2109:
514:
plants. Pectinesterase isoforms are encoded by a family of genes, some of which are constitutively expressed throughout the plant, whereas others are differentially expressed in specific tissues and at different developmental stages. Isoforms of pectinesterase differ in various biochemical parameters
489:
Pectinesterase is thought to be secreted to the apoplasm with highly methylated pectin although at some point along this secretory pathway the N-terminal pro-peptide is cleaved off. Currently, the role of the pro-region is unknown although it has been hypothesised that it may act as an intramolecular
493:
Recently, particular attention has been devoted to molecular studies of pectinesterase leading to the characterisation of several related isoforms in various higher plant species. Some of these pectinesterases were shown to be ubiquitously expressed, whereas others are specifically expressed during
412:
interactions. Pectinesterases can however display acidic isoelectric points as detected in soluble fractions of plant tissues. Until recently, it was generally assumed that plant pectinesterases remove methyl esters in a progressive block-wise fashion, giving rise to long contiguous stretches of
397:, methylesterified in the medial Golgi and substituted with side chains in the trans Golgi cisternae. Pectin biochemistry can be rather complicated but put simply, the pectin backbone comprises 3 types of polymer: homogalacturonan (HGA); rhamnogalacturonan I (RGI); rhamnogalacturonan II (RGII).
29:
743:
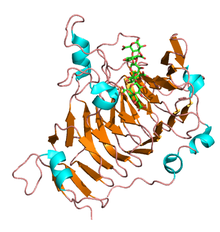
Prokaryotic and eukaryotic pectinesterases share a few regions of sequence similarity. The crystal structure of pectinesterase from
Erwinia chrysanthemi revealed a beta-helix structure similar to that found in pectinolytic enzymes, though it is different from most structures of esterases. The
400:
Homogalacturonan is highly methyl-esterified when exported into cell walls and is subsequently de-esterified by the action of pectinesterase and other pectic enzymes. Pectinesterase catalyses the de-esterification of methyl-esterified D-galactosiduronic acid units in pectic compounds yielding
494:
fruit ripening, germination of the pollen grain, or stem elongation. Such data suggests that pectinesterases are encoded by a family of genes that are differentially regulated in cell type in response to specific developmental or environmental cues.
417:. Alternatively it was thought that fungal pectinesterases had a random activity resulting in the de-esterification of single GalA residues per enzyme/substrate interactions. It has now been shown that some plant pectinesterase
365:
Pectinesterase action on the components of the plant cell wall can produce two diametrically opposite effects. The first being a contribution to the stiffening of the cell wall by producing blocks of unesterified
308:. Pectin is one of the main components of the plant cell wall. In plants, pectinesterase plays an important role in cell wall metabolism during fruit ripening. In plant bacterial pathogens such as
320:, pectinesterase is involved in maceration and soft-rotting of plant tissue. Plant pectinesterases are regulated by pectinesterase inhibitors, which are ineffective against microbial enzymes.
920:
Pickersgill RW, Smith D, Jenkins J, Mayans O, Worboys K (2001). "Three-dimensional structure of
Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site".
717:
catalytic region is highly conserved and constitutes the mature enzyme. The first three-dimensional structure solved for a plant pectinesterase was for an isoform from
460:
Spatial and temporal regulation of pectinesterase activity during plant development is based on a large family of isoforms. Recently, the systematic sequencing of the
612:
206:
225:
1183:
332:
can influence numerous physiological processes. In plants, pectinesterase plays a role in the modulation of cell wall mechanical stability during
1430:
713:
The N-terminal pro-peptides of pectinesterase are variable in size and sequence and show a low level of amino acid identity. Alternatively the
1344:
1445:
1439:
1240:
990:
1288:
1828:
1399:
1188:
218:
744:
putative catalytic residues are in a similar location to those of the active site and substrate-binding cleft of pectate lyase.
1506:
1394:
1315:
1225:
1178:
169:
145:
1984:
632:
1737:
1678:
810:
Giovane A, Tsernoglou D, Camardella L, Di Matteo A, Raiola A, Bonivento D, De
Lorenzo G, Cervone F, Bellincampi D (2005).
408:
Most of the purified plant pectinesterases have neutral or alkaline isoelectric points and are bound to the cell wall via
2099:
1782:
1349:
1339:
1742:
1634:
1259:
362:
movement protein and it has been shown that this interaction is required for cell-to-cell translocation of the virus.
1328:
1324:
1320:
1236:
1069:
1969:
515:
such as relative molecular mass, isoelectric point, optimum pH, substrate affinity, ion-requirement and location.
2085:
2072:
2059:
2046:
2033:
2020:
2007:
1590:
1536:
1496:
1455:
1359:
1198:
1166:
1052:
1016:
736:
domain and a pectin binding cleft. Similarly several pectinesterase structures have been elucidated in fungi and
1979:
163:
1933:
1876:
1244:
1093:
1007:
962:
56:
620:
490:
chaperone, ensuring correct folding or deactivating activity until PE insertion in the cell wall is complete.
150:
1881:
1113:
983:
257:
that facilitate plant cell wall modification and subsequent breakdown. It catalyzes the following reaction:
1669:
1308:
425:. The optimal pH of higher plants is usually between pH 7 and pH 8 although the pH of pectinesterase from
230:
1902:
1821:
1604:
1501:
1293:
1254:
1118:
1040:
475:
138:
1974:
616:
470:
that are annotated as pectinesterases, most of which are encoded as large pre-proproteins. The signal
449:. This terminal extension is eventually removed to yield a mature protein of 34-37 kDa. Most PEs lack
348:
yield and root development. Pectinesterase has also been shown to play a role in a plants response to
293:
1787:
1622:
1617:
1550:
1210:
1035:
812:"Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein"
462:
457:
in the mature protein, although at least one site is present in the amino-terminal extension region.
359:
73:
1938:
1642:
1612:
1416:
1411:
1335:
1271:
1057:
467:
450:
374:
forming a pectate gel. The other being that proton release may stimulate the activity of cell wall
166:
68:
90:
1871:
1775:
1627:
1108:
1098:
976:
310:
2129:
1576:
1521:
1489:
1364:
1083:
937:
910:
892:
859:
841:
792:
639:
607:
507:
354:
316:
157:
38:
1917:
1912:
1886:
1814:
1652:
1232:
1137:
1132:
1088:
929:
882:
831:
823:
782:
774:
503:
599:
126:
1964:
1948:
1861:
1754:
1568:
1484:
1479:
1474:
1387:
1382:
1142:
1020:
958:
454:
394:
329:
102:
61:
2113:
2002:
1943:
1727:
1722:
1717:
1103:
1078:
1074:
1030:
836:
811:
787:
762:
511:
333:
201:
887:
870:
181:
2123:
1907:
1866:
1468:
1377:
1127:
733:
483:
409:
176:
869:
Johansson K, El-Ahmad M, Friemann R, Jörnvall H, Markovic O, Eklund H (March 2002).
761:
Fries, M.; Ihrig, J.; Brocklehurst, K.; Shevchik, V. E.; Pickersgill, R. W. (2007).
549:
1856:
1595:
1526:
1249:
1170:
726:
595:
371:
421:
may exhibit both mechanisms and that such mechanisms are driven by alterations in
561:
2080:
2015:
1851:
1770:
1541:
1435:
1298:
1264:
1202:
968:
442:
337:
185:
2108:
445:
possessing a signal sequence and a large amino-terminal extension of around 22
1747:
714:
479:
375:
341:
778:
2054:
2028:
1660:
1459:
1372:
999:
763:"Molecular basis of the activity of the phytopathogen pectin methylesterase"
42:
941:
933:
896:
845:
827:
796:
725:) root and consists of a right-handed parallel β-helix as seen in all the
1705:
1700:
1695:
1516:
1151:
1003:
729:
556:
430:
418:
402:
367:
349:
305:
278:
254:
486:
sites and it is thought that these sites also play a role in targeting.
114:
1712:
1690:
1685:
1303:
573:
568:
471:
401:
substrates for depolymerising enzymes, particularly acidic pectins and
386:
301:
133:
914:
863:
699:
693:
687:
681:
675:
669:
663:
657:
651:
645:
2067:
1837:
1732:
1665:
1558:
1156:
1064:
718:
627:
414:
297:
250:
213:
109:
97:
85:
2041:
1674:
1425:
1421:
426:
390:
345:
328:
Recent studies have shown that the manipulation of pectinesterase
289:
of the cell wall resulting in alterations in cell wall integrity.
282:
1406:
1283:
1276:
1220:
1215:
589:
544:
121:
1810:
972:
285:. Pectinesterase functions primarily by altering the localised
28:
446:
422:
413:
un-esterified GalA residues in homogalacturonan domains of
286:
1806:
740:
and share most of the structural motifs seen in plants.
474:
pre-region is required for targeting the enzyme to the
441:
PE proteins are synthesised as pre-proteins of 540–580
358:
is involved in host cell receptor recognition for the
2097:
478:
and consists of about 25 amino acid residues. These
336:, cell wall extension during pollen germination and
277:
It is found in all higher plants as well as in some
1993:
1957:
1926:
1895:
1844:
1763:
1651:
1603:
1589:
1567:
1549:
1535:
1515:
1454:
1358:
1197:
1165:
1015:
638:
626:
606:
588:
583:
567:
555:
543:
535:
530:
525:
224:
212:
200:
195:
175:
156:
144:
132:
120:
108:
96:
84:
79:
67:
55:
50:
21:
871:"Crystal structure of plant pectin methylesterase"
1294:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
510:and biochemical activity have been identified in
352:attack. A cell wall-associated pectinesterase of
1822:
984:
502:Several pectinesterase isoforms differing in
8:
393:cell walls. They are polymerised in the cis
389:form approximately 35% of the dry weight of
466:genome has led to the identification of 66
1829:
1815:
1807:
1600:
1546:
1532:
991:
977:
969:
580:
192:
27:
961:at the U.S. National Library of Medicine
886:
835:
786:
2104:
1184:Ubiquitin carboxy-terminal hydrolase L1
753:
249:) is a ubiquitous cell-wall-associated
522:
18:
1764:either deoxy- or ribo-
378:contributing to cell wall loosening.
7:
1345:Protein serine/threonine phosphatase
1446:Cyclic nucleotide phosphodiesterase
1440:Clostridium perfringens alpha toxin
1241:Tartrate-resistant acid phosphatase
1289:Pyruvate dehydrogenase phosphatase
437:Molecular biology and biochemistry
14:
1189:4-hydroxybenzoyl-CoA thioesterase
433:is usually much lower than this.
2107:
314:and in fungal pathogens such as
37:in complex with hexasaccharide.
1507:N-acetylglucosamine-6-sulfatase
1395:Sphingomyelin phosphodiesterase
1316:Inositol-phosphate phosphatase
1179:Palmitoyl protein thioesterase
370:groups that can interact with
245:(EC 3.1.1.11; systematic name
1:
1679:RNA-induced silencing complex
888:10.1016/S0014-5793(02)02372-4
584:Available protein structures:
292:Pectinesterase catalyses the
1783:Serratia marcescens nuclease
1350:Dual-specificity phosphatase
1340:Protein tyrosine phosphatase
1260:Fructose 1,6-bisphosphatase
33:Pectin methylesterase from
2146:
1985:Michaelis–Menten kinetics
1497:Galactosamine-6 sulfatase
1053:6-phosphogluconolactonase
579:
526:Pectinesterase, catalytic
191:
26:
1877:Diffusion-limited enzyme
1245:Purple acid phosphatases
963:Medical Subject Headings
779:10.1038/sj.emboj.7601816
482:regions contain several
382:Esterification of pectin
1670:Microprocessor complex
1309:Beta-propeller phytase
934:10.1006/jmbi.2000.4324
828:10.1105/tpc.104.028886
253:that presents several
247:pectin pectylhydrolase
1970:Eadie–Hofstee diagram
1903:Allosteric regulation
1605:Endodeoxyribonuclease
1502:Iduronate-2-sulfatase
1255:Glucose 6-phosphatase
1041:Butyrylcholinesterase
476:endoplasmic reticulum
1980:Lineweaver–Burk plot
1788:Micrococcal nuclease
1623:Deoxyribonuclease IV
1618:Deoxyribonuclease II
1551:Exodeoxyribonuclease
1211:Alkaline phosphatase
1036:Acetylcholinesterase
463:Arabidopsis thaliana
360:tobacco mosaic virus
1643:UvrABC endonuclease
1613:Deoxyribonuclease I
1336:Protein phosphatase
1272:Protein phosphatase
1070:Bile salt-dependent
1058:PAF acetylhydrolase
468:open reading frames
451:consensus sequences
344:, stem elongation,
1939:Enzyme superfamily
1872:Enzyme promiscuity
1776:Mung bean nuclease
1635:Restriction enzyme
1628:Restriction enzyme
539:Pectinesterase_cat
311:Erwinia carotovora
273:methanol + pectate
2095:
2094:
1804:
1803:
1800:
1799:
1796:
1795:
1585:
1584:
1577:Oligonucleotidase
1522:deoxyribonuclease
1490:Steroid sulfatase
1365:Phosphodiesterase
1094:Hormone-sensitive
773:(17): 3879–3887.
711:
710:
707:
706:
633:structure summary
508:isoelectric point
355:Nicotiana tabacum
317:Aspergillus niger
294:de-esterification
240:
239:
236:
235:
139:metabolic pathway
2137:
2112:
2111:
2103:
1975:Hanes–Woolf plot
1918:Enzyme activator
1913:Enzyme inhibitor
1887:Enzyme catalysis
1831:
1824:
1817:
1808:
1653:Endoribonuclease
1639:
1633:
1601:
1547:
1533:
1233:Acid phosphatase
1114:Monoacylglycerol
1024:ester hydrolases
993:
986:
979:
970:
946:
945:
917:
907:
901:
900:
890:
866:
856:
850:
849:
839:
807:
801:
800:
790:
767:The EMBO Journal
758:
702:
696:
690:
684:
678:
672:
666:
660:
654:
648:
581:
523:
504:molecular weight
193:
45:
35:Dickeya dadantii
31:
19:
16:Class of enzymes
2145:
2144:
2140:
2139:
2138:
2136:
2135:
2134:
2120:
2119:
2118:
2106:
2098:
2096:
2091:
2003:Oxidoreductases
1989:
1965:Enzyme kinetics
1953:
1949:List of enzymes
1922:
1891:
1862:Catalytic triad
1840:
1835:
1805:
1792:
1759:
1647:
1637:
1631:
1594:
1581:
1569:Exoribonuclease
1563:
1540:
1524:
1520:
1511:
1485:Arylsulfatase L
1480:Arylsulfatase B
1475:Arylsulfatase A
1450:
1363:
1354:
1193:
1161:
1023:
1011:
997:
955:
950:
949:
919:
909:
908:
904:
868:
858:
857:
853:
809:
808:
804:
760:
759:
755:
750:
732:family CE-8, a
698:
692:
686:
680:
674:
668:
662:
656:
650:
644:
521:
500:
455:N-glycosylation
439:
384:
326:
268:
46:
41:
17:
12:
11:
5:
2143:
2141:
2133:
2132:
2122:
2121:
2117:
2116:
2093:
2092:
2090:
2089:
2076:
2063:
2050:
2037:
2024:
2011:
1997:
1995:
1991:
1990:
1988:
1987:
1982:
1977:
1972:
1967:
1961:
1959:
1955:
1954:
1952:
1951:
1946:
1941:
1936:
1930:
1928:
1927:Classification
1924:
1923:
1921:
1920:
1915:
1910:
1905:
1899:
1897:
1893:
1892:
1890:
1889:
1884:
1879:
1874:
1869:
1864:
1859:
1854:
1848:
1846:
1842:
1841:
1836:
1834:
1833:
1826:
1819:
1811:
1802:
1801:
1798:
1797:
1794:
1793:
1791:
1790:
1785:
1780:
1779:
1778:
1767:
1765:
1761:
1760:
1758:
1757:
1752:
1751:
1750:
1745:
1740:
1735:
1725:
1720:
1715:
1710:
1709:
1708:
1703:
1698:
1693:
1683:
1682:
1681:
1672:
1657:
1655:
1649:
1648:
1646:
1645:
1640:
1625:
1620:
1615:
1609:
1607:
1598:
1587:
1586:
1583:
1582:
1580:
1579:
1573:
1571:
1565:
1564:
1562:
1561:
1555:
1553:
1544:
1530:
1513:
1512:
1510:
1509:
1504:
1499:
1494:
1493:
1492:
1487:
1482:
1477:
1464:
1462:
1452:
1451:
1449:
1448:
1443:
1433:
1428:
1419:
1414:
1409:
1404:
1403:
1402:
1392:
1391:
1390:
1385:
1375:
1369:
1367:
1356:
1355:
1353:
1352:
1347:
1342:
1333:
1332:
1331:
1313:
1312:
1311:
1301:
1296:
1291:
1286:
1281:
1280:
1279:
1269:
1268:
1267:
1257:
1252:
1247:
1230:
1229:
1228:
1223:
1218:
1207:
1205:
1195:
1194:
1192:
1191:
1186:
1181:
1175:
1173:
1163:
1162:
1160:
1159:
1154:
1148:
1147:
1146:
1145:
1140:
1135:
1124:
1123:
1122:
1121:
1119:Diacylglycerol
1116:
1111:
1106:
1101:
1096:
1091:
1086:
1081:
1072:
1061:
1060:
1055:
1050:
1048:Pectinesterase
1045:
1044:
1043:
1038:
1031:Cholinesterase
1027:
1025:
1013:
1012:
998:
996:
995:
988:
981:
973:
967:
966:
959:pectinesterase
954:
953:External links
951:
948:
947:
928:(4): 951–960.
902:
881:(2–3): 243–9.
851:
822:(3): 849–858.
802:
752:
751:
749:
746:
709:
708:
705:
704:
642:
636:
635:
630:
624:
623:
610:
604:
603:
593:
586:
585:
577:
576:
571:
565:
564:
559:
553:
552:
547:
541:
540:
537:
533:
532:
528:
527:
520:
517:
512:dicotyledonous
499:
498:Plant isoforms
496:
438:
435:
383:
380:
334:fruit ripening
325:
322:
275:
274:
266:
243:Pectinesterase
238:
237:
234:
233:
228:
222:
221:
216:
210:
209:
204:
198:
197:
189:
188:
179:
173:
172:
161:
154:
153:
148:
142:
141:
136:
130:
129:
124:
118:
117:
112:
106:
105:
100:
94:
93:
88:
82:
81:
77:
76:
71:
65:
64:
59:
53:
52:
48:
47:
32:
24:
23:
22:pectinesterase
15:
13:
10:
9:
6:
4:
3:
2:
2142:
2131:
2128:
2127:
2125:
2115:
2110:
2105:
2101:
2087:
2083:
2082:
2077:
2074:
2070:
2069:
2064:
2061:
2057:
2056:
2051:
2048:
2044:
2043:
2038:
2035:
2031:
2030:
2025:
2022:
2018:
2017:
2012:
2009:
2005:
2004:
1999:
1998:
1996:
1992:
1986:
1983:
1981:
1978:
1976:
1973:
1971:
1968:
1966:
1963:
1962:
1960:
1956:
1950:
1947:
1945:
1944:Enzyme family
1942:
1940:
1937:
1935:
1932:
1931:
1929:
1925:
1919:
1916:
1914:
1911:
1909:
1908:Cooperativity
1906:
1904:
1901:
1900:
1898:
1894:
1888:
1885:
1883:
1880:
1878:
1875:
1873:
1870:
1868:
1867:Oxyanion hole
1865:
1863:
1860:
1858:
1855:
1853:
1850:
1849:
1847:
1843:
1839:
1832:
1827:
1825:
1820:
1818:
1813:
1812:
1809:
1789:
1786:
1784:
1781:
1777:
1774:
1773:
1772:
1769:
1768:
1766:
1762:
1756:
1753:
1749:
1746:
1744:
1741:
1739:
1736:
1734:
1731:
1730:
1729:
1726:
1724:
1721:
1719:
1716:
1714:
1711:
1707:
1704:
1702:
1699:
1697:
1694:
1692:
1689:
1688:
1687:
1684:
1680:
1676:
1673:
1671:
1667:
1664:
1663:
1662:
1659:
1658:
1656:
1654:
1650:
1644:
1641:
1636:
1629:
1626:
1624:
1621:
1619:
1616:
1614:
1611:
1610:
1608:
1606:
1602:
1599:
1597:
1592:
1588:
1578:
1575:
1574:
1572:
1570:
1566:
1560:
1557:
1556:
1554:
1552:
1548:
1545:
1543:
1538:
1534:
1531:
1528:
1523:
1518:
1514:
1508:
1505:
1503:
1500:
1498:
1495:
1491:
1488:
1486:
1483:
1481:
1478:
1476:
1473:
1472:
1471:
1470:
1469:arylsulfatase
1466:
1465:
1463:
1461:
1457:
1453:
1447:
1444:
1441:
1437:
1434:
1432:
1429:
1427:
1423:
1420:
1418:
1415:
1413:
1410:
1408:
1405:
1401:
1398:
1397:
1396:
1393:
1389:
1386:
1384:
1381:
1380:
1379:
1378:Phospholipase
1376:
1374:
1371:
1370:
1368:
1366:
1361:
1357:
1351:
1348:
1346:
1343:
1341:
1337:
1334:
1330:
1326:
1322:
1319:
1318:
1317:
1314:
1310:
1307:
1306:
1305:
1302:
1300:
1297:
1295:
1292:
1290:
1287:
1285:
1282:
1278:
1275:
1274:
1273:
1270:
1266:
1263:
1262:
1261:
1258:
1256:
1253:
1251:
1248:
1246:
1242:
1238:
1234:
1231:
1227:
1224:
1222:
1219:
1217:
1214:
1213:
1212:
1209:
1208:
1206:
1204:
1200:
1196:
1190:
1187:
1185:
1182:
1180:
1177:
1176:
1174:
1172:
1168:
1164:
1158:
1155:
1153:
1150:
1149:
1144:
1141:
1139:
1136:
1134:
1131:
1130:
1129:
1128:Phospholipase
1126:
1125:
1120:
1117:
1115:
1112:
1110:
1107:
1105:
1102:
1100:
1097:
1095:
1092:
1090:
1087:
1085:
1082:
1080:
1076:
1073:
1071:
1068:
1067:
1066:
1063:
1062:
1059:
1056:
1054:
1051:
1049:
1046:
1042:
1039:
1037:
1034:
1033:
1032:
1029:
1028:
1026:
1022:
1018:
1014:
1009:
1005:
1001:
994:
989:
987:
982:
980:
975:
974:
971:
964:
960:
957:
956:
952:
943:
939:
935:
931:
927:
923:
916:
912:
906:
903:
898:
894:
889:
884:
880:
876:
872:
865:
861:
855:
852:
847:
843:
838:
833:
829:
825:
821:
817:
813:
806:
803:
798:
794:
789:
784:
780:
776:
772:
768:
764:
757:
754:
747:
745:
741:
739:
735:
734:transmembrane
731:
728:
724:
723:Daucus carota
720:
716:
701:
695:
689:
683:
677:
671:
665:
659:
653:
647:
643:
641:
637:
634:
631:
629:
625:
622:
618:
614:
611:
609:
605:
601:
597:
594:
591:
587:
582:
578:
575:
572:
570:
566:
563:
560:
558:
554:
551:
548:
546:
542:
538:
534:
529:
524:
518:
516:
513:
509:
505:
497:
495:
491:
487:
485:
484:glycosylation
481:
477:
473:
469:
465:
464:
458:
456:
452:
448:
444:
436:
434:
432:
428:
424:
420:
416:
411:
410:electrostatic
406:
404:
398:
396:
392:
388:
381:
379:
377:
373:
369:
363:
361:
357:
356:
351:
347:
343:
339:
335:
331:
323:
321:
319:
318:
313:
312:
307:
303:
299:
295:
290:
288:
284:
280:
272:
264:
260:
259:
258:
256:
252:
248:
244:
232:
229:
227:
223:
220:
217:
215:
211:
208:
205:
203:
199:
194:
190:
187:
183:
180:
178:
177:Gene Ontology
174:
171:
168:
165:
162:
159:
155:
152:
149:
147:
143:
140:
137:
135:
131:
128:
125:
123:
119:
116:
115:NiceZyme view
113:
111:
107:
104:
101:
99:
95:
92:
89:
87:
83:
78:
75:
72:
70:
66:
63:
60:
58:
54:
49:
44:
40:
36:
30:
25:
20:
2081:Translocases
2078:
2065:
2052:
2039:
2026:
2016:Transferases
2013:
2000:
1857:Binding site
1638:}}
1632:{{
1596:Endonuclease
1527:ribonuclease
1467:
1250:Nucleotidase
1171:Thioesterase
1047:
925:
922:J. Mol. Biol
921:
905:
878:
874:
854:
819:
815:
805:
770:
766:
756:
742:
737:
727:carbohydrate
722:
712:
501:
492:
488:
461:
459:
440:
407:
399:
385:
372:calcium ions
364:
353:
327:
315:
309:
291:
276:
270:
262:
246:
242:
241:
103:BRENDA entry
34:
1852:Active site
1771:Nuclease S1
1542:Exonuclease
1436:Lecithinase
1265:Calcineurin
1203:Phosphatase
1109:Lipoprotein
1099:Endothelial
531:Identifiers
443:amino acids
338:pollen tube
91:IntEnz view
51:Identifiers
2055:Isomerases
2029:Hydrolases
1896:Regulation
1084:Pancreatic
1021:Carboxylic
816:Plant Cell
748:References
715:C-terminal
596:structures
480:N-terminal
376:hydrolases
342:abscission
330:expression
160:structures
127:KEGG entry
74:9025-98-3
1934:EC number
1661:RNase III
1519:(includes
1460:Sulfatase
1373:Autotaxin
1237:Prostatic
1089:Lysosomal
1004:esterases
1000:Hydrolase
918:;
875:FEBS Lett
867:;
697:,
691:,
685:,
679:,
673:,
667:,
661:,
649:,
574:PDOC00413
562:IPR000070
519:Structure
261:pectin +
80:Databases
2130:EC 3.1.1
2124:Category
1958:Kinetics
1882:Cofactor
1845:Activity
1755:RNase T1
1517:Nuclease
1152:Cutinase
942:11162105
897:11943159
846:15722470
797:17717531
730:esterase
655:
613:RCSB PDB
557:InterPro
431:bacteria
419:isoforms
403:methanol
368:carboxyl
350:pathogen
340:growth,
324:Function
306:methanol
279:bacteria
255:isoforms
231:proteins
219:articles
207:articles
164:RCSB PDB
62:3.1.1.11
2114:Biology
2068:Ligases
1838:Enzymes
1728:RNase E
1723:RNase Z
1718:RNase A
1713:RNase P
1686:RNase H
1304:Phytase
1104:Hepatic
1079:Lingual
1075:Gastric
837:1069703
788:2000356
703:
569:PROSITE
550:PF01095
472:peptide
387:Pectins
302:pectate
186:QuickGO
151:profile
134:MetaCyc
69:CAS no.
2100:Portal
2042:Lyases
1666:Drosha
1591:3.1.21
1559:RecBCD
1537:3.1.11
1157:PETase
1065:Lipase
965:(MeSH)
940:
895:
844:
834:
795:
785:
738:E.coli
719:carrot
628:PDBsum
602:
592:
536:Symbol
415:pectin
298:pectin
251:enzyme
214:PubMed
196:Search
182:AmiGO
170:PDBsum
110:ExPASy
98:BRENDA
86:IntEnz
57:EC no.
1994:Types
1675:Dicer
1630:;see
1456:3.1.6
1426:PDE4B
1422:PDE4A
1360:3.1.4
1329:IMPA3
1325:IMPA2
1321:IMPA1
1199:3.1.3
1167:3.1.2
1017:3.1.1
427:fungi
395:Golgi
391:dicot
346:tuber
300:into
283:fungi
146:PRIAM
2086:list
2079:EC7
2073:list
2066:EC6
2060:list
2053:EC5
2047:list
2040:EC4
2034:list
2027:EC3
2021:list
2014:EC2
2008:list
2001:EC1
1593:-31:
1539:-16:
1525:and
1431:PDE5
1417:PDE3
1412:PDE2
1407:PDE1
1299:PTEN
1284:OCRL
1277:PP2A
1226:ALPP
1221:ALPL
1216:ALPI
1010:3.1)
938:PMID
915:1QJV
893:PMID
864:1GQ8
842:PMID
793:PMID
700:2ntq
694:2ntp
688:2ntb
682:2nt9
676:2nt6
670:2nst
664:2nsp
658:1xg2
652:1qjv
646:1gq8
621:PDBj
617:PDBe
600:ECOD
590:Pfam
545:Pfam
453:for
429:and
304:and
281:and
269:O =
226:NCBI
167:PDBe
122:KEGG
43:2ntb
1748:4/5
930:doi
926:305
911:PDB
883:doi
879:514
860:PDB
832:PMC
824:doi
783:PMC
775:doi
640:PDB
608:PDB
447:kDa
296:of
202:PMC
158:PDB
39:PDB
2126::
1706:2C
1701:2B
1696:2A
1677::
1668::
1458::
1338::
1327:,
1323:,
1239:)/
1201::
1169::
1138:A2
1133:A1
1019::
1008:EC
1002::
936:.
924:.
913::
891:.
877:.
873:.
862::
840:.
830:.
820:17
818:.
814:.
791:.
781:.
771:26
769:.
765:.
619:;
615:;
598:/
506:,
423:pH
405:.
287:pH
184:/
2102::
2088:)
2084:(
2075:)
2071:(
2062:)
2058:(
2049:)
2045:(
2036:)
2032:(
2023:)
2019:(
2010:)
2006:(
1830:e
1823:t
1816:v
1743:3
1738:2
1733:1
1691:1
1529:)
1442:)
1438:(
1424:/
1400:1
1388:D
1383:C
1362::
1243:/
1235:(
1143:B
1077:/
1006:(
992:e
985:t
978:v
944:.
932::
899:.
885::
848:.
826::
799:.
777::
721:(
271:n
267:2
265:H
263:n
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.