332:
822:
32:
1603:
20:
27:
addition to being a LexA protease, the RecA protein also catalyzes a few novel DNA reactions such as annealing of single-stranded DNA and transfer of strands. The SOS system has enhanced DNA-repair capacity, including excision and post-replication repair, enhanced mutagenesis and prophage induction. The system can also inhibit cell division and cell respiration.
224:
115:) in the operator region for those genes. Some of these SOS genes are expressed at certain levels even in the repressed state, according to the affinity of LexA for their SOS box. Activation of the SOS genes occurs after DNA damage by the accumulation of single stranded (ssDNA) regions generated at replication forks, where
123:
Once the pool of LexA decreases, repression of the SOS genes goes down according to the level of LexA affinity for the SOS boxes. Operators that bind LexA weakly are the first to be fully expressed. In this way LexA can sequentially activate different mechanisms of repair. Genes having a weak SOS box
26:
SOS System: DNA can be damaged by cross-linking agents, UV irradiation, alkylating agents, etc. Once damaged, RecA, a LexA protease, senses that damaged DNA and becomes activated by removing its repressor. Once the LexA dimer repressor is removed, the expression of LexA operon is autoregulatory. In
172:, and the induction of UmuDC-dependent mutagenic repair. As a result of these properties, some genes may be partially induced in response to even endogenous levels of DNA damage, while other genes appear to be induced only when high or persistent DNA damage is present in the cell.
200:. Researchers are now targeting these proteins with the aim of creating drugs that prevent SOS repair. By doing so, the time needed for pathogenic bacteria to evolve antibiotic resistance could be extended, thus improving the long term viability of some antibiotic drugs.
331:
148:(NER), whose aim is to fix DNA damage without commitment to a full-fledged SOS response. If, however, NER does not suffice to fix the damage, the LexA concentration is further reduced, so the expression of genes with stronger LexA boxes (such as
119:
is blocked. RecA forms a filament around these ssDNA regions in an ATP-dependent fashion, and becomes activated. The activated form of RecA interacts with the LexA repressor to facilitate the LexA repressor's self-cleavage from the operator.
262:
mutation, which renders the bacteria lipopolysaccharide-deficient, allowing better diffusion of certain chemicals into the cell in order to induce the SOS response. Commercial kits which measures the primary response of the
203:
As well as genetic resistance the SOS response can also promote phenotypic resistance. Here, the genome is preserved whilst other non-genetic factors are altered to enable the bacteria to survive. The SOS dependent
243:
is possible. A lactose analog is added to the bacteria, which is then degraded by beta-galactosidase, thereby producing a colored compound which can be measured quantitatively through
1013:
Quillardet, Philippe; de
Bellecombe, Christine; Hofnung, Maurice (June 1985). "The SOS Chromotest, a colorimetric bacterial assay for genotoxins: validation study with 83 compounds".
99:
detailed the SOS response to UV radiation in bacteria. The SOS response to DNA damage was a seminal discovery because it was the first coordinated stress response to be elucidated.
79:) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
239:(responsible for producing beta-galactosidase, a protein which degrades lactose) under the control of an SOS-related protein, a simple colorimetric assay for
247:. The degree of color development is an indirect measure of the beta-galactosidase produced, which itself is directly related to the amount of DNA damage.
884:
Lee, AM; Ross, CT; Zeng, BB; Singleton, SF (July 2005). "A molecular target for suppression of the evolution of antibiotic resistance: Inhibition of the
1536:
1510:
235:, different classes of DNA-damaging agents can initiate the SOS response, as described above. Taking advantage of an operon fusion placing the
750:
1171:
1226:
1479:
1410:
258:
mutation which renders the strain deficient in excision repair, increasing the response to certain DNA-damaging agents, as well as an
1494:
205:
837:"Mutations for Worse or Better: Low Fidelity DNA Synthesis by SOS DNA Polymerase V is a Tightly-Regulated Double-Edged Sword"
144:) are fully induced in response to even weak SOS-inducing treatments. Thus the first SOS repair mechanism to be induced is
1469:
1376:
75:
in eukaryotes). The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (
1398:
1222:
1215:
145:
527:"The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction"
1050:"The role of biology in planetary evolution: cyanobacterial primary production in low‐oxygen Proterozoic oceans"
1627:
1393:
1315:
1164:
36:
821:
1505:
1415:
1210:
208:
1271:
181:
40:
184:
to antibiotics. The increased rate of mutation during the SOS response is caused by three low-fidelity
31:
1099:"Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications"
835:
Jaszczur, M; Bertram, JG; Robinson, A; van Oijen, AM; Woodgate, R; Cox, MM; Goodman, MF (April 2016).
1196:
697:
538:
337:
The SOS response inhibits septum formation until bacterial DNA can be repaired and is observable as
1632:
1606:
1293:
1157:
397:
1489:
1130:
1079:
1030:
995:
960:
905:
866:
810:
756:
746:
723:
657:
618:
566:
500:
448:
389:
372:
Little, John W.; Mount, David W. (May 1982). "The SOS regulatory system of
Escherichia coli".
244:
1120:
1110:
1069:
1061:
1022:
987:
950:
940:
897:
856:
848:
800:
790:
713:
705:
649:
608:
600:
584:
556:
546:
490:
486:
482:
438:
428:
381:
1205:
470:
292:
212:
96:
777:
Cirz, RT; Chin, JK; Andes, DR; De Crécy-Lagard, V; Craig, WA; Romesberg, FE (June 2005).
701:
542:
180:
Research has shown that the SOS response system can lead to mutations which can lead to
1125:
1098:
1074:
1049:
978:
Quillardet, Philippe; Hofnung, Maurice (October 1993). "The SOS chromotest: a review".
955:
924:
861:
836:
805:
778:
718:
685:
495:
443:
416:
312:
287:
185:
116:
111:
protein dimers. Under normal conditions, LexA binds to a 20-bp consensus sequence (the
108:
613:
588:
561:
526:
1621:
1517:
1338:
1026:
991:
385:
338:
298:
279:
169:
161:
88:
401:
1484:
1403:
1386:
1333:
1286:
604:
352:
290:, are major producers of the Earth’s oxygenic atmosphere. The marine cyanobacteria
240:
19:
945:
795:
653:
636:
Radman, M (1975). "Phenomenology of an inducible mutagenic DNA repair pathway in
433:
1048:
Hamilton, Trinity L.; Bryant, Donald A.; Macalady, Jennifer L. (February 2016).
852:
64:
223:
1420:
1241:
1188:
1180:
283:
236:
60:
56:
1115:
779:"Inhibition of mutation and combating the evolution of antibiotic resistance"
306:
like SOS system for repair of DNA, since they encode genes homologous to key
1097:
Cassier-Chauvat, Corinne; Veaudor, Théo; Chauvat, Franck (9 November 2016).
1065:
760:
268:
1134:
1083:
964:
909:
870:
814:
727:
551:
504:
452:
267:
cell to genetic damage are available and may be highly correlated with the
1034:
999:
661:
570:
469:
Fitzgerald, Devon M.; Hastings, P.J.; Rosenberg, Susan M. (6 March 2017).
393:
1501:
622:
925:"Ciprofloxacin Causes Persister Formation by Inducing the TisB toxin in
686:"The SOS system: A complex and tightly regulated response to DNA damage"
471:"Stress-Induced Mutagenesis: Implications in Cancer and Drug Resistance"
417:"After 30 Years of Study, the Bacterial SOS Response Still Surprises Us"
254:
are further modified in order to have a number of mutations including a
1448:
1200:
684:
Maslowska, K. H.; Makiela-Dzbenska, K.; Fijalkowska, I. J. (May 2019).
112:
901:
589:"Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli"
1573:
1328:
1281:
1276:
709:
193:
189:
1588:
1583:
1578:
1568:
1561:
1556:
1551:
1546:
1541:
1531:
1526:
1381:
1345:
1266:
1261:
1256:
1246:
222:
197:
72:
30:
18:
741:
Lehninger, Albert L.; Nelson, David Lee; Cox, Michael M. (2005).
227:
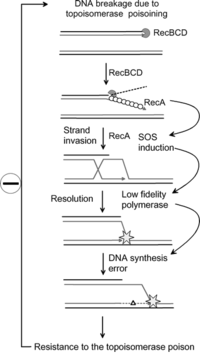
Overview of the use of the SOS response for genotoxicity testing.
1474:
1430:
1425:
1360:
1355:
1350:
1323:
1303:
1298:
1251:
1015:
Mutation
Research/Environmental Mutagenesis and Related Subjects
165:
107:
During normal growth, the SOS genes are negatively regulated by
76:
68:
1153:
1236:
1231:
52:
1149:
341:
when cells are examined by microscopy (top right of image).
91:. Later, by characterizing the phenotypes of mutagenised
745:(4th ed.). New York: W.H. Freeman. p. 1098.
211:
has, for example, been linked to DNA damage-dependent
168:, the initiating protein in this process. This causes
1462:
1441:
1369:
1314:
1187:
160:– these are expressed late) is induced. SulA stops
35:The SOS response has been proposed as a model for
980:Mutation Research/Reviews in Genetic Toxicology
531:Proceedings of the National Academy of Sciences
1165:
923:Dörr, T; Vulić, M; Lewis, K (February 2010).
8:
464:
462:
679:
677:
675:
673:
671:
1172:
1158:
1150:
1124:
1114:
1073:
954:
944:
860:
804:
794:
717:
612:
560:
550:
494:
442:
432:
690:Environmental and Molecular Mutagenesis
487:10.1146/annurev-cancerbio-050216-121919
364:
327:
888:RecA Protein by N6-(1-Naphthyl)-ADP".
772:
770:
67:are induced. The system involves the
7:
743:Lehninger principles of biochemistry
87:The SOS response was articulated by
1480:Proliferating Cell Nuclear Antigen
1411:Microhomology-mediated end joining
415:Michel, Bénédicte (12 July 2005).
353:Induction of lysis in lambda phage
14:
1602:
1601:
1495:Meiotic recombination checkpoint
820:
330:
95:, she and post doctoral student
475:Annual Review of Cancer Biology
890:Journal of Medicinal Chemistry
605:10.1128/MMBR.40.4.869-907.1976
1:
1377:Transcription-coupled repair
1027:10.1016/0165-1161(85)90021-4
992:10.1016/0165-1110(93)90019-J
946:10.1371/journal.pbio.1000317
796:10.1371/journal.pbio.0030176
654:10.1007/978-1-4684-2895-7_48
434:10.1371/journal.pbio.0030255
386:10.1016/0092-8674(82)90085-X
853:10.1021/acs.biochem.6b00117
286:capable of oxygen evolving
16:Cell response to DNA damage
1649:
1399:Non-homologous end joining
1223:Nucleotide excision repair
1216:Poly ADP ribose polymerase
1054:Environmental Microbiology
640:: SOS repair hypothesis".
146:nucleotide excision repair
1597:
1103:Frontiers in Microbiology
1394:Homology directed repair
1316:Homologous recombination
1116:10.3389/fmicb.2016.01809
525:Witkin, E M (May 1967).
51:is a global response to
1066:10.1111/1462-2920.13118
593:Bacteriological Reviews
271:for certain materials.
1416:Postreplication repair
1211:Uracil-DNA glycosylase
552:10.1073/pnas.57.5.1275
228:
209:toxin-antitoxin system
44:
28:
226:
176:Antibiotic resistance
41:antibiotic resistance
34:
22:
1522:core protein complex
1197:Base excision repair
219:Genotoxicity testing
55:damage in which the
39:of certain types of
1294:DNA mismatch repair
702:2019EnvMM..60..368M
642:Basic Life Sciences
543:1967PNAS...57.1275W
37:bacterial evolution
310:SOS genes such as
302:appear to have an
229:
45:
29:
1615:
1614:
1490:Adaptive response
902:10.1021/jm050113z
896:(17): 5408–5411.
752:978-0-7167-4339-2
324:Additional images
245:spectrophotometry
1640:
1605:
1604:
1174:
1167:
1160:
1151:
1139:
1138:
1128:
1118:
1094:
1088:
1087:
1077:
1045:
1039:
1038:
1010:
1004:
1003:
975:
969:
968:
958:
948:
927:Escherichia coli
920:
914:
913:
886:Escherichia coli
881:
875:
874:
864:
832:
826:
825:
824:
818:
808:
798:
774:
765:
764:
738:
732:
731:
721:
710:10.1002/em.22267
681:
666:
665:
638:Escherichia coli
633:
627:
626:
616:
581:
575:
574:
564:
554:
522:
516:
515:
513:
511:
498:
466:
457:
456:
446:
436:
412:
406:
405:
369:
334:
233:Escherichia coli
59:is arrested and
1648:
1647:
1643:
1642:
1641:
1639:
1638:
1637:
1628:1975 in science
1618:
1617:
1616:
1611:
1593:
1463:Other/ungrouped
1458:
1437:
1365:
1310:
1206:DNA glycosylase
1189:Excision repair
1183:
1178:
1148:
1143:
1142:
1096:
1095:
1091:
1047:
1046:
1042:
1012:
1011:
1007:
977:
976:
972:
939:(2): e1000317.
922:
921:
917:
883:
882:
878:
847:(16): 2309–18.
834:
833:
829:
819:
776:
775:
768:
753:
740:
739:
735:
683:
682:
669:
635:
634:
630:
583:
582:
578:
524:
523:
519:
509:
507:
468:
467:
460:
414:
413:
409:
371:
370:
366:
361:
349:
342:
335:
326:
293:Prochlorococcus
277:
221:
186:DNA polymerases
178:
105:
97:Miroslav Radman
85:
17:
12:
11:
5:
1646:
1644:
1636:
1635:
1630:
1620:
1619:
1613:
1612:
1610:
1609:
1598:
1595:
1594:
1592:
1591:
1586:
1581:
1576:
1571:
1566:
1565:
1564:
1559:
1554:
1549:
1544:
1539:
1534:
1529:
1514:
1513:
1508:
1498:
1497:
1492:
1487:
1482:
1477:
1472:
1466:
1464:
1460:
1459:
1457:
1456:
1451:
1445:
1443:
1439:
1438:
1436:
1435:
1434:
1433:
1428:
1418:
1413:
1408:
1407:
1406:
1396:
1391:
1390:
1389:
1384:
1373:
1371:
1370:Other pathways
1367:
1366:
1364:
1363:
1358:
1353:
1348:
1343:
1342:
1341:
1331:
1326:
1320:
1318:
1312:
1311:
1309:
1308:
1307:
1306:
1301:
1291:
1290:
1289:
1284:
1279:
1274:
1269:
1264:
1259:
1254:
1249:
1244:
1239:
1234:
1220:
1219:
1218:
1213:
1208:
1193:
1191:
1185:
1184:
1179:
1177:
1176:
1169:
1162:
1154:
1147:
1146:External links
1144:
1141:
1140:
1089:
1060:(2): 325–340.
1040:
1005:
986:(3): 235–279.
970:
915:
876:
827:
766:
751:
733:
696:(4): 368–384.
667:
628:
599:(4): 869–907.
576:
517:
481:(1): 119–140.
458:
407:
363:
362:
360:
357:
356:
355:
348:
345:
344:
343:
336:
329:
325:
322:
288:photosynthesis
276:
273:
220:
217:
213:persister cell
177:
174:
164:by binding to
117:DNA polymerase
109:LexA repressor
104:
101:
84:
81:
15:
13:
10:
9:
6:
4:
3:
2:
1645:
1634:
1631:
1629:
1626:
1625:
1623:
1608:
1600:
1599:
1596:
1590:
1587:
1585:
1582:
1580:
1577:
1575:
1572:
1570:
1567:
1563:
1560:
1558:
1555:
1553:
1550:
1548:
1545:
1543:
1540:
1538:
1535:
1533:
1530:
1528:
1525:
1524:
1523:
1519:
1518:FANC proteins
1516:
1515:
1512:
1509:
1507:
1503:
1500:
1499:
1496:
1493:
1491:
1488:
1486:
1483:
1481:
1478:
1476:
1473:
1471:
1468:
1467:
1465:
1461:
1455:
1452:
1450:
1447:
1446:
1444:
1440:
1432:
1429:
1427:
1424:
1423:
1422:
1419:
1417:
1414:
1412:
1409:
1405:
1402:
1401:
1400:
1397:
1395:
1392:
1388:
1385:
1383:
1380:
1379:
1378:
1375:
1374:
1372:
1368:
1362:
1359:
1357:
1354:
1352:
1349:
1347:
1344:
1340:
1339:RecQ helicase
1337:
1336:
1335:
1332:
1330:
1327:
1325:
1322:
1321:
1319:
1317:
1313:
1305:
1302:
1300:
1297:
1296:
1295:
1292:
1288:
1285:
1283:
1280:
1278:
1275:
1273:
1270:
1268:
1265:
1263:
1260:
1258:
1255:
1253:
1250:
1248:
1245:
1243:
1240:
1238:
1235:
1233:
1230:
1229:
1228:
1224:
1221:
1217:
1214:
1212:
1209:
1207:
1204:
1203:
1202:
1198:
1195:
1194:
1192:
1190:
1186:
1182:
1175:
1170:
1168:
1163:
1161:
1156:
1155:
1152:
1145:
1136:
1132:
1127:
1122:
1117:
1112:
1108:
1104:
1100:
1093:
1090:
1085:
1081:
1076:
1071:
1067:
1063:
1059:
1055:
1051:
1044:
1041:
1036:
1032:
1028:
1024:
1020:
1016:
1009:
1006:
1001:
997:
993:
989:
985:
981:
974:
971:
966:
962:
957:
952:
947:
942:
938:
934:
930:
928:
919:
916:
911:
907:
903:
899:
895:
891:
887:
880:
877:
872:
868:
863:
858:
854:
850:
846:
842:
838:
831:
828:
823:
816:
812:
807:
802:
797:
792:
788:
784:
780:
773:
771:
767:
762:
758:
754:
748:
744:
737:
734:
729:
725:
720:
715:
711:
707:
703:
699:
695:
691:
687:
680:
678:
676:
674:
672:
668:
663:
659:
655:
651:
647:
643:
639:
632:
629:
624:
620:
615:
610:
606:
602:
598:
594:
590:
586:
585:Witkin, E. M.
580:
577:
572:
568:
563:
558:
553:
548:
544:
540:
537:(5): 1275–9.
536:
532:
528:
521:
518:
506:
502:
497:
492:
488:
484:
480:
476:
472:
465:
463:
459:
454:
450:
445:
440:
435:
430:
426:
422:
418:
411:
408:
403:
399:
395:
391:
387:
383:
379:
375:
368:
365:
358:
354:
351:
350:
346:
340:
339:filamentation
333:
328:
323:
321:
319:
315:
314:
309:
305:
301:
300:
299:Synechococcus
295:
294:
289:
285:
281:
280:Cyanobacteria
275:Cyanobacteria
274:
272:
270:
266:
261:
257:
253:
248:
246:
242:
238:
234:
225:
218:
216:
214:
210:
207:
201:
199:
195:
191:
187:
183:
175:
173:
171:
170:filamentation
167:
163:
162:cell division
159:
155:
151:
147:
143:
139:
135:
131:
127:
121:
118:
114:
110:
102:
100:
98:
94:
90:
89:Evelyn Witkin
82:
80:
78:
74:
70:
66:
62:
58:
54:
50:
42:
38:
33:
25:
21:
1521:
1502:DNA helicase
1485:8-Oxoguanine
1454:SOS response
1453:
1334:RecF pathway
1287:Excinuclease
1106:
1102:
1092:
1057:
1053:
1043:
1021:(3): 79–95.
1018:
1014:
1008:
983:
979:
973:
936:
933:PLOS Biology
932:
926:
918:
893:
889:
885:
879:
844:
841:Biochemistry
840:
830:
786:
783:PLOS Biology
782:
742:
736:
693:
689:
645:
641:
637:
631:
596:
592:
579:
534:
530:
520:
508:. Retrieved
478:
474:
424:
421:PLoS Biology
420:
410:
380:(1): 11–22.
377:
373:
367:
317:
311:
307:
303:
297:
291:
278:
264:
259:
255:
251:
249:
241:genotoxicity
232:
230:
202:
179:
157:
153:
149:
141:
137:
133:
129:
125:
122:
106:
92:
86:
49:SOS response
48:
46:
23:
789:(6): e176.
648:: 355–367.
427:(7): e255.
284:prokaryotes
282:, the only
215:induction.
65:mutagenesis
1633:DNA repair
1622:Categories
1442:Regulation
1421:Photolyase
1181:DNA repair
359:References
237:lac operon
182:resistance
61:DNA repair
57:cell cycle
1262:XPG/ERCC5
1247:XPD/ERCC2
269:Ames Test
206:tisB-istR
124:(such as
103:Mechanism
83:Discovery
71:protein (
1607:Category
1257:XPF/DDB1
1252:XPE/DDB1
1135:27881980
1084:26549614
965:20186264
910:16107138
871:27043933
815:15869329
761:55476414
728:30447030
587:(1976).
505:29399660
453:16000023
402:12476812
347:See also
1449:SOS box
1201:AP site
1126:5101192
1075:5019231
1035:3923333
1000:7692273
956:2826370
862:4846499
806:1088971
719:6590174
698:Bibcode
662:1103845
571:5341236
539:Bibcode
510:6 March
496:5794033
444:1174825
394:7049397
308:E. coli
304:E. coli
265:E. coli
252:E. coli
113:SOS box
93:E. coli
24:E. coli
1574:FANCD2
1569:FANCD1
1329:RecBCD
1282:RAD23B
1277:RAD23A
1133:
1123:
1082:
1072:
1033:
998:
963:
953:
908:
869:
859:
813:
803:
759:
749:
726:
716:
660:
623:795416
621:
614:413988
611:
569:
562:224468
559:
503:
493:
451:
441:
400:
392:
194:Pol IV
190:Pol II
140:, and
1589:FANCN
1584:FANCJ
1579:FANCI
1562:FANCM
1557:FANCL
1552:FANCG
1547:FANCF
1542:FANCE
1537:FANCC
1532:FANCB
1527:FANCA
1387:ERCC8
1382:ERCC6
1346:RAD51
1267:ERCC1
398:S2CID
198:Pol V
73:Rad51
1475:PcrA
1431:CRY2
1426:CRY1
1361:LexA
1356:Slx4
1351:Sgs1
1324:RecA
1304:MSH2
1299:MLH1
1227:ERCC
1131:PMID
1080:PMID
1031:PMID
996:PMID
961:PMID
906:PMID
867:PMID
811:PMID
757:OCLC
747:ISBN
724:PMID
658:PMID
619:PMID
567:PMID
512:2023
501:PMID
449:PMID
390:PMID
374:Cell
318:sulA
316:and
313:lexA
296:and
256:uvrA
250:The
196:and
166:FtsZ
158:umuC
154:umuD
150:sulA
142:uvrD
138:uvrB
134:uvrA
130:recA
126:lexA
77:LexA
69:RecA
63:and
47:The
1511:WRN
1506:BLM
1470:Ogt
1272:RPA
1242:XPC
1237:XPB
1232:XPA
1121:PMC
1111:doi
1070:PMC
1062:doi
1023:doi
1019:147
988:doi
984:297
951:PMC
941:doi
898:doi
857:PMC
849:doi
801:PMC
791:doi
714:PMC
706:doi
650:doi
609:PMC
601:doi
557:PMC
547:doi
491:PMC
483:doi
439:PMC
429:doi
382:doi
260:rfa
231:In
53:DNA
1624::
1520::
1504::
1404:Ku
1129:.
1119:.
1109:.
1105:.
1101:.
1078:.
1068:.
1058:18
1056:.
1052:.
1029:.
1017:.
994:.
982:.
959:.
949:.
935:.
931:.
904:.
894:48
892:.
865:.
855:.
845:55
843:.
839:.
809:.
799:.
785:.
781:.
769:^
755:.
722:.
712:.
704:.
694:60
692:.
688:.
670:^
656:.
646:5A
644:.
617:.
607:.
597:40
595:.
591:.
565:.
555:.
545:.
535:57
533:.
529:.
499:.
489:.
477:.
473:.
461:^
447:.
437:.
423:.
419:.
396:.
388:.
378:29
376:.
320:.
192:,
188::
156:,
152:,
136:,
132:,
128:,
1225:/
1199:/
1173:e
1166:t
1159:v
1137:.
1113::
1107:7
1086:.
1064::
1037:.
1025::
1002:.
990::
967:.
943::
937:8
929:"
912:.
900::
873:.
851::
817:.
793::
787:3
763:.
730:.
708::
700::
664:.
652::
625:.
603::
573:.
549::
541::
514:.
485::
479:1
455:.
431::
425:3
404:.
384::
43:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.