307:. Antibiotics such as ciprofloxacin are able to prevent the action of these molecules by attaching themselves to the gyrate–DNA complex, leading to replication fork stall and the induction of the SOS response. The expression of error-prone polymerases under the SOS response increases the basal mutation rate of bacteria. While mutations are often lethal to the cell, they can also enhance survival. In the specific case of topoisomerases, some bacteria have mutated one of their amino acids so that the ciprofloxacin can only create a weak bond to the topoisomerase. This is one of the methods that bacteria use to become
1956:
1982:
42:
311:
to antibiotics. Ciprofloxacin treatment can therefore potentially lead to the generation of mutations that may render bacteria resistant to ciprofloxacin. In addition, ciprofloxacin has also been shown to induce via the SOS response dissemination of
236:, which senses DNA damage at stalled replication forks, forming monofilaments and acquiring an active conformation capable of binding to LexA and causing LexA to cleave itself, in a process called
1168:
1028:
154:
768:
1889:
251:
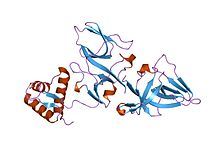
domain. The dimerization domain binds to other LexA polypeptides to form dumbbell shaped dimers. The DNA-binding domain is a variant form of the
1863:
102:
1524:
1579:
761:
1832:
1763:
540:"Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci"
1233:
473:
1847:
579:
Beaber JW, Hochhut B, Waldor MK (January 2004). "SOS response promotes horizontal dissemination of antibiotic resistance genes".
229:
754:
1389:
174:
1972:
1822:
1729:
988:
2007:
746:
1751:
1575:
1568:
1374:
1490:
1477:
1464:
1451:
1438:
1425:
1412:
903:
888:
788:
1384:
2012:
2002:
1746:
1668:
1517:
1338:
1281:
785:
330:
Impaired LexA proteolysis has been shown to interfere with ciprofloxacin resistance. This offers potential for
327:, potentially increasing the likelihood of acquisition and dissemination of antibiotic resistance by bacteria.
199:
162:
1286:
1178:
1143:
1138:
1858:
1768:
1563:
1624:
1307:
1226:
1133:
983:
317:
308:
1379:
158:
1549:
1123:
643:
588:
335:
115:
2022:
1959:
1646:
1510:
1343:
1023:
1013:
830:
825:
331:
2017:
1276:
667:
612:
399:
244:
491:"Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy"
1842:
1005:
718:
659:
604:
561:
520:
469:
443:
391:
287:. Of potential clinical interest is the induction of the SOS response by antibiotics, such as
149:
630:
Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, et al. (May 2009).
1322:
1317:
1291:
1219:
797:
708:
698:
651:
596:
551:
510:
502:
435:
381:
373:
313:
300:
252:
338:
with strategies aimed at interfering with the action of LexA, either directly or via RecA.
141:
1558:
1369:
1353:
1266:
1038:
1033:
805:
781:
304:
647:
592:
386:
361:
1986:
1407:
1348:
713:
686:
506:
210:
515:
490:
1996:
1870:
1691:
1312:
1271:
1183:
1056:
1045:
777:
556:
539:
439:
292:
288:
280:
256:
248:
221:
107:
671:
489:
Fogh RH, Ottleben G, Rüterjans H, Schnarr M, Boelens R, Kaptein R (September 1994).
403:
70:
1837:
1806:
1756:
1739:
1686:
1639:
1261:
1079:
973:
815:
685:
Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE (June 2005).
616:
206:
137:
468:(Fifth ed.). Hoboken, NJ : Washington, D.C: John Wiley & Sons, Inc.
83:
742:
703:
95:
1485:
1420:
1256:
1198:
1193:
842:
237:
1981:
1773:
1594:
1541:
1533:
1160:
1102:
883:
878:
858:
424:"Aeons of distress: an evolutionary perspective on the bacterial SOS response"
377:
296:
276:
260:
214:
687:"Inhibition of mutation and combating the evolution of antibiotic resistance"
1459:
1433:
1188:
1174:
993:
873:
863:
655:
324:
267:
upstream of SOS response genes until DNA damage stimulates autoproteolysis.
195:
722:
663:
608:
565:
447:
395:
524:
111:
41:
1854:
1074:
1069:
1018:
868:
853:
820:
738:
321:
284:
202:
90:
600:
538:
Ubeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penadés JR (May 2005).
1801:
1553:
1128:
1118:
1064:
978:
810:
264:
1926:
1681:
1634:
1629:
1472:
1242:
1155:
1097:
217:
169:
631:
423:
17:
1941:
1936:
1931:
1921:
1914:
1909:
1904:
1899:
1894:
1884:
1879:
1734:
1698:
1619:
1614:
1609:
1599:
1446:
1164:
1150:
1089:
963:
958:
953:
948:
943:
938:
1827:
1783:
1778:
1713:
1708:
1703:
1676:
1656:
1651:
1604:
933:
928:
923:
918:
913:
908:
898:
893:
734:
233:
131:
77:
65:
1506:
1215:
750:
1589:
1584:
1502:
464:
Henkin TM, Peters JE (2020). "DNA Repair and
Mutagenesis".
1211:
733:
This article incorporates text from the public domain
1970:
194:(Locus for X-ray sensitivity A) is a transcriptional
1815:
1794:
1722:
1667:
1540:
1398:
1362:
1331:
1300:
1249:
1111:
1088:
1054:
1004:
841:
796:
466:
Snyder and
Champness molecular genetics of bacteria
168:
148:
130:
125:
101:
89:
76:
64:
56:
51:
34:
632:"The SOS response controls integron recombination"
360:Butala M, Žgur-Bertok D, Busby SJ (January 2009).
362:"The bacterial LexA transcriptional repressor"
1518:
1227:
762:
459:
457:
275:DNA damage can be inflicted by the action of
8:
422:Erill I, Campoy S, Barbé J (November 2007).
355:
353:
351:
243:LexA polypeptides contains a two domains: a
320:determinants, as well as the activation of
263:of the protein. This domain is bound to an
1525:
1511:
1503:
1234:
1220:
1212:
769:
755:
747:
122:
40:
712:
702:
555:
514:
385:
1977:
347:
209:genes coding primarily for error-prone
782:serine proteases/serine endopeptidases
417:
415:
413:
31:
7:
366:Cellular and Molecular Life Sciences
1833:Proliferating Cell Nuclear Antigen
1764:Microhomology-mediated end joining
507:10.1002/j.1460-2075.1994.tb06709.x
25:
1980:
1955:
1954:
1848:Meiotic recombination checkpoint
557:10.1111/j.1365-2958.2005.04584.x
440:10.1111/j.1574-6976.2007.00082.x
259:, and is usually located at the
230:two-component regulatory system
1:
994:Urinary plasminogen activator
126:Available protein structures:
1730:Transcription-coupled repair
989:Tissue plasminogen activator
704:10.1371/journal.pbio.0030176
2039:
1752:Non-homologous end joining
1576:Nucleotide excision repair
1569:Poly ADP ribose polymerase
732:
1950:
1390:Michaelis–Menten kinetics
1080:Proteinase 3/Myeloblastin
428:FEMS Microbiology Reviews
378:10.1007/s00018-008-8378-6
121:
39:
1747:Homology directed repair
1669:Homologous recombination
1282:Diffusion-limited enzyme
656:10.1126/science.1172914
224:inhibitors. LexA forms
35:LexA DNA binding domain
1769:Postreplication repair
1564:Uracil-DNA glycosylase
1134:Proprotein convertases
544:Molecular Microbiology
1375:Eadie–Hofstee diagram
1308:Allosteric regulation
984:Plasminogen activator
318:antibiotic resistance
271:Clinical significance
1875:core protein complex
1550:Base excision repair
1385:Lineweaver–Burk plot
1124:Prolyl endopeptidase
1647:DNA mismatch repair
648:2009Sci...324.1034G
601:10.1038/nature02241
593:2004Natur.427...72B
332:combination therapy
291:. Bacteria require
27:Prokaryotic protein
2008:Bacterial proteins
1344:Enzyme superfamily
1277:Enzyme promiscuity
245:DNA-binding domain
1968:
1967:
1843:Adaptive response
1500:
1499:
1209:
1208:
1006:Complement system
798:Digestive enzymes
501:(17): 3936–3944.
314:virulence factors
205:) that represses
184:
183:
180:
179:
175:structure summary
46:lexa s119a mutant
16:(Redirected from
2030:
1985:
1984:
1976:
1958:
1957:
1527:
1520:
1513:
1504:
1380:Hanes–Woolf plot
1323:Enzyme activator
1318:Enzyme inhibitor
1292:Enzyme catalysis
1236:
1229:
1222:
1213:
771:
764:
757:
748:
727:
726:
716:
706:
682:
676:
675:
627:
621:
620:
576:
570:
569:
559:
535:
529:
528:
518:
495:The EMBO Journal
486:
480:
479:
461:
452:
451:
419:
408:
407:
389:
357:
301:topoisomerase IV
253:helix-turn-helix
123:
44:
32:
21:
2038:
2037:
2033:
2032:
2031:
2029:
2028:
2027:
2013:DNA replication
2003:Protein domains
1993:
1992:
1991:
1979:
1971:
1969:
1964:
1946:
1816:Other/ungrouped
1811:
1790:
1718:
1663:
1559:DNA glycosylase
1542:Excision repair
1536:
1531:
1501:
1496:
1408:Oxidoreductases
1394:
1370:Enzyme kinetics
1358:
1354:List of enzymes
1327:
1296:
1267:Catalytic triad
1245:
1240:
1210:
1205:
1107:
1084:
1050:
1000:
837:
806:Enteropeptidase
792:
775:
745:
731:
730:
684:
683:
679:
629:
628:
624:
587:(6969): 72–74.
578:
577:
573:
537:
536:
532:
488:
487:
483:
476:
463:
462:
455:
421:
420:
411:
359:
358:
349:
344:
305:DNA replication
273:
238:autoproteolysis
211:DNA polymerases
47:
28:
23:
22:
15:
12:
11:
5:
2036:
2034:
2026:
2025:
2020:
2015:
2010:
2005:
1995:
1994:
1990:
1989:
1966:
1965:
1963:
1962:
1951:
1948:
1947:
1945:
1944:
1939:
1934:
1929:
1924:
1919:
1918:
1917:
1912:
1907:
1902:
1897:
1892:
1887:
1882:
1867:
1866:
1861:
1851:
1850:
1845:
1840:
1835:
1830:
1825:
1819:
1817:
1813:
1812:
1810:
1809:
1804:
1798:
1796:
1792:
1791:
1789:
1788:
1787:
1786:
1781:
1771:
1766:
1761:
1760:
1759:
1749:
1744:
1743:
1742:
1737:
1726:
1724:
1723:Other pathways
1720:
1719:
1717:
1716:
1711:
1706:
1701:
1696:
1695:
1694:
1684:
1679:
1673:
1671:
1665:
1664:
1662:
1661:
1660:
1659:
1654:
1644:
1643:
1642:
1637:
1632:
1627:
1622:
1617:
1612:
1607:
1602:
1597:
1592:
1587:
1573:
1572:
1571:
1566:
1561:
1546:
1544:
1538:
1537:
1532:
1530:
1529:
1522:
1515:
1507:
1498:
1497:
1495:
1494:
1481:
1468:
1455:
1442:
1429:
1416:
1402:
1400:
1396:
1395:
1393:
1392:
1387:
1382:
1377:
1372:
1366:
1364:
1360:
1359:
1357:
1356:
1351:
1346:
1341:
1335:
1333:
1332:Classification
1329:
1328:
1326:
1325:
1320:
1315:
1310:
1304:
1302:
1298:
1297:
1295:
1294:
1289:
1284:
1279:
1274:
1269:
1264:
1259:
1253:
1251:
1247:
1246:
1241:
1239:
1238:
1231:
1224:
1216:
1207:
1206:
1204:
1203:
1202:
1201:
1196:
1186:
1181:
1172:
1158:
1153:
1148:
1147:
1146:
1141:
1131:
1126:
1121:
1115:
1113:
1109:
1108:
1106:
1105:
1100:
1094:
1092:
1086:
1085:
1083:
1082:
1077:
1072:
1067:
1061:
1059:
1052:
1051:
1049:
1048:
1043:
1042:
1041:
1036:
1026:
1021:
1016:
1010:
1008:
1002:
1001:
999:
998:
997:
996:
991:
981:
969:
968:
967:
966:
961:
956:
951:
946:
941:
936:
931:
926:
921:
916:
911:
906:
901:
896:
891:
881:
876:
871:
866:
861:
856:
847:
845:
839:
838:
836:
835:
834:
833:
828:
818:
813:
808:
802:
800:
794:
793:
778:Endopeptidases
776:
774:
773:
766:
759:
751:
729:
728:
677:
642:(5930): 1034.
622:
571:
550:(3): 836–844.
530:
481:
474:
453:
434:(6): 637–656.
409:
346:
345:
343:
340:
334:that combines
293:topoisomerases
281:bacteriophages
272:
269:
188:LexA repressor
182:
181:
178:
177:
172:
166:
165:
152:
146:
145:
135:
128:
127:
119:
118:
105:
99:
98:
93:
87:
86:
81:
74:
73:
68:
62:
61:
58:
54:
53:
49:
48:
45:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2035:
2024:
2021:
2019:
2016:
2014:
2011:
2009:
2006:
2004:
2001:
2000:
1998:
1988:
1983:
1978:
1974:
1961:
1953:
1952:
1949:
1943:
1940:
1938:
1935:
1933:
1930:
1928:
1925:
1923:
1920:
1916:
1913:
1911:
1908:
1906:
1903:
1901:
1898:
1896:
1893:
1891:
1888:
1886:
1883:
1881:
1878:
1877:
1876:
1872:
1871:FANC proteins
1869:
1868:
1865:
1862:
1860:
1856:
1853:
1852:
1849:
1846:
1844:
1841:
1839:
1836:
1834:
1831:
1829:
1826:
1824:
1821:
1820:
1818:
1814:
1808:
1805:
1803:
1800:
1799:
1797:
1793:
1785:
1782:
1780:
1777:
1776:
1775:
1772:
1770:
1767:
1765:
1762:
1758:
1755:
1754:
1753:
1750:
1748:
1745:
1741:
1738:
1736:
1733:
1732:
1731:
1728:
1727:
1725:
1721:
1715:
1712:
1710:
1707:
1705:
1702:
1700:
1697:
1693:
1692:RecQ helicase
1690:
1689:
1688:
1685:
1683:
1680:
1678:
1675:
1674:
1672:
1670:
1666:
1658:
1655:
1653:
1650:
1649:
1648:
1645:
1641:
1638:
1636:
1633:
1631:
1628:
1626:
1623:
1621:
1618:
1616:
1613:
1611:
1608:
1606:
1603:
1601:
1598:
1596:
1593:
1591:
1588:
1586:
1583:
1582:
1581:
1577:
1574:
1570:
1567:
1565:
1562:
1560:
1557:
1556:
1555:
1551:
1548:
1547:
1545:
1543:
1539:
1535:
1528:
1523:
1521:
1516:
1514:
1509:
1508:
1505:
1492:
1488:
1487:
1482:
1479:
1475:
1474:
1469:
1466:
1462:
1461:
1456:
1453:
1449:
1448:
1443:
1440:
1436:
1435:
1430:
1427:
1423:
1422:
1417:
1414:
1410:
1409:
1404:
1403:
1401:
1397:
1391:
1388:
1386:
1383:
1381:
1378:
1376:
1373:
1371:
1368:
1367:
1365:
1361:
1355:
1352:
1350:
1349:Enzyme family
1347:
1345:
1342:
1340:
1337:
1336:
1334:
1330:
1324:
1321:
1319:
1316:
1314:
1313:Cooperativity
1311:
1309:
1306:
1305:
1303:
1299:
1293:
1290:
1288:
1285:
1283:
1280:
1278:
1275:
1273:
1272:Oxyanion hole
1270:
1268:
1265:
1263:
1260:
1258:
1255:
1254:
1252:
1248:
1244:
1237:
1232:
1230:
1225:
1223:
1218:
1217:
1214:
1200:
1197:
1195:
1192:
1191:
1190:
1187:
1185:
1184:Streptokinase
1182:
1180:
1176:
1173:
1170:
1166:
1162:
1159:
1157:
1154:
1152:
1149:
1145:
1142:
1140:
1137:
1136:
1135:
1132:
1130:
1127:
1125:
1122:
1120:
1117:
1116:
1114:
1110:
1104:
1101:
1099:
1096:
1095:
1093:
1091:
1087:
1081:
1078:
1076:
1073:
1071:
1068:
1066:
1063:
1062:
1060:
1058:
1057:immune system
1053:
1047:
1046:C3-convertase
1044:
1040:
1037:
1035:
1032:
1031:
1030:
1027:
1025:
1022:
1020:
1017:
1015:
1012:
1011:
1009:
1007:
1003:
995:
992:
990:
987:
986:
985:
982:
980:
977:
975:
971:
970:
965:
962:
960:
957:
955:
952:
950:
947:
945:
942:
940:
937:
935:
932:
930:
927:
925:
922:
920:
917:
915:
912:
910:
907:
905:
902:
900:
897:
895:
892:
890:
887:
886:
885:
882:
880:
877:
875:
872:
870:
867:
865:
862:
860:
857:
855:
852:
849:
848:
846:
844:
840:
832:
829:
827:
824:
823:
822:
819:
817:
814:
812:
809:
807:
804:
803:
801:
799:
795:
790:
787:
783:
779:
772:
767:
765:
760:
758:
753:
752:
749:
744:
740:
736:
724:
720:
715:
710:
705:
700:
696:
692:
688:
681:
678:
673:
669:
665:
661:
657:
653:
649:
645:
641:
637:
633:
626:
623:
618:
614:
610:
606:
602:
598:
594:
590:
586:
582:
575:
572:
567:
563:
558:
553:
549:
545:
541:
534:
531:
526:
522:
517:
512:
508:
504:
500:
496:
492:
485:
482:
477:
475:9781555819750
471:
467:
460:
458:
454:
449:
445:
441:
437:
433:
429:
425:
418:
416:
414:
410:
405:
401:
397:
393:
388:
383:
379:
375:
371:
367:
363:
356:
354:
352:
348:
341:
339:
337:
333:
328:
326:
323:
319:
315:
310:
306:
302:
298:
294:
290:
289:ciprofloxacin
286:
282:
278:
270:
268:
266:
262:
258:
254:
250:
246:
241:
239:
235:
231:
227:
223:
222:cell division
219:
216:
212:
208:
204:
201:
197:
193:
189:
176:
173:
171:
167:
164:
160:
156:
153:
151:
147:
143:
139:
136:
133:
129:
124:
120:
117:
113:
109:
106:
104:
100:
97:
94:
92:
88:
85:
82:
79:
75:
72:
69:
67:
63:
60:LexA_DNA_bind
59:
55:
50:
43:
38:
33:
30:
19:
1874:
1855:DNA helicase
1838:8-Oxoguanine
1807:SOS response
1687:RecF pathway
1640:Excinuclease
1486:Translocases
1483:
1470:
1457:
1444:
1431:
1421:Transferases
1418:
1405:
1262:Binding site
974:fibrinolysis
972:
850:
816:Chymotrypsin
694:
691:PLOS Biology
690:
680:
639:
635:
625:
584:
580:
574:
547:
543:
533:
498:
494:
484:
465:
431:
427:
372:(1): 82–93.
369:
365:
329:
274:
255:DNA binding
249:dimerization
242:
225:
207:SOS response
191:
187:
185:
29:
1257:Active site
879:Factor XIIa
859:Factor VIIa
843:Coagulation
697:(6): e176.
277:antibiotics
52:Identifiers
2023:DNA repair
1997:Categories
1795:Regulation
1774:Photolyase
1534:DNA repair
1460:Isomerases
1434:Hydrolases
1301:Regulation
1161:Subtilisin
1103:Batroxobin
884:Kallikrein
874:Factor XIa
864:Factor IXa
831:Pancreatic
826:Neutrophil
342:References
336:quinolones
325:integrases
297:DNA gyrase
261:N-terminus
215:DNA repair
138:structures
2018:EC 3.4.21
1615:XPG/ERCC5
1600:XPD/ERCC2
1339:EC number
1189:Cathepsin
1175:Sedolisin
1151:Prostasin
869:Factor Xa
743:IPR006199
309:resistant
203:3.4.21.88
196:repressor
96:IPR006199
1960:Category
1610:XPF/DDB1
1605:XPE/DDB1
1363:Kinetics
1287:Cofactor
1250:Activity
1090:Venombin
1075:Tryptase
1070:Granzyme
1024:Factor I
1019:Factor D
1014:Factor B
854:Thrombin
851:factors:
821:Elastase
739:InterPro
723:15869329
672:42334786
664:19460999
609:14688795
566:15819636
448:17883408
404:29537019
396:18726173
387:11131485
322:integron
295:such as
285:UV light
226:de facto
155:RCSB PDB
91:InterPro
1987:Biology
1802:SOS box
1554:AP site
1473:Ligases
1243:Enzymes
1129:Pronase
1119:Acrosin
1065:Chymase
979:Plasmin
811:Trypsin
714:1088971
644:Bibcode
636:Science
617:4300746
589:Bibcode
525:8076591
265:SOS box
218:enzymes
71:PF01726
1973:Portal
1927:FANCD2
1922:FANCD1
1682:RecBCD
1635:RAD23B
1630:RAD23A
1447:Lyases
1156:Reelin
1098:Ancrod
1055:Other
789:3.4.21
721:
711:
670:
662:
615:
607:
581:Nature
564:
523:
516:395313
513:
472:
446:
402:
394:
384:
283:, and
247:and a
170:PDBsum
144:
134:
116:SUPFAM
84:CL0123
57:Symbol
1942:FANCN
1937:FANCJ
1932:FANCI
1915:FANCM
1910:FANCL
1905:FANCG
1900:FANCF
1895:FANCE
1890:FANCC
1885:FANCB
1880:FANCA
1740:ERCC8
1735:ERCC6
1699:RAD51
1620:ERCC1
1399:Types
1165:Furin
1112:Other
1039:MASP2
1034:MASP1
964:KLK15
959:KLK14
954:KLK13
949:KLK12
944:KLK11
939:KLK10
668:S2CID
613:S2CID
400:S2CID
257:motif
232:with
112:SCOPe
103:SCOP2
1828:PcrA
1784:CRY2
1779:CRY1
1714:LexA
1709:Slx4
1704:Sgs1
1677:RecA
1657:MSH2
1652:MLH1
1580:ERCC
1491:list
1484:EC7
1478:list
1471:EC6
1465:list
1458:EC5
1452:list
1445:EC4
1439:list
1432:EC3
1426:list
1419:EC2
1413:list
1406:EC1
1179:TPP1
1029:MASP
934:KLK9
929:KLK8
924:KLK7
919:KLK6
914:KLK5
909:KLK4
904:KLK3
899:KLK2
894:KLK1
737:and
735:Pfam
719:PMID
660:PMID
605:PMID
562:PMID
521:PMID
470:ISBN
444:PMID
392:PMID
316:and
303:for
234:RecA
220:and
192:LexA
186:The
163:PDBj
159:PDBe
142:ECOD
132:Pfam
108:1leb
80:clan
78:Pfam
66:Pfam
18:LexA
1864:WRN
1859:BLM
1823:Ogt
1625:RPA
1595:XPC
1590:XPB
1585:XPA
1169:S1P
889:PSA
709:PMC
699:doi
652:doi
640:324
597:doi
585:427
552:doi
511:PMC
503:doi
436:doi
382:PMC
374:doi
299:or
190:or
150:PDB
1999::
1873::
1857::
1757:Ku
786:EC
780::
741::
717:.
707:.
693:.
689:.
666:.
658:.
650:.
638:.
634:.
611:.
603:.
595:.
583:.
560:.
548:56
546:.
542:.
519:.
509:.
499:13
497:.
493:.
456:^
442:.
432:31
430:.
426:.
412:^
398:.
390:.
380:.
370:66
368:.
364:.
350:^
279:,
240:.
228:a
213:,
200:EC
161:;
157:;
140:/
114:/
110:/
1975::
1578:/
1552:/
1526:e
1519:t
1512:v
1493:)
1489:(
1480:)
1476:(
1467:)
1463:(
1454:)
1450:(
1441:)
1437:(
1428:)
1424:(
1415:)
1411:(
1235:e
1228:t
1221:v
1199:G
1194:A
1177:/
1171:4
1167:/
1163:/
1144:2
1139:1
976::
791:)
784:(
770:e
763:t
756:v
725:.
701::
695:3
674:.
654::
646::
619:.
599::
591::
568:.
554::
527:.
505::
478:.
450:.
438::
406:.
376::
198:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.