428:. It was assumed that divalent metal ions like Mg were thought to have two roles: Promote proper folding of RNA and to form the catalytic core. Since RNA itself did not contain enough variation in the functional groups, metal ions were thought to play a role at the active site, as was known about proteins. The proposed mechanism for the Mg2+ ion was: the deprotonation of the 2'-OH group by a Magnesium.aqua.hydroxy complex bound by the pro-R oxygen at the phosphate-cleavage site, followed by nucleophilic attack of the resultant 2'-alkaoxide on the scissile phosphate forming a pentacoordinate phosphate intermediate. The last step is the departing of the 5' leaving group, yielding a 2',3'-cyclic phosphate with an inverted configuration.
894:
in acid/base catalysis. G12 is within hydrogen bonding distance to the 2’–O of C17, the nucleophile in the cleavage reaction, and the ribose of G8 hydrogen bonds to the leaving group 5’-O. (see below), while the nucleotide base of G8 forms a Watson-Crick pair with the invariant C3. This arrangement permits one to suggest that G12 is the general base in the cleavage reaction, and that G8 may function as the general acid, consistent with previous biochemical observations. G5 hydrogen bonds to the furanose oxygen of C17, helping to position it for in-line attack. U4 and U7, as a consequence of the base-pair formation between G8 and C3, are now positioned such that an NOE between their bases is easily explained.
340:
794:
881:
17:
933:
771:
of the sequence contribute to stem I then it is a type I hammerhead ribozyme, to stem II is a type II and to stem III then it is a type III hammerhead ribozyme. Of the three possible topological types, type I can be found in the genomes of prokaryotes, eukaryotes and RNA plant pathogens, whereas type II have been only described in prokaryotes and type III are mostly found in plants, plant pathogens and prokaryotes.
663:
546:
166:
762:
645:
261:
940:
If the invariant G8 is changed to C8, hammerhead catalysis is abolished. However, a G8C + C3G double-mutant that maintains the G8-C3 base pair found in the full-length hammerhead restores most of the catalytic activity. The 2'-OH of G8 has also been observed to be essential for catalysis; replacement
893:
phosphates are observed to be 4.3 Å apart, consistent with the idea that, when modified, these phosphates could bind a single thiophilic metal ion. The structure also reveals how two invariant residues, G-12 and G-8, are positioned within the active site consistent with their previously proposed role
888:
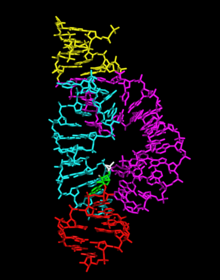
In 2006 a 2.2 Å resolution crystal structure of the full-length hammerhead ribozyme was obtained. This new structure (shown on the right) appears to resolve the most worrisome of the previous discrepancies. In particular, C17 is now positioned for in-line attack, and the invariant residues C3, G5, G8
770:
Structurally the hammerhead ribozyme is composed of three base paired helices, separated by short linkers of conserved sequences. These helices are called I, II and III. Hammerhead ribozymes can be classified into three types based on which helix the 5' and 3' ends are found in. If the 5' and 3' ends
489:
is present to permit the RNA to fold. This discovery suggested that the RNA itself, rather than serving as an inert, passive scaffold for the binding of chemically active divalent metal ions, is instead itself intimately involved in the chemistry of catalysis. The latest structural results, described
274:
and a few mammals like rodents and the platypus. In 2010, it was found that the hammerhead ribozyme occurs in a wide variety of bacterial and eukaryotic genomes, including in humans. Similar reports confirmed and extended these observations, unveiling the hammerhead ribozyme as a ubiquitous catalytic
920:
2(P) reaction). G12 is positioned within hydrogen bonding distance of this nucleophile, and therefore would be able to abstract a proton from the 2'-oxygen if G12 itself becomes deprotonated. The 2'-OH of G8 forms a hydrogen bond to the 5'-leaving group oxygen, and therefore potentially may supply a
871:
For these reasons, the two sets of experiments (biochemical vs. crystallographic) appeared not only to be at odds, but to be completely and hopelessly irreconcilable, generating a substantial amount of discord in the field. No compelling evidence for dismissing either set of experimental results was
835:
Despite the observations of hammerhead ribozyme catalysis in a crystal of the minimal hammerhead sequence in which the crystal lattice packing contacts by necessity confined the global positions of the distal termini of all three flanking helical stems, many biochemical experiments designed to probe
801:
The minimal hammerhead ribozyme has been exhaustively studied by biochemists and enzymologists as well as by X-ray crystallographers, NMR spectroscopists, and other practitioners of biophysical techniques. The first detailed three-dimensional structural information for a hammerhead ribozyme appeared
412:
minute at pH 7.5 in 10 mM Mg (so-called “standard reaction conditions” for the minimal hammerhead RNA sequence), depending upon the sequence of the particular hammerhead ribozyme construct measured. This represents an approximately 10,000-fold rate enhancement over the nonenzymatic cleavage of RNA.
343:
In-line transition-state for the hammerhead ribozyme reaction. A general base (B) abstracts a proton from the 2'-O, and a general acid (A), supplies a proton to the 5'-O leaving group as negative charge accumulates. The reaction product is a 2',3'-cyclic phosphate. The bonds breaking (shown in red)
301:
virus genomes of circular RNA from diverse animals were found to encode hammerhead ribozymes similar to those present in plant viroids and viral satellites. A massive bioinformatic search of deltavirus-like agents around the globe has uncovered hundreds of examples of circular RNA genomes carrying
843:
A particularly striking and only recently observed example consisted of G8 and G12, which were identified as possible participants in acid/base catalysis. Once it was demonstrated that the hammerhead RNA does not require divalent metal ions for catalysis, it gradually became apparent that the RNA
867:
phosphates must come within about 4 Å of one another in the transition-state, based upon double phosphorothioate substitution and soft metal ion rescue experiments; the distance between these phosphates in the minimal hammerhead crystal structure was about 18 Å, with no clear mechanism for close
855:
characterization, which suggested that these nucleotide bases must approach one another closer than about 6 Å, although close approach of U7 to U4 did not appear to be possible from the crystal structure. Finally, as previously discussed, the attacking nucleophile in the original structures, the
839:
For example, the invariant core residues G5, G8, G12 and C3 in the minimal hammerhead ribozyme were each observed to be so fragile that changing even a single exocyclic functional group on any one of these nucleotides results in a dramatic reduction or abolition of catalytic activity, yet few of
411:
or a similar exogenous energy source. The hammerhead ribozyme-catalyzed reaction, unlike the formally identical non-enzymatic alkaline cleavage of RNA, is a highly sequence-specific cleavage reaction with a typical turnover rate of approximately 1 molecule of substrate per molecule of enzyme per
910:
The active site of the full-length hammerhead ribozyme. G12 is positioned consistent with a role as a general base in the cleavage reaction, and the 2'-OH of G8 is positioned for acid catalysis. Potentially "active" hydrogen bonds are shown as orange dotted lines. The 2'-O of C17 is shown to be
844:
itself, rather than passively bound divalent metal ions, must play a direct chemical role in any acid-base chemistry in the hammerhead ribozyme active site. It was however completely unclear how G12 and G8 could accomplish this, given the original structures of the minimal hammerhead ribozyme.
505:
Secondary structures and sequences of the minimal (A) and full-length (B) hammerhead ribozymes. Conserved and invariant nucleotides are shown explicitly. Watson-Crick base-paired helical stems are represented as ladder-like drawings. The red arrow depicts the cleavage site, 3' to C17, on each
523:
but are otherwise not constrained with respect to sequence. The catalytic turnover rate of minimal hammerhead ribozymes is ~ 1/min (a range of 0.1/min to 10/min is commonly observed, depending upon the nonconserved sequences and the lengths of the three helical stems) under standard reaction
136:
The hammerhead ribozyme is arguably the best-characterized ribozyme. Its small size, thoroughly-investigated cleavage chemistry, known crystal structure, and its biological relevance make the hammerhead ribozyme particularly well-suited for biochemical and biophysical investigations into the
269:
In 1986, the first hammerhead ribozymes were found in RNA plant pathogens like viroids and viral satellites. One year later, a hammerhead ribozyme was also reported in the satellite DNA of newt genomes. New examples of this ribozyme were then found in the genomes of unrelated organisms like
907:
91:
hammerhead constructs can be engineered such that they consist of two interacting RNA strands, with one strand composing a hammerhead ribozyme that cleaves the other strand. The strand that gets cleaved can be supplied in excess, and multiple turnover can be demonstrated and shown to obey
868:
approach if the Stem II and Stem I A-form helices were treated as rigid bodies. Taken together, these results appeared to suggest that a fairly large-scale conformational change must have taken place in order to reach the transition-state within the minimal hammerhead ribozyme structure.
779:
The full-length hammerhead ribozyme consists of additional sequence elements in stems I and II that permit additional tertiary contacts to form. The tertiary interactions stabilize the active conformation of the ribozyme, resulting in cleavage rates up to 1000-fold greater than those for
948:
phosphates requires the presence of a high concentration of positive charge. This is probably the source of the observation that divalent metal ions are required at low ionic strength, but can be dispensed with at higher concentrations of monovalent cations.
817:. These helices are called I, II and III. The conserved uridine-turn links helix I to helix II and usually contains the sequence CUGA. Helix II and III are linked by a sequence GAAA. The cleavage reaction occurs between helix III and I, and is usually a C.
965:
Modified hammerhead ribozymes are being tested as therapeutic agents. Synthetic RNAs containing sequences complementary to the mutant SOD1 mRNA and sequences necessary to form the hammerhead catalytic structure are being studied as a possible therapy for
915:
The tertiary interactions in the full-length hammerhead ribozyme stabilize what strongly appears to be the active conformation. The nucleophile, the 2'-oxygen of the cleavage-site nucleotide, C17, is aligned almost perfectly for an in-line attack (the
402:
The 5’-product, as a result of this cleavage reaction mechanism, possesses a 2’,3’-cyclic phosphate terminus, and the 3’-product possesses a 5’-OH terminus, as with nonenzymatic alkaline cleavage of RNA. The reaction is therefore reversible, as the
928:
of the hammerhead ribozyme is 8.5, whereas the pKa of guanosine is about 9.5. It is possible that the pKa of G12 is perturbed from 9.5 to 8.5 in the hammerhead catalytic core; this hypothesis is currently the subject of intense investigation.
310:
The hammerhead ribozyme carries out a very simple chemical reaction that results in the breakage of the substrate strand of RNA, specifically at C17, the cleavage-site nucleotide. Although RNA cleavage is often referred to as
956:
phosphorus and the 5'-O leaving group begins to break, a proton is supplied from the ribose of G8, which then likely reprotonates at the expense of a water molecule observed to hydrogen bond to it in the crystal structure.
897:
The crystal structure of the full-length hammerhead ribozyme thus clearly addresses all of the major concerns that appeared irreconcilable with the previous crystal structures of the minimal hammerhead ribozyme.
515:
The minimal hammerhead sequence that is required for the self-cleavage reaction includes approximately 13 conserved or invariant "core" nucleotides, most of which are not involved in forming canonical
840:
these appeared to form hydrogen bonds involving the Watson-Crick faces of these nucleotide bases in any of the minimal hammerhead structures, apart from a G-5 interaction in the product structure.
993:
and therapeutic derivatives are likely to complement other nucleic acid hybridizing therapeutic strategies. Already there are hammerhead ribozymes which are close to clinical application.
371:-like reaction, although it is not known whether this proton is removed prior to or during the chemical step of the hammerhead cleavage reaction. (The cleavage reaction is technically not
42:
and properties of RNA, and is used for targeted RNA cleavage experiments, some with proposed therapeutic applications. Named for the resemblance of early secondary structure diagrams to a
2163:
Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF, de la Peña M, Korobeynikov A, Chikhi R, Babaian A (2022).
111:
The minimal trans-acting hammerhead ribozyme sequence that is catalytically active consists of three base-paired stems flanking a central core of 15 conserved (mostly invariant)
2725:
Fei Q, Zhang H, Fu L, Dai X, Gao B, Ni M, Ge C, Li J, Ding X, Ke Y, Yao X, Zhu J (June 2008). "Experimental cancer gene therapy by multiple anti-survivin hammerhead ribozymes".
490:
below, indeed confirm that two invariant nucleotides, G12 and G8, are positioned consistent with roles as the general base and general acid in the hammerhead cleavage reaction.
502:
952:
The reaction thus likely involves abstraction of the 2'-proton from C17, followed by nucleophilic attack upon the adjacent phosphate. As the bond between the
344:
and forming (dashed lines) must be in the axial positions and reside approximately 180° apart, as shown. Electron movements are represented by curly arrows.
115:, as shown. The conserved central bases, with few exceptions, are essential for ribozyme's catalytic activity. Such hammerhead ribozyme constructs exhibit
2435:
Khvorova A, Lescoute A, Westhof E, Jayasena SD (2003). "Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity".
924:
The most likely explanation is then that G12, in the deprotonated form, is the general base, and the ribose of G8 is the general acid. The apparent
974:-resistant lines of T-cells. Modified hammerhead ribozyme adenoviruses have been shown to be potent in treating cancer both in vitro and in vivo.
34:
motif that catalyzes reversible cleavage and ligation reactions at a specific site within an RNA molecule. It is one of several catalytic RNAs (
1428:
286:, which express as small circular RNAs. However, an exceptional group of strikingly conserved hammerheads can be found in the genomes of all
1417:"The Hammerhead Ribozyme Revisited: New Biological Insights for the Development of Therapeutic Agents and for Reverse Genomics Applications"
1054:"Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs"
302:
hammerhead motifs, indicating that not only this ribozyme but small circular RNAs with ribozymes are ubiquitous molecules in the biosphere.
813:
The minimal hammerhead ribozyme is composed of three base paired helices, separated by short linkers of conserved sequence as shown in the
69:
mechanism. The hammerhead sequence is sufficient for self-cleavage and acts by forming a conserved three-dimensional tertiary structure.
85:
in its natural state, as it is consumed by the reaction (i.e. performs self-cleavage) and therefore cannot catalyze multiple turnovers.
806:
by Pley, Flaherty and McKay. Subsequently, an all-RNA minimal hammerhead ribozyme structure was published by Scott, Finch and Klug in
77:
In its natural state, a hammerhead RNA motif is a single strand of RNA. Although the cleavage takes place in the absence of protein
407:
phosphate remains a phosphodiester, and may thus act as a substrate for hammerhead RNA-mediated ligation without a requirement for
297:
occur in the introns of a few specific genes and point to a preserved biological role during pre-mRNA biosynthesis. In 2021, novel
528:(~10 mM), pH 7.5 and 25 °C. Much of the experimental work carried out on hammerhead ribozymes has used a minimal construct.
1009:
Forster AC, Symons RH (1987). "Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites".
1228:
Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G (1986). "Autolytic
Processing of Dimeric Plant Virus Satellite RNA".
941:
of G8 with deoxyG8 greatly reduces the rate of catalysis, suggesting the 2'-OH is indeed crucial to the catalytic mechanism.
820:
The structure of a full-length ribozyme shows that there are extensive interactions between the loop of stem II and stem I.
802:
in 1994 in the form of an X-ray crystal structure of a hammerhead ribozyme bound to a DNA substrate analogue, published in
967:
140:
Hammerhead ribozymes may play an important role as therapeutic agents; as enzymes which tailor defined RNA sequences, as
836:
transition-state interactions and the chemistry of catalysis appeared to be irreconcilable with the crystal structures.
339:
104:
experiments, and the term "hammerhead RNA" has become in practice synonymous with the more frequently used "hammerhead
1331:
Forster AC, Symons RH (1987). "Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site".
459:
447:
443:
501:
380:
331:-mediated RNA degradation, except that it is highly site-specific and the rate is accelerated 10,000-fold or more.
66:
848:
93:
2061:
García-Robles I, Sánchez-Navarro J, de la Peña M (2012). "Intronic hammerhead ribozymes in mRNA biogenesis".
2680:
Citti L, Rainaldi G (2005). "Synthetic hammerhead ribozymes as therapeutic tools to control disease genes".
1536:
Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R (2000).
793:
2689:
2583:"The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage"
408:
39:
482:, do not require the presence of metal ions for catalysis, provided a sufficiently high concentration of
2773:
672:
555:
175:
2116:"Hepatitis delta virus-like circular RNAs from diverse metazoans encode conserved hammerhead ribozymes"
749:
632:
248:
2582:
2386:
1538:"Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets"
1377:
Usman, Nassim; Beigelman, Leonid; McSwiggen, James A (1996-08-01). "Hammerhead ribozyme engineering".
2536:
2282:
2271:"A two-metal ion mechanism operates in the hammerhead ribozyme-mediated cleavage of an RNA substrate"
2176:
2011:"Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes"
1911:
1647:
1237:
392:
2694:
62:. The hammerhead ribozyme motif has been ubiquitously reported in lineages across the tree of life.
2387:"The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone"
1900:"Identification of hammerhead ribozymes in all domains of life reveals novel structural variations"
880:
668:
551:
439:
171:
145:
47:
2480:"Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity"
2857:
2750:
2613:
2560:
2460:
2417:
2202:
2096:
1469:
1356:
1261:
1034:
925:
932:
906:
2527:
Pley HW, Flaherty KM, McKay DB (1994). "Three-dimensional structure of a hammerhead ribozyme".
2742:
2707:
2662:
2605:
2552:
2509:
2452:
2409:
2367:
2359:
2318:
2300:
2251:
2194:
2145:
2088:
2040:
1991:
1939:
1877:
1823:
1774:
1722:
1673:
1616:
1567:
1518:
1461:
1424:
1394:
1348:
1313:
1253:
1207:
1189:
1150:
1132:
1093:
1075:
1026:
889:
and G12 all appear involved in vital interactions relevant to catalysis. Moreover, the A9 and
814:
756:
744:
639:
627:
255:
243:
16:
2734:
2699:
2652:
2644:
2597:
2544:
2499:
2491:
2444:
2401:
2349:
2308:
2290:
2241:
2233:
2184:
2135:
2127:
2078:
2070:
2030:
2022:
1981:
1973:
1929:
1919:
1898:
Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR (May 2011).
1867:
1857:
1813:
1805:
1764:
1756:
1712:
1704:
1663:
1655:
1606:
1598:
1557:
1549:
1508:
1500:
1453:
1386:
1340:
1303:
1295:
1245:
1197:
1181:
1140:
1124:
1083:
1065:
1018:
824:
803:
728:
611:
519:. The core region is flanked by Stems I, II and III, which are in general made of canonical
479:
463:
384:
328:
231:
43:
2354:
2337:
46:, hammerhead ribozymes were originally discovered in two classes of plant virus-like RNAs:
2777:
127:
120:
97:
55:
1587:"Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana"
872:
ever made successfully, although many claims to the contrary were made in favor of each.
2540:
2286:
2180:
1915:
1651:
1444:
Epstein LM, Gall JG (1987). "Self-cleaving transcripts of satellite DNA from the newt".
1241:
851:
between U4 and U7 of the cleaved hammerhead ribozyme that had also been observed during
2657:
2632:
2246:
2221:
2140:
2115:
2035:
2010:
1986:
1961:
1934:
1899:
1872:
1845:
1818:
1793:
1769:
1744:
1717:
1692:
1668:
1635:
1611:
1586:
1202:
1169:
1145:
1112:
1088:
1053:
807:
483:
425:
349:
324:
2601:
2504:
2479:
2405:
1562:
1537:
1513:
1488:
1390:
1308:
1283:
2846:
2738:
2313:
2270:
2206:
1457:
1344:
1022:
953:
945:
890:
864:
857:
404:
388:
352:
320:
279:
2754:
2464:
2421:
2100:
1473:
1265:
1168:
Hammann, Christian; Luptak, Andrej; Perreault, Jonathan; de la Peña, Marcos (2012).
1038:
981:-cleaving hammerhead ribozymes has been severely hampered by its low-level activity
2852:
2617:
2564:
1585:
Przybilski R, Gräf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C (2005).
1360:
458:. An additional role for divalent metal ions has also been proposed in the form of
88:
1249:
936:
Possible transition-state interactions as extrapolated from the crystal structure.
493:
Strictly speaking, therefore, the hammerhead ribozyme cannot be a metallo-enzyme.
1924:
856:
2’-OH of C17, was not in a position amenable to in-line attack upon the adjacent
278:
Most eukaryotic hammerhead ribozymes are related to a kind of short interspersed
475:
372:
360:
298:
112:
2648:
2633:"Tertiary contacts distant from the active site prime a ribozyme for catalysis"
2275:
Proceedings of the
National Academy of Sciences of the United States of America
2189:
2164:
1070:
921:
proton as negative charge accumulates on the 5'-oxygen of the ribose of A1.1.
451:
312:
2703:
2363:
2304:
1846:"Structure-based search reveals hammerhead ribozymes in the human microbiome"
1693:"Ubiquitous presence of the hammerhead ribozyme motif along the tree of life"
1553:
1284:"Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid"
1193:
1136:
1113:"Ubiquitous presence of the hammerhead ribozyme motif along the tree of life"
1079:
2833:
2822:
2811:
2800:
2789:
1862:
1299:
737:
662:
620:
545:
525:
520:
516:
432:
421:
283:
236:
165:
141:
59:
38:) known to occur in nature. It serves as a model system for research on the
2746:
2711:
2666:
2513:
2495:
2456:
2255:
2198:
2149:
2092:
2044:
1995:
1977:
1943:
1881:
1827:
1778:
1760:
1726:
1677:
1636:"A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA"
1620:
1602:
1571:
1257:
1211:
1185:
1154:
1097:
970:. Work is also underway to find out whether they could be used to engineer
2609:
2556:
2413:
2371:
2322:
2295:
2074:
2026:
1522:
1504:
1465:
1398:
1352:
1317:
1030:
2131:
721:
604:
287:
224:
105:
82:
35:
2269:
Lott, William B.; Pontius, Brian W.; von Hippel, Peter H. (1998-01-20).
1809:
1708:
1659:
1128:
359:
2’-hydroxyl proton from the 2’-oxygen, which then becomes the attacking
2083:
1421:
RNA and the
Regulation of Gene Expression: A Hidden Layer of Complexity
474:
In 1998 it was discovered that the hammerhead ribozyme, as well as the
455:
2385:
J.B. Murray; A.A. Seyhan; N.G. Walter; J.M. Burke; W.G. Scott (1998).
2237:
1745:"Intronic hammerhead ribozymes are ultraconserved in the human genome"
911:
aligned for nucleophilic attack along the blue dotted line trajectory.
2548:
1416:
733:
616:
486:
356:
78:
51:
2338:"The structure, function and application of the hammerhead ribozyme"
2448:
863:
Perhaps most worrisome were experiments that suggested the A-9 and
387:
consisting of a pentacoordinated oxyphosphrane.) The attacking and
327:
bond. It is the same reaction, chemically, that occurs with random
65:
The self-cleavage reactions, first reported in 1986, are part of a
1794:"From alpaca to zebrafish: hammerhead ribozymes wherever you look"
931:
905:
884:
Three-dimensional structure of the full-length hammerhead ribozyme
879:
792:
500:
316:
315:, the mechanism employed does not in fact involve the addition of
15:
699:
582:
294:
291:
202:
2837:
2826:
2815:
2804:
2793:
717:
694:
600:
577:
435:
220:
197:
178:
of the HH9 ribozyme found conserved from lizard to human genomes
1489:"Schistosome satellite DNA encodes active hammerhead ribozymes"
355:
reaction that is initiated by abstraction of the cleavage-site
971:
852:
711:
594:
364:
214:
31:
2770:
2165:"Petabase-scale sequence alignment catalyses viral discovery"
1052:
Cervera, Amelia; Urbina, Denisse; de la Peña, Marcos (2016).
2222:"Efficient ligation of the Schistosoma hammerhead ribozyme"
1792:
Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C (2011).
823:
Similar structures are observed in other ribozymes such as
2782:
2336:
Birikh, K. R.; Heaton, P. A.; Eckstein, F. (1997-04-01).
391:
oxygens will both occupy the two axial positions in the
2783:
Marcos de la Peña's lab page on the hammerhead ribozyme
1960:
Hammann C, Luptak A, Perreault J, de la Peña M (2012).
1282:
Hutchins CJ, Rathjen PD, Forster AC, Symons RH (1986).
1111:
de la Peña, Marcos; García-Robles, Inmaculada (2010).
797:
The crystal structure of a minimal hammerhead ribozyme
2114:
de la Peña M, Ceprian R, Casey J, Cervera A (2021).
755:
743:
727:
710:
705:
693:
685:
680:
655:
638:
626:
610:
593:
588:
576:
568:
563:
538:
254:
242:
230:
213:
208:
196:
188:
183:
158:
395:transition-state structure as is required for an S
2771:Bill Scott's lab pages on the hammerhead ribozyme
1634:Martick M, Horan LH, Noller HF, Scott WG (2008).
2056:
2054:
989:-cleaving hammerhead ribozymes may be recouped
323:that consists of rearrangement of the linking
319:. Rather, the cleavage reaction is simply an
100:. Such constructs are typically employed for
8:
1893:
1891:
1738:
1736:
780:corresponding minimal hammerhead sequences.
450:, in a way analogous to those played by two
532:Type I, type II and type III hammerhead RNA
1955:
1953:
1839:
1837:
1487:Ferbeyre G, Smith JM, Cedergren R (1998).
1277:
1275:
661:
544:
164:
2693:
2656:
2503:
2478:De la Peña M; Gago S.; Flores R. (2003).
2437:Nature Structural & Molecular Biology
2353:
2312:
2294:
2245:
2188:
2139:
2082:
2034:
1985:
1933:
1923:
1871:
1861:
1817:
1768:
1716:
1667:
1610:
1561:
1512:
1307:
1201:
1144:
1087:
1069:
375:, but behaves in the same way a genuine S
54:. They are also known in some classes of
2576:
2574:
1844:Jimenez RM, Delwart E, Lupták A (2011).
338:
335:Cleavage by phosphodiester isomerization
1223:
1221:
1001:
2355:10.1111/j.1432-1033.1997.t01-3-00001.x
1743:De la Peña M, García-Robles I (2010).
1691:De la Peña M, García-Robles I (2010).
1410:
1408:
652:
535:
155:
2220:Canny MD, Jucker FM, Pardi A (2007).
1379:Current Opinion in Structural Biology
383:subsequent to forming an associative
137:fundamental nature of RNA catalysis.
81:, the hammerhead RNA itself is not a
20:Stylized rendering of the full-length
7:
2727:Acta Biochimica et Biophysica Sinica
1962:"The ubiquitous hammerhead ribozyme"
1372:
1370:
1170:"The ubiquitous hammerhead ribozyme"
2581:Scott WG, Finch JT, Klug A (1995).
416:Requirement for divalent metal ions
126:) of about 1 molecule/minute and a
985:. The true catalytic potential of
438:, which exist in equilibrium with
431:It was presumed that hexahydrated
14:
944:The close approach of the A9 and
675:of Hammerhead ribozyme (type III)
379:2(P) reaction does; it undergoes
2739:10.1111/j.1745-7270.2008.00430.x
2342:European Journal of Biochemistry
2009:Cervera A, de la Peña M (2020).
22:hammerhead ribozyme RNA molecule
497:Primary and secondary structure
656:Hammerhead ribozyme (type III)
424:were originally thought to be
133:on the order of 10 nanomolar.
1:
2602:10.1016/S0092-8674(05)80004-2
2406:10.1016/S1074-5521(98)90116-8
1391:10.1016/S0959-440X(96)80119-9
1250:10.1126/science.231.4745.1577
968:amyotrophic lateral sclerosis
290:. These hammerhead ribozymes
270:schistosomes, cave crickets,
2812:Page for Hammerhead type III
2631:Martick M, Scott WG (2006).
1925:10.1371/journal.pcbi.1002031
1458:10.1016/0092-8674(87)90204-2
1415:Hean J, Weinberg MS (2008).
1345:10.1016/0092-8674(87)90657-X
1023:10.1016/0092-8674(87)90562-9
539:Hammerhead ribozyme (type I)
2801:Page for Hammerhead type II
847:Other concerns included an
460:electrostatic stabilization
399:2-like reaction mechanism.
348:The cleavage reaction is a
2874:
2790:Page for Hammerhead type I
2649:10.1016/j.cell.2006.06.036
2190:10.1038/s41586-021-04332-2
1904:PLOS Computational Biology
1423:. Caister Academic Press.
442:, could play the roles of
381:inversion of configuration
275:RNA in all life kingdoms.
144:, and for applications in
67:rolling circle replication
1071:10.1186/s13059-016-1002-4
660:
543:
163:
94:Michaelis-Menten kinetics
2834:Page for Hammerhead HH10
2704:10.2174/1566523052997541
961:Therapeutic applications
2823:Page for Hammerhead HH9
1863:10.1074/jbc.C110.209288
977:The therapeutic use of
902:Structure and catalysis
521:Watson-Crick base-pairs
517:Watson-Crick base-pairs
159:Hammerhead ribozyme HH9
98:protein enzyme kinetics
2015:Nucleic Acids Research
1978:10.1261/rna.031401.111
1761:10.1038/embor.2010.100
1603:10.1105/tpc.105.032730
1554:10.1093/nar/28.20.4037
1186:10.1261/rna.031401.111
937:
912:
885:
798:
507:
345:
306:Chemistry of catalysis
23:
2296:10.1073/pnas.95.2.542
2075:10.1515/hsz-2012-0223
1505:10.1128/MCB.18.7.3880
1300:10.1093/nar/14.9.3627
935:
909:
883:
796:
673:sequence conservation
556:sequence conservation
504:
342:
176:sequence conservation
19:
2682:Current Gene Therapy
2496:10.1093/emboj/cdg530
2021:(9): (9):5054–5064.
775:Full-length ribozyme
470:Not a metallo-enzyme
393:trigonal bipyramidal
272:Arabidopsis thaliana
152:Species distribution
148:and gene discovery.
2541:1994Natur.372...68P
2287:1998PNAS...95..542L
2181:2022Natur.602..142E
2027:10.1093/nar/gkaa187
1916:2011PLSCB...7E2031P
1810:10.1261/rna.2429911
1709:10.1261/rna.2130310
1660:10.1038/nature07117
1652:2008Natur.454..899M
1242:1986Sci...231.1577P
1236:(4745): 1577–1580.
1129:10.1261/rna.2130310
669:secondary structure
552:secondary structure
524:conditions of high
440:magnesium hydroxide
363:in an “in-line” or
292:(the so-called HH9
172:secondary structure
146:functional genomics
28:hammerhead ribozyme
2776:2011-09-29 at the
2132:10.1093/ve/veab016
938:
913:
886:
831:Structure-function
799:
784:Tertiary structure
508:
346:
24:
2490:(20): 5561–5570.
2238:10.1021/bi062077r
2232:(12): 3826–3834.
2175:(7895): 142–147.
2069:(11): 1317–1326.
1856:(10): 7737–7743.
1703:(10): 1943–1950.
1646:(7206): 899–902.
1548:(20): 4037–4043.
1542:Nucleic Acids Res
1430:978-1-904455-25-7
1288:Nucleic Acids Res
1123:(10): 1943–1950.
815:crystal structure
768:
767:
651:
650:
267:
266:
119:a turnover rate (
58:, including the
2865:
2759:
2758:
2722:
2716:
2715:
2697:
2677:
2671:
2670:
2660:
2628:
2622:
2621:
2587:
2578:
2569:
2568:
2549:10.1038/372068a0
2524:
2518:
2517:
2507:
2475:
2469:
2468:
2432:
2426:
2425:
2391:
2382:
2376:
2375:
2357:
2333:
2327:
2326:
2316:
2298:
2266:
2260:
2259:
2249:
2217:
2211:
2210:
2192:
2160:
2154:
2153:
2143:
2111:
2105:
2104:
2086:
2058:
2049:
2048:
2038:
2006:
2000:
1999:
1989:
1957:
1948:
1947:
1937:
1927:
1895:
1886:
1885:
1875:
1865:
1841:
1832:
1831:
1821:
1789:
1783:
1782:
1772:
1740:
1731:
1730:
1720:
1688:
1682:
1681:
1671:
1631:
1625:
1624:
1614:
1597:(7): 1877–1885.
1582:
1576:
1575:
1565:
1533:
1527:
1526:
1516:
1499:(7): 3880–3888.
1484:
1478:
1477:
1441:
1435:
1434:
1412:
1403:
1402:
1374:
1365:
1364:
1328:
1322:
1321:
1311:
1294:(9): 3627–3640.
1279:
1270:
1269:
1225:
1216:
1215:
1205:
1165:
1159:
1158:
1148:
1108:
1102:
1101:
1091:
1073:
1049:
1043:
1042:
1006:
665:
653:
548:
536:
511:Minimal ribozyme
480:hairpin ribozyme
464:transition-state
385:transition-state
168:
156:
56:retrotransposons
44:hammerhead shark
2873:
2872:
2868:
2867:
2866:
2864:
2863:
2862:
2843:
2842:
2778:Wayback Machine
2767:
2762:
2724:
2723:
2719:
2695:10.1.1.333.3872
2679:
2678:
2674:
2630:
2629:
2625:
2596:(7): 991–1002.
2585:
2580:
2579:
2572:
2535:(6501): 68–74.
2526:
2525:
2521:
2477:
2476:
2472:
2434:
2433:
2429:
2400:(10): 587–595.
2389:
2384:
2383:
2379:
2335:
2334:
2330:
2268:
2267:
2263:
2219:
2218:
2214:
2162:
2161:
2157:
2120:Virus Evolution
2113:
2112:
2108:
2060:
2059:
2052:
2008:
2007:
2003:
1959:
1958:
1951:
1910:(5): e1002031.
1897:
1896:
1889:
1843:
1842:
1835:
1791:
1790:
1786:
1742:
1741:
1734:
1690:
1689:
1685:
1633:
1632:
1628:
1584:
1583:
1579:
1535:
1534:
1530:
1493:Mol. Cell. Biol
1486:
1485:
1481:
1443:
1442:
1438:
1431:
1414:
1413:
1406:
1376:
1375:
1368:
1330:
1329:
1325:
1281:
1280:
1273:
1227:
1226:
1219:
1167:
1166:
1162:
1110:
1109:
1105:
1051:
1050:
1046:
1008:
1007:
1003:
999:
963:
919:
904:
878:
833:
810:in early 1995.
791:
786:
777:
676:
559:
558:of Hammerhead_1
534:
513:
499:
472:
426:metallo-enzymes
418:
398:
378:
368:
337:
308:
282:(SINEs) called
179:
154:
131:
124:
75:
21:
12:
11:
5:
2871:
2869:
2861:
2860:
2855:
2845:
2844:
2841:
2840:
2831:
2829:
2820:
2818:
2809:
2807:
2798:
2796:
2787:
2785:
2780:
2766:
2765:External links
2763:
2761:
2760:
2717:
2672:
2643:(2): 309–320.
2623:
2570:
2519:
2470:
2449:10.1038/nsb959
2443:(9): 708–712.
2427:
2377:
2328:
2281:(2): 542–547.
2261:
2212:
2155:
2126:(1): veab016.
2106:
2050:
2001:
1972:(5): 871–885.
1949:
1887:
1833:
1784:
1755:(9): 711–716.
1732:
1683:
1626:
1577:
1528:
1479:
1452:(3): 535–543.
1436:
1429:
1404:
1385:(4): 527–533.
1366:
1323:
1271:
1217:
1180:(5): 871–885.
1160:
1103:
1058:Genome Biology
1044:
1017:(2): 211–220.
1000:
998:
995:
962:
959:
917:
903:
900:
877:
874:
832:
829:
790:
787:
785:
782:
776:
773:
766:
765:
760:
753:
752:
747:
741:
740:
731:
725:
724:
715:
708:
707:
703:
702:
697:
691:
690:
687:
683:
682:
678:
677:
666:
658:
657:
649:
648:
643:
636:
635:
630:
624:
623:
614:
608:
607:
598:
591:
590:
586:
585:
580:
574:
573:
570:
566:
565:
561:
560:
549:
541:
540:
533:
530:
512:
509:
498:
495:
471:
468:
417:
414:
396:
376:
366:
350:phosphodiester
336:
333:
325:phosphodiester
307:
304:
265:
264:
259:
252:
251:
246:
240:
239:
234:
228:
227:
218:
211:
210:
206:
205:
200:
194:
193:
190:
186:
185:
181:
180:
169:
161:
160:
153:
150:
129:
122:
74:
71:
48:satellite RNAs
13:
10:
9:
6:
4:
3:
2:
2870:
2859:
2856:
2854:
2851:
2850:
2848:
2839:
2835:
2832:
2830:
2828:
2824:
2821:
2819:
2817:
2813:
2810:
2808:
2806:
2802:
2799:
2797:
2795:
2791:
2788:
2786:
2784:
2781:
2779:
2775:
2772:
2769:
2768:
2764:
2756:
2752:
2748:
2744:
2740:
2736:
2733:(6): 466–77.
2732:
2728:
2721:
2718:
2713:
2709:
2705:
2701:
2696:
2691:
2687:
2683:
2676:
2673:
2668:
2664:
2659:
2654:
2650:
2646:
2642:
2638:
2634:
2627:
2624:
2619:
2615:
2611:
2607:
2603:
2599:
2595:
2591:
2584:
2577:
2575:
2571:
2566:
2562:
2558:
2554:
2550:
2546:
2542:
2538:
2534:
2530:
2523:
2520:
2515:
2511:
2506:
2501:
2497:
2493:
2489:
2485:
2481:
2474:
2471:
2466:
2462:
2458:
2454:
2450:
2446:
2442:
2438:
2431:
2428:
2423:
2419:
2415:
2411:
2407:
2403:
2399:
2395:
2388:
2381:
2378:
2373:
2369:
2365:
2361:
2356:
2351:
2347:
2343:
2339:
2332:
2329:
2324:
2320:
2315:
2310:
2306:
2302:
2297:
2292:
2288:
2284:
2280:
2276:
2272:
2265:
2262:
2257:
2253:
2248:
2243:
2239:
2235:
2231:
2227:
2223:
2216:
2213:
2208:
2204:
2200:
2196:
2191:
2186:
2182:
2178:
2174:
2170:
2166:
2159:
2156:
2151:
2147:
2142:
2137:
2133:
2129:
2125:
2121:
2117:
2110:
2107:
2102:
2098:
2094:
2090:
2085:
2080:
2076:
2072:
2068:
2064:
2057:
2055:
2051:
2046:
2042:
2037:
2032:
2028:
2024:
2020:
2016:
2012:
2005:
2002:
1997:
1993:
1988:
1983:
1979:
1975:
1971:
1967:
1963:
1956:
1954:
1950:
1945:
1941:
1936:
1931:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1894:
1892:
1888:
1883:
1879:
1874:
1869:
1864:
1859:
1855:
1851:
1847:
1840:
1838:
1834:
1829:
1825:
1820:
1815:
1811:
1807:
1803:
1799:
1795:
1788:
1785:
1780:
1776:
1771:
1766:
1762:
1758:
1754:
1750:
1746:
1739:
1737:
1733:
1728:
1724:
1719:
1714:
1710:
1706:
1702:
1698:
1694:
1687:
1684:
1679:
1675:
1670:
1665:
1661:
1657:
1653:
1649:
1645:
1641:
1637:
1630:
1627:
1622:
1618:
1613:
1608:
1604:
1600:
1596:
1592:
1588:
1581:
1578:
1573:
1569:
1564:
1559:
1555:
1551:
1547:
1543:
1539:
1532:
1529:
1524:
1520:
1515:
1510:
1506:
1502:
1498:
1494:
1490:
1483:
1480:
1475:
1471:
1467:
1463:
1459:
1455:
1451:
1447:
1440:
1437:
1432:
1426:
1422:
1418:
1411:
1409:
1405:
1400:
1396:
1392:
1388:
1384:
1380:
1373:
1371:
1367:
1362:
1358:
1354:
1350:
1346:
1342:
1338:
1334:
1327:
1324:
1319:
1315:
1310:
1305:
1301:
1297:
1293:
1289:
1285:
1278:
1276:
1272:
1267:
1263:
1259:
1255:
1251:
1247:
1243:
1239:
1235:
1231:
1224:
1222:
1218:
1213:
1209:
1204:
1199:
1195:
1191:
1187:
1183:
1179:
1175:
1171:
1164:
1161:
1156:
1152:
1147:
1142:
1138:
1134:
1130:
1126:
1122:
1118:
1114:
1107:
1104:
1099:
1095:
1090:
1085:
1081:
1077:
1072:
1067:
1063:
1059:
1055:
1048:
1045:
1040:
1036:
1032:
1028:
1024:
1020:
1016:
1012:
1005:
1002:
996:
994:
992:
988:
984:
980:
975:
973:
969:
960:
958:
955:
950:
947:
942:
934:
930:
927:
922:
908:
901:
899:
895:
892:
882:
875:
873:
869:
866:
861:
859:
854:
850:
845:
841:
837:
830:
828:
826:
821:
818:
816:
811:
809:
805:
795:
788:
783:
781:
774:
772:
764:
761:
758:
754:
751:
748:
746:
742:
739:
735:
732:
730:
726:
723:
719:
716:
713:
709:
704:
701:
698:
696:
692:
688:
684:
679:
674:
670:
664:
659:
654:
647:
644:
641:
637:
634:
631:
629:
625:
622:
618:
615:
613:
609:
606:
602:
599:
596:
592:
587:
584:
581:
579:
575:
571:
567:
562:
557:
553:
547:
542:
537:
531:
529:
527:
522:
518:
510:
503:
496:
494:
491:
488:
485:
481:
477:
469:
467:
465:
461:
457:
453:
449:
445:
441:
437:
434:
429:
427:
423:
415:
413:
410:
406:
400:
394:
390:
389:leaving group
386:
382:
374:
370:
362:
358:
354:
353:isomerization
351:
341:
334:
332:
330:
326:
322:
321:isomerization
318:
314:
305:
303:
300:
296:
293:
289:
285:
281:
280:retroelements
276:
273:
263:
260:
257:
253:
250:
247:
245:
241:
238:
235:
233:
229:
226:
222:
219:
216:
212:
207:
204:
201:
199:
195:
191:
187:
182:
177:
173:
167:
162:
157:
151:
149:
147:
143:
138:
134:
132:
125:
118:
114:
109:
107:
103:
99:
96:, typical of
95:
90:
86:
84:
80:
72:
70:
68:
63:
61:
57:
53:
49:
45:
41:
37:
33:
29:
18:
2730:
2726:
2720:
2688:(1): 11–24.
2685:
2681:
2675:
2640:
2636:
2626:
2593:
2589:
2532:
2528:
2522:
2487:
2483:
2473:
2440:
2436:
2430:
2397:
2393:
2380:
2345:
2341:
2331:
2278:
2274:
2264:
2229:
2226:Biochemistry
2225:
2215:
2172:
2168:
2158:
2123:
2119:
2109:
2066:
2062:
2018:
2014:
2004:
1969:
1965:
1907:
1903:
1853:
1849:
1804:(1): 21–26.
1801:
1797:
1787:
1752:
1748:
1700:
1696:
1686:
1643:
1639:
1629:
1594:
1590:
1580:
1545:
1541:
1531:
1496:
1492:
1482:
1449:
1445:
1439:
1420:
1382:
1378:
1336:
1332:
1326:
1291:
1287:
1233:
1229:
1177:
1173:
1163:
1120:
1116:
1106:
1061:
1057:
1047:
1014:
1010:
1004:
990:
986:
982:
978:
976:
964:
951:
943:
939:
923:
914:
896:
887:
870:
862:
846:
842:
838:
834:
822:
819:
812:
800:
778:
769:
689:Hammerhead_3
572:Hammerhead_1
514:
492:
473:
448:general base
444:general acid
430:
419:
401:
347:
309:
277:
271:
268:
139:
135:
116:
110:
101:
89:Trans-acting
87:
76:
64:
27:
25:
2348:(1): 1–16.
2084:10251/34564
1850:J Biol Chem
1339:(1): 9–16.
926:kinetic pKa
876:Full-length
860:phosphate.
827:ribozymes.
681:Identifiers
564:Identifiers
476:VS ribozyme
373:bimolecular
361:nucleophile
299:Hepatitis D
184:Identifiers
113:nucleotides
2847:Categories
1591:Plant Cell
1064:(1): 135.
997:References
759:structures
750:SO:0000380
706:Other data
667:Predicted
642:structures
633:SO:0000380
589:Other data
550:Predicted
506:construct.
484:monovalent
452:histidines
313:hydrolysis
284:retrozymes
258:structures
249:SO:0000380
209:Other data
170:Predicted
142:biosensors
60:retrozymes
2858:Ribozymes
2690:CiteSeerX
2394:Chem Biol
2364:0014-2956
2305:0027-8424
2207:246297430
2063:Biol Chem
1194:1355-8382
1137:1355-8382
1080:1474-760X
738:Eukaryota
729:Domain(s)
621:Eukaryota
612:Domain(s)
433:magnesium
422:ribozymes
295:and HH10)
237:Eukaryota
232:Domain(s)
73:Catalysis
40:structure
36:ribozymes
2774:Archived
2755:31532433
2747:18535745
2712:15638708
2667:16859740
2514:14532128
2465:52859654
2457:12881719
2422:17025877
2256:17319693
2199:35082445
2150:33708415
2101:16402212
2093:23109545
2045:32198887
1996:22454536
1944:21573207
1882:21257745
1828:21081661
1779:20651741
1749:EMBO Rep
1727:20705646
1678:18615019
1621:15937227
1572:11024185
1474:24110291
1266:21563490
1258:17833317
1212:22454536
1155:20705646
1098:27339130
1039:33415709
954:scissile
946:scissile
891:scissile
865:scissile
858:scissile
722:ribozyme
605:ribozyme
405:scissile
288:amniotes
225:ribozyme
117:in vitro
106:ribozyme
102:in vitro
83:catalyst
2658:4447102
2618:6965437
2610:7541315
2565:4333072
2557:7969422
2537:Bibcode
2414:9818150
2372:9128718
2323:9435228
2283:Bibcode
2247:3203546
2177:Bibcode
2141:7936874
2036:7229834
1987:3334697
1935:3088659
1912:Bibcode
1873:3048661
1819:3004062
1770:2933863
1718:2941103
1669:2612532
1648:Bibcode
1612:1167538
1523:9632772
1466:2433049
1399:8794164
1361:7231363
1353:3594567
1318:3714492
1238:Bibcode
1230:Science
1203:3334697
1146:2941103
1089:4918200
1031:2436805
991:in vivo
983:in vivo
825:hatchet
789:Minimal
734:Viroids
700:RF00008
617:Viroids
583:RF00163
462:of the
456:RNase A
203:RF02275
79:enzymes
52:viroids
2753:
2745:
2710:
2692:
2665:
2655:
2616:
2608:
2563:
2555:
2529:Nature
2512:
2505:213784
2502:
2484:EMBO J
2463:
2455:
2420:
2412:
2370:
2362:
2321:
2311:
2303:
2254:
2244:
2205:
2197:
2169:Nature
2148:
2138:
2099:
2091:
2043:
2033:
1994:
1984:
1942:
1932:
1880:
1870:
1826:
1816:
1777:
1767:
1725:
1715:
1676:
1666:
1640:Nature
1619:
1609:
1570:
1563:110794
1560:
1521:
1514:108972
1511:
1472:
1464:
1427:
1397:
1359:
1351:
1316:
1309:339804
1306:
1264:
1256:
1210:
1200:
1192:
1153:
1143:
1135:
1096:
1086:
1078:
1037:
1029:
804:Nature
686:Symbol
569:Symbol
487:cation
357:ribose
189:Symbol
30:is an
2751:S2CID
2614:S2CID
2586:(PDF)
2561:S2CID
2461:S2CID
2418:S2CID
2390:(PDF)
2314:18456
2203:S2CID
2097:S2CID
1470:S2CID
1357:S2CID
1262:S2CID
1035:S2CID
987:trans
979:trans
317:water
2838:Rfam
2827:Rfam
2816:Rfam
2805:Rfam
2794:Rfam
2743:PMID
2708:PMID
2663:PMID
2637:Cell
2606:PMID
2590:Cell
2553:PMID
2510:PMID
2453:PMID
2410:PMID
2368:PMID
2360:ISSN
2319:PMID
2301:ISSN
2252:PMID
2195:PMID
2146:PMID
2089:PMID
2041:PMID
1992:PMID
1940:PMID
1878:PMID
1824:PMID
1775:PMID
1723:PMID
1674:PMID
1617:PMID
1568:PMID
1519:PMID
1462:PMID
1446:Cell
1425:ISBN
1395:PMID
1349:PMID
1333:Cell
1314:PMID
1254:PMID
1208:PMID
1190:ISSN
1151:PMID
1133:ISSN
1094:PMID
1076:ISSN
1027:PMID
1011:Cell
808:Cell
763:PDBe
718:Gene
714:type
695:Rfam
671:and
646:PDBe
601:Gene
597:type
578:Rfam
554:and
478:and
446:and
436:ions
420:All
369:2(P)
329:base
262:PDBe
221:Gene
217:type
198:Rfam
174:and
50:and
26:The
2853:RNA
2836:at
2825:at
2814:at
2803:at
2792:at
2735:doi
2700:doi
2653:PMC
2645:doi
2641:126
2598:doi
2545:doi
2533:372
2500:PMC
2492:doi
2445:doi
2402:doi
2350:doi
2346:245
2309:PMC
2291:doi
2242:PMC
2234:doi
2185:doi
2173:602
2136:PMC
2128:doi
2079:hdl
2071:doi
2067:393
2031:PMC
2023:doi
1982:PMC
1974:doi
1966:RNA
1930:PMC
1920:doi
1868:PMC
1858:doi
1854:286
1814:PMC
1806:doi
1798:RNA
1765:PMC
1757:doi
1713:PMC
1705:doi
1697:RNA
1664:PMC
1656:doi
1644:454
1607:PMC
1599:doi
1558:PMC
1550:doi
1509:PMC
1501:doi
1454:doi
1387:doi
1341:doi
1304:PMC
1296:doi
1246:doi
1234:231
1198:PMC
1182:doi
1174:RNA
1141:PMC
1125:doi
1117:RNA
1084:PMC
1066:doi
1019:doi
972:HIV
853:NMR
849:NOE
757:PDB
712:RNA
640:PDB
595:RNA
454:in
409:ATP
256:PDB
215:RNA
192:HH9
123:cat
108:".
32:RNA
2849::
2749:.
2741:.
2731:40
2729:.
2706:.
2698:.
2684:.
2661:.
2651:.
2639:.
2635:.
2612:.
2604:.
2594:81
2592:.
2588:.
2573:^
2559:.
2551:.
2543:.
2531:.
2508:.
2498:.
2488:22
2486:.
2482:.
2459:.
2451:.
2441:10
2439:.
2416:.
2408:.
2396:.
2392:.
2366:.
2358:.
2344:.
2340:.
2317:.
2307:.
2299:.
2289:.
2279:95
2277:.
2273:.
2250:.
2240:.
2230:46
2228:.
2224:.
2201:.
2193:.
2183:.
2171:.
2167:.
2144:.
2134:.
2122:.
2118:.
2095:.
2087:.
2077:.
2065:.
2053:^
2039:.
2029:.
2019:48
2017:.
2013:.
1990:.
1980:.
1970:18
1968:.
1964:.
1952:^
1938:.
1928:.
1918:.
1906:.
1902:.
1890:^
1876:.
1866:.
1852:.
1848:.
1836:^
1822:.
1812:.
1802:17
1800:.
1796:.
1773:.
1763:.
1753:11
1751:.
1747:.
1735:^
1721:.
1711:.
1701:16
1699:.
1695:.
1672:.
1662:.
1654:.
1642:.
1638:.
1615:.
1605:.
1595:17
1593:.
1589:.
1566:.
1556:.
1546:28
1544:.
1540:.
1517:.
1507:.
1497:18
1495:.
1491:.
1468:.
1460:.
1450:48
1448:.
1419:.
1407:^
1393:.
1381:.
1369:^
1355:.
1347:.
1337:50
1335:.
1312:.
1302:.
1292:14
1290:.
1286:.
1274:^
1260:.
1252:.
1244:.
1232:.
1220:^
1206:.
1196:.
1188:.
1178:18
1176:.
1172:.
1149:.
1139:.
1131:.
1121:16
1119:.
1115:.
1092:.
1082:.
1074:.
1062:17
1060:.
1056:.
1033:.
1025:.
1015:49
1013:.
745:SO
736:;
720:;
628:SO
619:;
603:;
526:Mg
466:.
244:SO
223:;
2757:.
2737::
2714:.
2702::
2686:5
2669:.
2647::
2620:.
2600::
2567:.
2547::
2539::
2516:.
2494::
2467:.
2447::
2424:.
2404::
2398:5
2374:.
2352::
2325:.
2293::
2285::
2258:.
2236::
2209:.
2187::
2179::
2152:.
2130::
2124:7
2103:.
2081::
2073::
2047:.
2025::
1998:.
1976::
1946:.
1922::
1914::
1908:7
1884:.
1860::
1830:.
1808::
1781:.
1759::
1729:.
1707::
1680:.
1658::
1650::
1623:.
1601::
1574:.
1552::
1525:.
1503::
1476:.
1456::
1433:.
1401:.
1389::
1383:6
1363:.
1343::
1320:.
1298::
1268:.
1248::
1240::
1214:.
1184::
1157:.
1127::
1100:.
1068::
1041:.
1021::
918:N
916:S
397:N
377:N
367:N
365:S
130:m
128:K
121:k
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.