638:
3600:
3599:
3595:
3558:(1359): 245–9.
3549:
3548:
3544:
3513:
3512:
3508:
3478:
3477:
3473:
3446:Tissue Antigens
3442:
3441:
3437:
3410:Tissue Antigens
3407:
3406:
3402:
3372:
3371:
3367:
3342:
3335:
3298:Tissue Antigens
3291:
3290:
3271:
3240:
3239:
3228:
3189:
3188:
3167:
3140:Tissue Antigens
3137:
3136:
3127:
3114:
3113:
3109:
3096:
3095:
3091:
3047:
3046:
3042:
3032:
3030:
3016:
3015:
3004:
2967:Tissue Antigens
2960:
2959:
2950:
2927:(3–4): 162–77.
2914:
2913:
2909:
2882:Tissue Antigens
2879:
2878:
2874:
2828:
2827:
2823:
2785:(5258): 67–74.
2776:
2775:
2771:
2764:
2751:
2750:
2746:
2736:
2734:
2727:hla.alleles.org
2721:
2720:
2716:
2664:
2663:
2659:
2615:
2614:
2610:
2566:
2565:
2561:
2517:
2516:
2512:
2485:Tissue Antigens
2481:
2480:
2476:
2469:
2456:
2450:
2443:
2435:
2422:
2420:
2397:
2361:
2360:
2356:
2312:
2311:
2304:
2267:Transplantation
2260:
2259:
2255:
2211:
2210:
2206:
2176:
2175:
2171:
2125:
2124:
2120:
2090:
2089:
2085:
2047:
2046:
2042:
1996:
1995:
1991:
1981:
1979:
1978:. December 2020
1970:
1969:
1965:
1921:
1920:
1911:
1907:
1890:
1874:
1868:
1852:
1839:
1826:
1822:
1818:
1814:
1810:
1806:
1795:(By serotyping
1792:
1788:
1784:
1780:
1776:
1772:
1759:
1744:
1740:
1736:
1732:
1728:
1724:
1720:
1716:
1712:
1708:
1704:
1700:
1690:
1682:Tissue Antigens
1669:
1667:Gene sequencing
1660:
1658:Cellular typing
1647:
1641:
1623:at a given HLA
1611:
1606:
1202:
1197:
1192:
1187:
1182:
1177:
1172:
1167:
1162:
1157:
1144:
1131:
1118:
951:Minor Antigens
922:Major Antigens
899:
879:gene conversion
861:
856:
846:
746:
701:
685:
676:
624:type I diabetes
616:
614:In autoimmunity
603:Coeliac disease
579:
401:
393:MHC restriction
337:When a foreign
335:
319:
301:
145:stretch within
135:type I diabetes
47:
28:
23:
22:
15:
12:
11:
5:
4153:
4151:
4143:
4142:
4137:
4132:
4127:
4122:
4117:
4115:Gene complexes
4107:
4106:
4100:
4099:
4097:
4096:
4091:
4086:
4081:
4076:
4071:
4066:
4061:
4055:
4053:
4049:
4048:
4046:
4045:
4044:
4043:
4038:
4033:
4028:
4023:
4013:
4012:
4011:
4006:
4001:
3996:
3991:
3981:
3980:
3979:
3974:
3964:
3963:
3962:
3957:
3947:
3946:
3945:
3940:
3929:
3927:
3921:
3920:
3918:
3917:
3912:
3907:
3902:
3897:
3892:
3886:
3884:
3878:
3877:
3871:
3869:
3868:
3861:
3854:
3846:
3840:
3839:
3834:
3829:
3819:
3814:
3809:
3804:
3799:
3790:
3785:
3774:
3773:External links
3771:
3770:
3769:
3754:
3733:
3730:
3728:
3727:
3706:(24): 3130–3.
3686:
3636:
3593:
3542:
3522:(2): 179–224.
3506:
3471:
3435:
3400:
3365:
3333:
3304:(4): 194–203.
3269:
3250:(5): 392–417.
3226:
3165:
3125:
3107:
3089:
3040:
3002:
2973:(4): 291–455.
2948:
2921:Human Heredity
2907:
2872:
2821:
2769:
2762:
2744:
2714:
2677:(5): 281–289.
2657:
2608:
2585:10.1086/318799
2559:
2510:
2474:
2467:
2441:
2433:
2421:Table 5-7 in:
2395:
2354:
2302:
2273:(8): 904–912.
2253:
2204:
2169:
2118:
2083:
2040:
1989:
1963:
1908:
1906:
1903:
1902:
1901:
1896:
1889:
1886:
1872:Savior sibling
1870:Main article:
1867:
1864:
1851:
1848:
1838:
1835:
1758:
1755:
1689:
1686:
1668:
1665:
1659:
1656:
1640:
1637:
1610:
1607:
1605:
1602:
1599:
1598:
1595:
1592:
1589:
1586:
1583:
1580:
1577:
1574:
1571:
1567:
1566:
1563:
1560:
1557:
1554:
1551:
1548:
1545:
1542:
1539:
1533:
1532:
1529:
1526:
1523:
1520:
1517:
1514:
1511:
1508:
1505:
1499:
1498:
1495:
1492:
1489:
1486:
1483:
1480:
1477:
1474:
1471:
1465:
1464:
1461:
1458:
1455:
1452:
1449:
1446:
1443:
1440:
1437:
1431:
1430:
1427:
1425:
1423:
1421:
1419:
1416:
1413:
1410:
1407:
1401:
1400:
1397:
1395:
1393:
1391:
1389:
1386:
1383:
1380:
1377:
1371:
1370:
1367:
1365:
1363:
1361:
1359:
1356:
1353:
1350:
1347:
1341:
1340:
1337:
1334:
1331:
1328:
1325:
1322:
1319:
1316:
1313:
1307:
1306:
1303:
1300:
1297:
1294:
1291:
1288:
1285:
1282:
1279:
1273:
1272:
1269:
1266:
1263:
1260:
1257:
1254:
1251:
1248:
1245:
1239:
1238:
1235:
1232:
1229:
1226:
1223:
1220:
1217:
1214:
1211:
1205:
1204:
1199:
1194:
1189:
1184:
1179:
1174:
1169:
1164:
1159:
1143:
1140:
1130:
1127:
1117:
1114:
1111:
1110:
1106:
1105:
1102:
1099:
1096:
1093:
1089:
1088:
1085:
1083:
1080:
1077:
1073:
1072:
1069:
1067:
1064:
1061:
1057:
1056:
1053:
1051:
1048:
1045:
1041:
1040:
1037:
1035:
1032:
1029:
1025:
1024:
1021:
1018:
1015:
1012:
1008:
1007:
1004:
1001:
998:
995:
991:
990:
977:
976:
973:
969:
968:
965:
961:
960:
957:
953:
952:
948:
947:
944:
940:
939:
936:
932:
931:
928:
924:
923:
919:
918:
915:
911:
910:
898:
895:
860:
857:
845:
842:
834:
833:
832:
831:
828:
820:
819:
818:
811:
799:
798:
797:
790:
747:-microglobulin
744:
727:
726:
721:
716:
700:
699:Classification
697:
684:
681:
675:
672:
668:celiac disease
636:celiac disease
615:
612:
609:
608:
605:
600:
590:
589:
586:
581:
580:-DR4 combined
575:
574:
571:
565:
564:
561:
556:
550:
549:
546:
540:
539:
536:
530:
529:
526:
519:
518:
515:
510:
504:
503:
500:
495:
489:
488:
483:
477:
476:
473:
468:
462:
461:
458:
452:
451:
448:
442:
441:
438:
433:
427:
426:
421:
418:
400:
397:
334:
331:
300:
297:
251:T-helper cells
208:killer T-cells
171:as factors in
139:celiac disease
86:
85:
80:
74:
73:
68:
62:
61:
58:
54:
53:
49:
48:
45:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4152:
4141:
4138:
4136:
4133:
4131:
4128:
4126:
4123:
4121:
4120:Immune system
4118:
4116:
4113:
4112:
4110:
4095:
4092:
4090:
4087:
4085:
4082:
4080:
4077:
4075:
4072:
4070:
4067:
4065:
4062:
4060:
4057:
4056:
4054:
4050:
4042:
4039:
4037:
4034:
4032:
4029:
4027:
4024:
4022:
4019:
4018:
4017:
4014:
4010:
4007:
4005:
4002:
4000:
3997:
3995:
3992:
3990:
3987:
3986:
3985:
3982:
3978:
3975:
3973:
3970:
3969:
3968:
3965:
3961:
3958:
3956:
3953:
3952:
3951:
3948:
3944:
3941:
3939:
3936:
3935:
3934:
3931:
3930:
3928:
3926:
3922:
3916:
3913:
3911:
3908:
3906:
3903:
3901:
3898:
3896:
3893:
3891:
3888:
3887:
3885:
3883:
3879:
3874:
3867:
3862:
3860:
3855:
3853:
3848:
3847:
3844:
3838:
3835:
3833:
3830:
3827:
3823:
3820:
3818:
3815:
3813:
3810:
3808:
3805:
3803:
3800:
3798:
3794:
3791:
3789:
3786:
3784:
3780:
3777:
3776:
3772:
3767:
3766:0-246-11760-5
3763:
3759:
3755:
3752:
3748:
3744:
3743:Penguin Books
3740:
3736:
3735:
3731:
3723:
3719:
3714:
3709:
3705:
3701:
3697:
3690:
3687:
3682:
3678:
3673:
3668:
3664:
3660:
3656:
3652:
3648:
3640:
3637:
3632:
3628:
3624:
3620:
3616:
3612:
3609:(3): 183–90.
3608:
3604:
3597:
3594:
3589:
3585:
3581:
3577:
3573:
3569:
3565:
3561:
3557:
3553:
3546:
3543:
3537:
3533:
3529:
3525:
3521:
3517:
3510:
3507:
3502:
3498:
3494:
3490:
3486:
3482:
3475:
3472:
3467:
3463:
3459:
3455:
3451:
3447:
3439:
3436:
3431:
3427:
3423:
3419:
3416:(2): 107–13.
3415:
3411:
3404:
3401:
3396:
3392:
3388:
3384:
3380:
3376:
3369:
3366:
3362:
3361:0-8493-0134-3
3358:
3354:
3350:
3346:
3340:
3338:
3334:
3329:
3325:
3320:
3315:
3311:
3307:
3303:
3299:
3295:
3288:
3286:
3284:
3282:
3280:
3278:
3276:
3274:
3270:
3265:
3261:
3257:
3253:
3249:
3245:
3237:
3235:
3233:
3231:
3227:
3222:
3218:
3214:
3210:
3206:
3202:
3198:
3194:
3186:
3184:
3182:
3180:
3178:
3176:
3174:
3172:
3170:
3166:
3161:
3157:
3153:
3149:
3145:
3141:
3134:
3132:
3130:
3126:
3121:
3117:
3111:
3108:
3103:
3099:
3093:
3090:
3085:
3081:
3076:
3071:
3067:
3063:
3060:(4): 707–19.
3059:
3055:
3051:
3044:
3041:
3028:
3024:
3023:www.ebi.ac.uk
3020:
3013:
3011:
3009:
3007:
3003:
2998:
2994:
2989:
2984:
2980:
2976:
2972:
2968:
2964:
2957:
2955:
2953:
2949:
2944:
2940:
2935:
2930:
2926:
2922:
2918:
2911:
2908:
2903:
2899:
2895:
2891:
2888:(5): 307–23.
2887:
2883:
2876:
2873:
2868:
2864:
2859:
2854:
2849:
2844:
2840:
2836:
2832:
2825:
2822:
2816:
2812:
2808:
2804:
2800:
2796:
2792:
2788:
2784:
2780:
2773:
2770:
2765:
2759:
2755:
2748:
2745:
2732:
2728:
2724:
2718:
2715:
2710:
2706:
2702:
2698:
2693:
2688:
2684:
2680:
2676:
2672:
2668:
2661:
2658:
2653:
2649:
2644:
2639:
2635:
2631:
2627:
2623:
2619:
2612:
2609:
2604:
2600:
2595:
2590:
2586:
2582:
2579:(3): 686–99.
2578:
2574:
2570:
2563:
2560:
2555:
2551:
2546:
2541:
2537:
2533:
2529:
2525:
2521:
2514:
2511:
2506:
2502:
2498:
2494:
2490:
2486:
2478:
2475:
2470:
2464:
2460:
2454:
2448:
2446:
2442:
2436:
2430:
2426:
2418:
2416:
2414:
2412:
2410:
2408:
2406:
2404:
2402:
2400:
2396:
2391:
2387:
2382:
2377:
2373:
2369:
2365:
2358:
2355:
2350:
2346:
2341:
2336:
2332:
2328:
2324:
2320:
2316:
2309:
2307:
2303:
2298:
2294:
2290:
2286:
2281:
2276:
2272:
2268:
2264:
2257:
2254:
2249:
2245:
2240:
2235:
2231:
2227:
2223:
2219:
2215:
2208:
2205:
2200:
2196:
2192:
2188:
2185:(7): 579–96.
2184:
2180:
2173:
2170:
2165:
2161:
2156:
2151:
2146:
2141:
2137:
2133:
2129:
2122:
2119:
2114:
2110:
2106:
2102:
2098:
2094:
2087:
2084:
2079:
2075:
2071:
2067:
2063:
2059:
2055:
2051:
2044:
2041:
2036:
2032:
2027:
2022:
2017:
2012:
2008:
2004:
2000:
1993:
1990:
1977:
1973:
1967:
1964:
1959:
1955:
1950:
1945:
1941:
1937:
1933:
1929:
1925:
1918:
1916:
1914:
1910:
1904:
1900:
1897:
1895:
1892:
1891:
1887:
1885:
1883:
1879:
1873:
1865:
1863:
1859:
1856:
1849:
1847:
1843:
1836:
1834:
1830:
1803:
1800:
1798:
1769:
1767:
1762:
1756:
1754:
1750:
1747:
1697:
1694:
1687:
1685:
1683:
1677:
1673:
1666:
1664:
1657:
1655:
1651:
1646:
1638:
1636:
1634:
1630:
1626:
1622:
1617:
1608:
1603:
1596:
1593:
1590:
1587:
1584:
1581:
1578:
1575:
1572:
1569:
1568:
1564:
1561:
1558:
1555:
1552:
1549:
1546:
1543:
1540:
1538:
1535:
1534:
1530:
1527:
1524:
1521:
1518:
1515:
1512:
1509:
1506:
1504:
1501:
1500:
1496:
1493:
1490:
1487:
1484:
1481:
1478:
1475:
1472:
1470:
1467:
1466:
1462:
1459:
1456:
1453:
1450:
1447:
1444:
1441:
1438:
1436:
1433:
1432:
1428:
1426:
1424:
1422:
1420:
1417:
1414:
1411:
1408:
1406:
1403:
1402:
1398:
1396:
1394:
1392:
1390:
1387:
1384:
1381:
1378:
1376:
1373:
1372:
1368:
1366:
1364:
1362:
1360:
1357:
1354:
1351:
1348:
1346:
1343:
1342:
1338:
1335:
1332:
1329:
1326:
1323:
1320:
1317:
1314:
1312:
1309:
1308:
1304:
1301:
1298:
1295:
1292:
1289:
1286:
1283:
1280:
1278:
1275:
1274:
1270:
1267:
1264:
1261:
1258:
1255:
1252:
1249:
1246:
1244:
1241:
1240:
1236:
1233:
1230:
1227:
1224:
1221:
1218:
1215:
1212:
1210:
1207:
1206:
1200:
1195:
1191:No. very rare
1190:
1185:
1180:
1175:
1170:
1165:
1160:
1155:
1154:
1151:
1149:
1141:
1139:
1135:
1128:
1126:
1124:
1115:
1107:
1103:
1100:
1097:
1094:
1091:
1090:
1086:
1084:
1081:
1078:
1075:
1074:
1070:
1068:
1065:
1062:
1059:
1058:
1054:
1052:
1049:
1046:
1043:
1042:
1038:
1036:
1033:
1030:
1027:
1026:
1023:combinations
1019:
1016:
1013:
1010:
1009:
992:
989:MHC class II
987:
982:
974:
971:
970:
966:
963:
962:
958:
955:
954:
950:
949:
945:
942:
941:
937:
934:
933:
929:
926:
925:
921:
920:
912:
907:
902:
896:
894:
890:
886:
882:
880:
875:
865:
858:
855:
850:
843:
841:
839:
829:
826:
825:
824:
821:
816:
812:
809:
805:
804:
803:
800:
795:
791:
788:
787:
786:
783:
782:
781:
779:
775:
771:
766:
764:
754:
750:
748:
740:
736:
732:
725:
722:
720:
717:
715:
712:
711:
710:
707:
704:
698:
696:
694:
690:
682:
680:
673:
671:
669:
665:
659:
657:
653:
649:
645:
641:
637:
633:
629:
625:
621:
613:
606:
604:
601:
599:
595:
592:
591:
587:
585:
582:
577:
576:
572:
570:
567:
566:
562:
560:
557:
555:
551:
547:
545:
542:
541:
537:
535:
532:
531:
527:
525:
521:
520:
516:
514:
511:
509:
505:
501:
499:
496:
494:
491:
490:
487:
484:
482:
479:
478:
474:
472:
469:
467:
464:
463:
459:
457:
454:
453:
449:
447:
444:
443:
439:
437:
434:
432:
428:
425:
424:Relative risk
422:
419:
416:
415:
409:
407:
398:
396:
394:
389:
385:
383:
379:
374:
372:
368:
364:
360:
356:
352:
348:
344:
340:
332:
326:
322:
316:
312:
310:
309:immune system
306:
298:
296:
294:
288:
286:
281:
279:
275:
274:MHC class III
270:
268:
264:
260:
256:
253:(also called
252:
248:
244:
240:
236:
232:
228:
223:
221:
220:chromosome 15
217:
213:
209:
205:
201:
197:
193:
189:
185:
181:
176:
174:
170:
166:
165:
160:
156:
152:
148:
144:
140:
136:
132:
127:
125:
121:
120:immune system
117:
113:
109:
105:
101:
92:
84:
81:
79:
75:
72:
69:
67:
63:
59:
55:
50:
43:
38:
33:
30:
19:
4058:
3925:MHC class II
3757:
3738:
3732:Bibliography
3703:
3699:
3689:
3654:
3650:
3639:
3606:
3603:Autoimmunity
3602:
3596:
3555:
3551:
3545:
3519:
3515:
3509:
3487:(5): 562–6.
3484:
3480:
3474:
3452:(5): 581–7.
3449:
3445:
3438:
3413:
3409:
3403:
3378:
3374:
3368:
3348:
3344:
3301:
3297:
3247:
3243:
3196:
3192:
3146:(6): 480–5.
3143:
3139:
3120:the original
3110:
3092:
3057:
3053:
3043:
3031:. Retrieved
3022:
2970:
2966:
2924:
2920:
2910:
2885:
2881:
2875:
2838:
2834:
2824:
2782:
2778:
2772:
2753:
2747:
2735:. Retrieved
2726:
2717:
2674:
2670:
2660:
2625:
2621:
2611:
2576:
2572:
2562:
2530:(2): 170–9.
2527:
2523:
2513:
2491:(6): 562–7.
2488:
2484:
2477:
2458:
2439:8th edition.
2424:
2371:
2367:
2357:
2322:
2318:
2270:
2266:
2256:
2221:
2217:
2207:
2182:
2178:
2172:
2138:(10): 2307.
2135:
2131:
2121:
2096:
2092:
2086:
2053:
2049:
2043:
2009:(18): 4544.
2006:
2002:
1992:
1980:. Retrieved
1975:
1966:
1934:(1): 11–23.
1931:
1927:
1875:
1860:
1857:
1853:
1844:
1840:
1831:
1825: ; DQB1
1821: ; DQA1
1817: ; DRB1
1804:
1801:
1791: ; DQB1
1787: ; DQA1
1783: ; DRB1
1770:
1765:
1763:
1760:
1751:
1748:
1698:
1695:
1691:
1681:
1678:
1674:
1670:
1661:
1652:
1648:
1633:MHC class II
1612:
1203:categorized
1145:
1136:
1132:
1119:
980:
909:MHC class I
900:
891:
887:
883:
870:
847:
844:Nomenclature
835:
767:
759:
728:
708:
705:
702:
686:
677:
660:
617:
402:
390:
386:
382:CD8+ T cells
375:
363:MHC class II
351:phagocytosis
336:
302:
289:
282:
271:
227:MHC class II
224:
177:
162:
128:
112:chromosome 6
103:
99:
97:
29:
4079:Calgranulin
3882:MHC class I
3381:(1): 4–18.
3199:(1): 60–5.
2099:(1): 11–6.
1688:Phenotyping
1629:MHC class I
1196:% very rare
1003:-B3 to -B5
859:Variability
689:mate choice
578:HLA-DR3 and
378:MHC class I
261:-producing
204:amino acids
196:proteasomes
180:MHC class I
151:polymorphic
52:Identifiers
4109:Categories
3741:, London,
1905:References
1850:Antibodies
1766:haplotypes
1757:Haplotypes
1639:Serotyping
1161:No. common
664:serotyping
656:narcolepsy
417:HLA allele
78:Membranome
3353:CRC Press
3221:205192491
2709:220947699
2179:Cytometry
1813: ; B
1809: ; C
1779: ; B
1775: ; C
1201:% alleles
674:In cancer
299:Functions
71:IPR037055
4074:Arrestin
3745:, 2014 (
3722:11427142
3681:32764139
3623:12389643
3588:34971350
3501:12691707
3466:15496201
3430:16029430
3328:23510415
3264:17462507
3213:23198982
3160:19793314
3102:Archived
3084:19933168
3027:Archived
2997:20356336
2943:24861861
2902:24738646
2867:33013857
2841:: 2008.
2815:22209086
2731:Archived
2701:32747723
2671:Heredity
2652:32811307
2603:11179016
2554:23288628
2505:15140032
2390:28603069
2349:21727137
2297:32428268
2248:17118924
2164:34065814
2113:16297661
2035:31540289
1976:MS Trust
1958:17326240
1888:See also
1645:Serotype
1570:All Loci
1181:No. rare
1166:% common
815:HLA-DQB1
808:HLA-DQA1
794:HLA-DPB1
522:Primary
359:peptides
355:Proteins
339:pathogen
305:proteins
259:antibody
200:polymers
164:antigens
133:such as
66:InterPro
3875:classes
3672:7461684
3631:5690960
3580:7630893
3560:Bibcode
3536:9094452
3395:1399721
3319:3634360
3075:2807365
2988:2848993
2858:7494848
2807:8600539
2787:Bibcode
2779:Science
2692:7553942
2643:7482290
2594:1274481
2545:3655138
2453:Page 61
2340:3130422
2289:5696819
2239:1764843
2199:1782829
2155:8150281
2132:Cancers
2078:1323878
2058:Bibcode
2050:Science
2026:6769817
1949:2628004
1616:HLA-B27
1198:alleles
1193:alleles
1188:alleles
1183:alleles
1178:alleles
1173:alleles
1168:alleles
1163:alleles
1104:11,431
1087:34,528
1071:16,036
778:T cells
774:B cells
598:HLA-DQ8
594:HLA-DQ2
554:HLA-DR4
548:2 to 3
508:HLA-DR3
502:2 to 3
493:HLA-DR2
481:HLA-B51
466:HLA-B47
431:HLA-B27
345:called
328:
263:B-cells
155:alleles
4016:HLA-DR
3984:HLA-DQ
3967:HLA-DP
3950:HLA-DO
3933:HLA-DM
3828:(MeSH)
3764:
3749:
3720:
3679:
3669:
3629:
3621:
3586:
3578:
3534:
3499:
3464:
3428:
3393:
3359:
3326:
3316:
3262:
3219:
3211:
3158:
3082:
3072:
2995:
2985:
2941:
2900:
2865:
2855:
2813:
2805:
2760:
2707:
2699:
2689:
2650:
2640:
2601:
2591:
2552:
2542:
2503:
2465:
2431:
2388:
2347:
2337:
2295:
2287:
2246:
2236:
2197:
2162:
2152:
2111:
2076:
2033:
2023:
1982:7 June
1956:
1946:
1827:*06:02
1823:*01:02
1819:*15:01
1815:*07:02
1811:*07:02
1807:*03:01
1793:*02:01
1789:*05:01
1785:*03:01
1781:*08:01
1777:*07:01
1773:*01:01
1745:*06:02
1741:*02:01
1739:, DQB1
1737:*01:02
1733:*05:01
1731:, DQA1
1729:*15:01
1725:*03:01
1723:, DRB1
1721:*08:01
1717:*07:02
1713:*07:02
1709:*07:01
1705:*03:01
1701:*01:01
1621:allele
1186:% rare
1011:locus
972:HLA G
964:HLA F
956:HLA E
946:3,930
943:HLA C
938:5,212
935:HLA B
930:4,340
927:HLA A
914:locus
823:HLA-DR
802:HLA-DQ
785:HLA-DP
776:, and
654:, and
245:, and
190:, and
137:, and
57:Symbol
4052:Other
3915:HLA-G
3910:HLA-F
3905:HLA-E
3900:HLA-C
3895:HLA-B
3890:HLA-A
3627:S2CID
3584:S2CID
3217:S2CID
3033:2 May
2811:S2CID
2737:2 May
2705:S2CID
2293:S2CID
1625:locus
1597:~76%
1594:40.8%
1588:20.6%
1565:~90%
1562:32.8%
1556:22.7%
1544:28.8%
1531:~88%
1528:55.6%
1522:14.8%
1510:17.6%
1497:~91%
1494:45.2%
1488:28.9%
1476:12.5%
1463:~88%
1460:20.6%
1454:26.5%
1442:31.9%
1429:~40%
1418:15.0%
1412:25.0%
1399:~53%
1388:13.3%
1382:40.0%
1369:~21%
1358:12.1%
1339:~77%
1336:35.2%
1330:22.7%
1324:12.7%
1305:~74%
1302:42.8%
1296:21.4%
1271:~75%
1268:43.5%
1262:17.6%
1237:~75%
1234:41.6%
1228:21.5%
1158:Locus
1098:2,593
1082:1,257
1066:1,014
817:locus
810:locus
796:locus
739:HLA-G
735:HLA-F
731:HLA-E
724:HLA-C
719:HLA-B
714:HLA-A
343:cells
108:genes
3762:ISBN
3747:ISBN
3718:PMID
3700:JAMA
3677:PMID
3619:PMID
3576:PMID
3532:PMID
3497:PMID
3462:PMID
3426:PMID
3391:PMID
3357:ISBN
3324:PMID
3260:PMID
3209:PMID
3156:PMID
3080:PMID
3035:2018
2993:PMID
2939:PMID
2898:PMID
2863:PMID
2803:PMID
2758:ISBN
2739:2018
2697:PMID
2648:PMID
2599:PMID
2550:PMID
2501:PMID
2463:ISBN
2429:ISBN
2386:PMID
2345:PMID
2285:PMID
2244:PMID
2195:PMID
2160:PMID
2109:PMID
2074:PMID
2031:PMID
1984:2021
1954:PMID
1894:HCP5
1631:and
1591:1214
1582:9.0%
1576:5.3%
1550:9.0%
1537:DPB1
1516:0.0%
1503:DPA1
1482:4.5%
1469:DQB1
1448:8.5%
1435:DQA1
1405:DRB5
1375:DRB4
1352:8.6%
1345:DRB3
1318:6.8%
1311:DRB1
1290:6.6%
1284:2.8%
1256:9.3%
1250:4.8%
1222:8.8%
1216:3.4%
1101:312
1092:DR-
1076:DQ-
1060:DP-
1055:156
1044:DO-
1028:DM-
1000:-B1
997:-A1
994:HLA
737:and
596:and
325:2G9H
303:The
293:loci
98:The
3795:at
3781:at
3708:doi
3704:285
3667:PMC
3659:doi
3611:doi
3568:doi
3556:260
3524:doi
3489:doi
3454:doi
3418:doi
3383:doi
3314:PMC
3306:doi
3252:doi
3201:doi
3148:doi
3070:PMC
3062:doi
2983:PMC
2975:doi
2929:doi
2890:doi
2853:PMC
2843:doi
2795:doi
2783:272
2687:PMC
2679:doi
2675:125
2638:PMC
2630:doi
2626:287
2589:PMC
2581:doi
2540:PMC
2532:doi
2493:doi
2376:doi
2335:PMC
2327:doi
2323:366
2275:doi
2234:PMC
2226:doi
2222:361
2187:doi
2150:PMC
2140:doi
2101:doi
2066:doi
2054:257
2021:PMC
2011:doi
1944:PMC
1936:doi
1715:, B
1707:, C
1585:613
1579:707
1573:415
1333:206
1327:133
1321:147
1299:154
1287:102
1265:468
1259:190
1253:242
1247:125
1231:280
1225:145
1219:178
1156:HLA
1039:91
975:61
967:31
959:27
640:SLE
588:15
528:10
517:14
475:15
460:15
450:14
440:12
369:to
321:PDB
255:CD4
212:CD8
143:Mbp
110:on
104:HLA
60:HLA
4111::
4041:β5
4036:β4
4031:β3
4026:β1
4009:β3
4004:β2
3999:β1
3994:α2
3989:α1
3977:β1
3972:α1
3768:).
3753:).
3716:.
3702:.
3698:.
3675:.
3665:.
3655:31
3653:.
3649:.
3625:.
3617:.
3607:35
3605:.
3582:.
3574:.
3566:.
3554:.
3530:.
3520:17
3518:.
3495:.
3485:64
3483:.
3460:.
3450:64
3448:.
3424:.
3414:66
3412:.
3389:.
3379:34
3377:.
3355:,
3345:In
3336:^
3322:.
3312:.
3302:81
3300:.
3296:.
3272:^
3258:.
3248:68
3246:.
3229:^
3215:.
3207:.
3197:40
3195:.
3168:^
3154:.
3144:74
3142:.
3128:^
3100:.
3078:.
3068:.
3058:19
3056:.
3052:.
3025:.
3021:.
3005:^
2991:.
2981:.
2971:75
2969:.
2965:.
2951:^
2937:.
2925:76
2923:.
2919:.
2896:.
2886:83
2884:.
2861:.
2851:.
2839:11
2837:.
2833:.
2809:.
2801:.
2793:.
2781:.
2729:.
2725:.
2703:.
2695:.
2685:.
2673:.
2669:.
2646:.
2636:.
2624:.
2620:.
2597:.
2587:.
2577:68
2575:.
2571:.
2548:.
2538:.
2528:45
2526:.
2522:.
2499:.
2489:63
2487:.
2444:^
2398:^
2384:.
2372:23
2370:.
2366:.
2343:.
2333:.
2321:.
2317:.
2305:^
2291:.
2283:.
2269:.
2265:.
2242:.
2232:.
2220:.
2216:.
2193:.
2183:12
2181:.
2158:.
2148:.
2136:13
2134:.
2130:.
2107:.
2097:27
2095:.
2072:.
2064:.
2052:.
2029:.
2019:.
2007:20
2005:.
2001:.
1974:.
1952:.
1942:.
1932:48
1930:.
1926:.
1912:^
1880:,
1799:)
1768::
1684:.
1559:29
1553:29
1547:14
1541:40
1525:15
1491:42
1485:26
1473:22
1439:15
1315:79
1293:77
1281:44
1213:68
1079:95
1063:67
1050:13
1047:12
1034:13
1020:#
917:#
840:.
780:.
772:,
765:.
741:.
733:,
695:.
658:.
650:,
646:,
638:,
634:,
630:,
626:,
607:7
573:6
563:4
538:5
384:.
353:.
323::
280:.
247:DR
243:DQ
241:,
239:DO
237:,
235:DM
233:,
231:DP
222:.
186:,
175:.
83:63
4021:α
3960:β
3955:α
3943:β
3938:α
3865:e
3858:t
3851:v
3724:.
3710::
3683:.
3661::
3633:.
3613::
3590:.
3570::
3562::
3540:.
3538:.
3526::
3503:.
3491::
3468:.
3456::
3432:.
3420::
3397:.
3385::
3363:.
3330:.
3308::
3266:.
3254::
3223:.
3203::
3162:.
3150::
3086:.
3064::
3037:.
2999:.
2977::
2945:.
2931::
2904:.
2892::
2869:.
2845::
2819:.
2817:.
2797::
2789::
2766:.
2741:.
2711:.
2681::
2654:.
2632::
2605:.
2583::
2556:.
2534::
2507:.
2495::
2471:.
2437:.
2392:.
2378::
2351:.
2329::
2299:.
2277::
2271:6
2250:.
2228::
2201:.
2189::
2166:.
2142::
2115:.
2103::
2080:.
2068::
2060::
2037:.
2013::
1986:.
1960:.
1938::
1805:A
1771:A
1743:/
1735:/
1727:/
1719:/
1711:/
1703:/
1699:A
1519:4
1513:0
1507:6
1479:8
1457:7
1451:9
1445:4
1415:3
1409:5
1385:2
1379:6
1355:7
1349:5
1277:C
1243:B
1209:A
1095:7
1031:7
1017:#
1014:#
745:2
743:β
229:(
192:C
188:B
184:A
182:(
102:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.