377:
90:
24:
158:
InChI=1S/C45H48O21/c1-6-8-42(60)40(58)32(54)24(46)14-11-12-10-13-16(25(47)15(12)38(56)44(14,42)61)26(48)20(37(65-5)34(13)63-3)19-27(49)17-18(30(52)36(19)64-4)28(50)21-22(29(17)51)39(57)45(62)23-31(53)33(55)41(59)43(45,9-7-2)66-35(21)23/h10,14,23-24,31-33,35,40-41,46-48,50-51,53-55,58-62H,6-9,11H2,1-5H3/t14-,23+,24+,31+,32-,33-,35+,40+,41+,42+,43+,44-,45-/m1/s1
331:
The biosynthesis of hibarimicinone was initially hypothesized to be produced from a polyketide synthase. Through 13C-labelling and blocked mutants of the TP-A0121 strain, Oki et al. demonstrated that hibarimicinone is produced by an oxidative coupling of two tetracyclic polyketides, which undergo an
340:
derived two carbon ketide unit. The KS/CLF binds to the starting butyryl-CoA, freeing the ACP domain to bind to another malonate group from malonyl-CoA. Thioester exchange occurs between the KS/CLF and the ACP; the cycle then repeats nine more times to produce the full carbon chain
319:, with steric hindrance imposed by the four ortho substituents. The absolute configuration around the axis was elucidated by Romaine et al., utilizing a C2-symmetric shunt metabolite HMP-Y6, a glycosylated structural analogue of hibarimicinone, in tandem with
332:
oxidative cyclization to generate the ether ring. The hibarimicinone monomer is formed initially by a Type II polyketide synthase. The 22 carbon chain is initiated by butyryl-CoA and its
525:
295:
backbone, which are now known as the hibarimicins. Hibarimicinone and its derivatives were initially extracted for their potential inhibitory properties of various
530:
505:
174:
300:
315:(PTK). The atropisomerism that the hibarimicin family possess arises from the hindered rotation about the biaryl axis connecting to the two
411:
Kajiura, T.; Furumai, T.; Igarashi, Y.; Hori, H.; Higashi, K.; Ishiyama, T.; Uramoto, M.; Uehara, Y.; Oki, T. J. Antibiot. 1998, 51, 394-401
453:
Romaine, I. M.; Hempel, J. E.; Shanmugam, G.; Hori, H.; Igarashi, Y.; Polavarpu, P. L.; Sulikowshi, G. A. Org. Lett. 2011, 13, 4538-4541
432:
Hori, H.; Kajiura, T.; Igarashi, Y.; Furumai, T; Higashi, K.; Ishiyama, T; Uramoto, M; Uehara, Y; Oki, T. J. Antibiot., 2002, 55, 46-52
420:
Hori, H.; Kajiura, T.; Igarashi, Y.; Hori, H.; Higashi, K; Ishiyama, T.; Uramoto, M; Uehara, Y.; Oki, T. J. Antibiot. 2002, 55, 53-60
149:
471:
Tatsuta, K.; Fukuda, T.; Ishimori, T.; Yachi, R.; Yoshida, S.; Hashimoto, H.; Hosokawa, S. Tetrahedron Lett., 2012, 53, 422-425
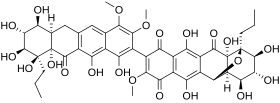
65:)-15--5,6,7,9,12,19-hexahydroxy-16-methoxy-4-propyl ;-3-oxapentacyclononadeca-1(11),12,15,18-tetraene-10,14,17-trione
246:
182:
CCC1((((21(C(=O)c3c(cc4c(c3O)c(c(c(c4OC)OC)C5=C(C(=O)c6c(c(c7c(c6O)89(((C(9(C7=O)O)(O8)CCC)O)O)O)O)C5=O)OC)O)C2)O)O)O)O)O
515:
535:
323:
homocoupling. Recent work by multiple groups have shown the total synthesis of hibarimicinone and its derivatives.
312:
353:. The CLF/KS subunit is cleaved off by a thioesterase and modified by numerous steps (four oxidations, two
397:
Hori, H.; Higashi, K.; Ishiyama, T.; Uramoto, M.; Uehara, Y.; Oki, T. Tetrahedron Lett. 1996 37, 2785-2788
275:
280:
36:
369:. Finally, the hydroquinone on the same side of the previously mentioned ether oxidizes to form the
520:
270:
376:
110:
357:
dependent reductions, two methylations and decarboxylation) to form the final monomeric unit
510:
308:
304:
197:
333:
296:
346:
240:
135:
499:
462:
B. B. Liau, B. C. Milgram and M. D. Shair, J. Am. Chem. Soc., 2012, 134, 16765-16772
365:. Oxidative cyclization occurs on one end of the molecule to form the ether ring
337:
262:
361:. Two monomer units undergo oxidative coupling to form the atropisomeric axis
320:
292:
225:
101:
489:
Dewick, Paul M. (2002). Medicinal
Natural Products. John Wiley and Sons, LTD
23:
316:
288:
266:
370:
122:
441:
Smyth, J.; Butler, N.; Keller, P. Nat. Prod. Rep. 2015, 32, 1562-1583
291:
identified a new class of molecule containing a dimeric-tetracyclic
239:
Except where otherwise noted, data are given for materials in their
375:
354:
284:
89:
79:
349:
occur to form four six-membered cyclization and aromization
273:, which are isolated from the microbial culture strain of
480:
Tatsuta, K.; Hosokawa, S. Chem. Rec., 2014, 14, 28-40
134:
8:
407:
405:
403:
109:
15:
526:3-Hydroxypropenals within hydroxyquinones
449:
447:
428:
426:
380:Predicted Biosynthesis of Habarimicinone
390:
301:calmodulin-dependent protein kinase III
179:
154:
161:Key: SSVUTDACAKUVQH-RNRCTZQLSA-N
7:
531:Heterocyclic compounds with 4 rings
506:Heterocyclic compounds with 5 rings
125:
14:
373:species, giving hibarimicinone.
209:
22:
243:(at 25 °C , 100 kPa).
215:
203:
1:
552:
237:
190:
170:
145:
71:
35:
30:
21:
313:protein tyrosine kinase
381:
379:
37:Preferred IUPAC name
233: g·mol
18:
516:Dimers (chemistry)
382:
287:. Analysis of the
276:Microbispora rosea
247:Infobox references
16:
536:Methoxy compounds
255:Chemical compound
253:
252:
91:Interactive image
543:
490:
487:
481:
478:
472:
469:
463:
460:
454:
451:
442:
439:
433:
430:
421:
418:
412:
409:
398:
395:
309:protein kinase C
305:protein kinase A
232:
217:
211:
205:
198:Chemical formula
138:
127:
113:
93:
26:
19:
551:
550:
546:
545:
544:
542:
541:
540:
496:
495:
494:
493:
488:
484:
479:
475:
470:
466:
461:
457:
452:
445:
440:
436:
431:
424:
419:
415:
410:
401:
396:
392:
387:
347:aldol additions
334:decarboxylative
329:
297:protein kinases
269:, derived from
256:
249:
244:
230:
220:
214:
208:
200:
186:
183:
178:
177:
166:
163:
162:
159:
153:
152:
141:
128:
116:
96:
83:
67:
66:
17:Hibarimicinone
12:
11:
5:
549:
547:
539:
538:
533:
528:
523:
518:
513:
508:
498:
497:
492:
491:
482:
473:
464:
455:
443:
434:
422:
413:
399:
389:
388:
386:
383:
336:addition to a
328:
325:
261:is an organic
259:Hibarimicinone
254:
251:
250:
245:
241:standard state
238:
235:
234:
228:
222:
221:
218:
212:
206:
201:
196:
193:
192:
188:
187:
185:
184:
181:
173:
172:
171:
168:
167:
165:
164:
160:
157:
156:
148:
147:
146:
143:
142:
140:
139:
131:
129:
121:
118:
117:
115:
114:
106:
104:
98:
97:
95:
94:
86:
84:
77:
74:
73:
69:
68:
40:
39:
33:
32:
28:
27:
13:
10:
9:
6:
4:
3:
2:
548:
537:
534:
532:
529:
527:
524:
522:
519:
517:
514:
512:
509:
507:
504:
503:
501:
486:
483:
477:
474:
468:
465:
459:
456:
450:
448:
444:
438:
435:
429:
427:
423:
417:
414:
408:
406:
404:
400:
394:
391:
384:
378:
374:
372:
368:
364:
360:
356:
352:
348:
344:
339:
335:
326:
324:
322:
318:
314:
310:
306:
302:
298:
294:
290:
286:
282:
278:
277:
272:
268:
264:
263:atropisomeric
260:
248:
242:
236:
229:
227:
224:
223:
202:
199:
195:
194:
189:
180:
176:
169:
155:
151:
144:
137:
133:
132:
130:
124:
120:
119:
112:
108:
107:
105:
103:
100:
99:
92:
88:
87:
85:
81:
76:
75:
70:
64:
60:
56:
52:
48:
44:
38:
34:
29:
25:
20:
485:
476:
467:
458:
437:
416:
393:
366:
362:
358:
350:
342:
330:
327:Biosynthesis
274:
271:hibarimicins
258:
257:
72:Identifiers
62:
58:
54:
50:
46:
42:
345:. Multiple
338:malonyl-CoA
311:(PKC), and
303:(CAMKIII),
191:Properties
521:Triketones
500:Categories
385:References
321:biomimetic
299:, such as
293:polyketide
283:region of
226:Molar mass
102:ChemSpider
78:3D model (
279:from the
136:101157796
317:monomers
289:bacteria
267:molecule
111:28289394
511:Phenols
371:quinone
307:(PKA),
231:924.858
123:PubChem
281:Hibari
265:small
175:SMILES
31:Names
355:NADPH
285:Japan
150:InChI
80:JSmol
367:(5)
363:(4)
359:(3)
351:(2)
343:(1)
126:CID
502::
446:^
425:^
402:^
219:21
213:48
207:45
61:,9
57:,8
53:,7
49:,6
45:,5
41:(2
216:O
210:H
204:C
82:)
63:S
59:S
55:S
51:R
47:S
43:R
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.