482:. The 3’ region of a transcript contains many polyadenylation signals (PAS). When more proximal (closer towards 5’ end) PAS sites are utilized, this shortens the length of the 3’ untranslated region (3' UTR) of a transcript. Studies in both humans and flies have shown tissue specific APA. With neuronal tissues preferring distal PAS usage, leading to longer 3’ UTRs and testis tissues preferring proximal PAS leading to shorter 3’ UTRs. Studies have shown there is a correlation between a gene's conservation level and its tendency to do alternative polyadenylation, with highly conserved genes exhibiting more APA. Similarly, highly expressed genes follow this same pattern.
578:
466:
680:, two closely related complexes that recycle RNA into nucleotides. This enzyme degrades RNA by attacking the bond between the 3′-most nucleotides with a phosphate, breaking off a diphosphate nucleotide. This reaction is reversible, and so the enzyme can also extend RNA with more nucleotides. The heteropolymeric tail added by polynucleotide phosphorylase is very rich in adenine. The choice of adenine is most likely the result of higher
399:
reduces deadenylation. The rate of deadenylation may also be regulated by RNA-binding proteins. Additionally, RNA triple helix structures and RNA motifs such as the poly(A) tail 3’ end binding pocket retard deadenylation process and inhibit poly(A) tail removal. Once the poly(A) tail is removed, the decapping complex removes the 5′ cap, leading to a degradation of the RNA. Several other proteins are involved in deadenylation in
124:
350:. Poly(A)-binding protein promotes export from the nucleus and translation, and inhibits degradation. This protein binds to the poly(A) tail prior to mRNA export from the nucleus and in yeast also recruits poly(A) nuclease, an enzyme that shortens the poly(A) tail and allows the export of the mRNA. Poly(A)-binding protein is exported to the cytoplasm with the RNA. mRNAs that are not exported are degraded by the
22:
760:
protection of the 3′ end of the RNA from nucleases, but later the specific roles of polyadenylation in nuclear export and translation were identified. The polymerases responsible for polyadenylation were first purified and characterized in the 1960s and 1970s, but the large number of accessory proteins that control this process were discovered only in the early 1990s.
5750:
375:, the poly(A) tails of most mRNAs in the cytoplasm gradually get shorter, and mRNAs with shorter poly(A) tail are translated less and degraded sooner. However, it can take many hours before an mRNA is degraded. This deadenylation and degradation process can be accelerated by microRNAs complementary to the
438:
In the early mouse embryo, cytoplasmic polyadenylation of maternal RNAs from the egg cell allows the cell to survive and grow even though transcription does not start until the middle of the 2-cell stage (4-cell stage in human). In the brain, cytoplasmic polyadenylation is active during learning and
329:
long the enzyme can no longer bind to CPSF and polyadenylation stops, thus determining the length of the poly(A) tail. CPSF is in contact with RNA polymerase II, allowing it to signal the polymerase to terminate transcription. When RNA polymerase II reaches a "termination sequence" (⁵'TTTATT' on the
497:
in the 3′ UTR. MicroRNAs tend to repress translation and promote degradation of the mRNAs they bind to, although there are examples of microRNAs that stabilise transcripts. Alternative polyadenylation can also shorten the coding region, thus making the mRNA code for a different protein, but this is
305:
The RNA is typically cleaved before transcription termination, as CstF also binds to RNA polymerase II. Through a poorly understood mechanism (as of 2002), it signals for RNA polymerase II to slip off of the transcript. Cleavage also involves the protein CFII, though it is unknown how. The cleavage
616:
contain both stabilising and destabilising poly(A) tails. Destabilising polyadenylation targets both mRNA and noncoding RNAs. The poly(A) tails are 43 nucleotides long on average. The stabilising ones start at the stop codon, and without them the stop codon (UAA) is not complete as the genome only
398:
and remove nucleotides from the poly(A) tail. The level of access to the 5′ cap and poly(A) tail is important in controlling how soon the mRNA is degraded. PARN deadenylates less if the RNA is bound by the initiation factors 4E (at the 5′ cap) and 4G (at the poly(A) tail), which is why translation
652:
of all living organisms, it is presumed, had some form of polyadenylation system. A few organisms do not polyadenylate mRNA, which implies that they have lost their polyadenylation machineries during evolution. Although no examples of eukaryotes that lack polyadenylation are known, mRNAs from the
688:
as an energy currency, making it more likely to be incorporated in this tail in early lifeforms. It has been suggested that the involvement of adenine-rich tails in RNA degradation prompted the later evolution of polyadenylate polymerases (the enzymes that produce poly(A) tails with no other
362:
ribosomal subunit. However, a poly(A) tail is not required for the translation of all mRNAs. Further, poly(A) tailing (oligo-adenylation) can determine the fate of RNA molecules that are usually not poly(A)-tailed (such as (small) non-coding (sn)RNAs etc.) and thereby induce their RNA decay.
759:
in extracts made from cell nuclei that could polymerise ATP, but not ADP, into polyadenine. Although identified in many types of cells, this activity had no known function until 1971, when poly(A) sequences were found in mRNAs. The only function of these sequences was thought at first to be
301:
exist. Two other proteins add specificity to the binding to an RNA: CstF and CFI. CstF binds to a GU-rich region further downstream of CPSF's site. CFI recognises a third site on the RNA (a set of UGUAA sequences in mammals) and can recruit CPSF even if the AAUAAA sequence is missing. The
97:
The poly(A) tail is important for the nuclear export, translation and stability of mRNA. The tail is shortened over time, and, when it is short enough, the mRNA is enzymatically degraded. However, in a few cell types, mRNAs with short poly(A) tails are stored for later activation by
203:
Many eukaryotic non-coding RNAs are always polyadenylated at the end of transcription. There are small RNAs where the poly(A) tail is seen only in intermediary forms and not in the mature RNA as the ends are removed during processing, the notable ones being
161:
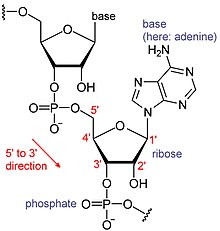
template. By convention, RNA sequences are written in a 5′ to 3′ direction. The 5′ end is the part of the RNA molecule that is transcribed first, and the 3′ end is transcribed last. The 3′ end is also where the poly(A) tail is found on polyadenylated RNAs.
517:(a group of bacterial compounds that trigger an immune response). This results in the selection of weak poly(A) sites and thus shorter transcripts. This removes regulatory elements in the 3′ untranslated regions of mRNAs for defense-related products like
302:
polyadenylation signal – the sequence motif recognised by the RNA cleavage complex – varies between groups of eukaryotes. Most human polyadenylation sites contain the AAUAAA sequence, but this sequence is less common in plants and fungi.
86:; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (
636:). It can synthesise a 3′ extension where the vast majority of the bases are adenines. Like in bacteria, polyadenylation by polynucleotide phosphorylase promotes degradation of the RNA in plastids and likely also archaea.
5195:
Evguenieva-Hackenberg E, Roppelt V, Finsterseifer P, Klug G (December 2008). "Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome".
525:. These mRNAs then have longer half-lives and produce more of these proteins. RNA-binding proteins other than those in the polyadenylation machinery can also affect whether a polyadenylation site is used, as can
3910:
Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D (June 1980). "Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends".
4047:
Ogorodnikov A, Levin M, Tattikota S, Tokalov S, Hoque M, Scherzinger D, Marini F, Poetsch A, Binder H, Macher-Göppinger S, Probst HC, Tian B, Schaefer M, Lackner KJ, Westermann F, Danckwardt S (December 2018).
383:, mRNAs with shortened poly(A) tails are not degraded, but are instead stored and translationally inactive. These short tailed mRNAs are activated by cytoplasmic polyadenylation after fertilisation, during
704:. It is presumed that the horizontal transfer of bacterial CCA-adding enzyme to eukaryotes allowed the archaeal-like CCA-adding enzyme to switch function to a poly(A) polymerase. Some lineages, like
6169:
192:
and aids in transcription termination, export of the mRNA from the nucleus, and translation. Almost all eukaryotic mRNAs are polyadenylated, with the exception of animal replication-dependent
644:
Although polyadenylation is seen in almost all organisms, it is not universal. However, the wide distribution of this modification and the fact that it is present in organisms from all three
175:, tune how active the mRNA is. There are also many RNAs that are not translated, called non-coding RNAs. Like the untranslated regions, many of these non-coding RNAs have regulatory roles.
4008:"Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection"
501:
The choice of poly(A) site can be influenced by extracellular stimuli and depends on the expression of the proteins that take part in polyadenylation. For example, the expression of
2472:
Tefferi A, Wieben ED, Dewald GW, Whiteman DA, Bernard ME, Spelsberg TC (August 2002). "Primer on medical genomics part II: Background principles and methods in molecular genetics".
3592:
Smibert, Peter; Miura, Pedro; Westholm, Jakub O.; Shenker, Sol; May, Gemma; Duff, Michael O.; Zhang, Dayu; Eads, Brian D.; Carlson, Joe; Brown, James B.; Eisman, Robert C. (2012).
3156:
Sakurai T, Sato M, Kimura M (November 2005). "Diverse patterns of poly(A) tail elongation and shortening of murine maternal mRNAs from fully grown oocyte to 2-cell embryo stages".
188:
In nuclear polyadenylation, a poly(A) tail is added to an RNA at the end of transcription. On mRNAs, the poly(A) tail protects the mRNA molecule from enzymatic degradation in the
617:
encodes the U or UA part. Plant mitochondria have only destabilising polyadenylation. Mitochondrial polyadenylation has never been observed in either budding or fission yeast.
5017:"Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins"
325:. Another protein, PAB2, binds to the new, short poly(A) tail and increases the affinity of polyadenylate polymerase for the RNA. When the poly(A) tail is approximately 250
105:
mRNA molecules in both prokaryotes and eukaryotes have polyadenylated 3′-ends, with the prokaryotic poly(A) tails generally shorter and fewer mRNA molecules polyadenylated.
2999:
Torabi, Seyed-Fakhreddin; Vaidya, Anand T.; Tycowski, Kazimierz T.; DeGregorio, Suzanne J.; Wang, Jimin; Shu, Mei-Di; Steitz, Thomas A.; Steitz, Joan A. (2021-02-05).
605:
to overcome these secondary structures. The poly(A) tail can also recruit RNases that cut the RNA in two. These bacterial poly(A) tails are about 30 nucleotides long.
529:
near the polyadenylation signal. In addition, numerous other components involved in transcription, splicing or other mechanisms regulating RNA biology can affect APA.
458:. Depending on the cell type, the polymerase can be the same type of polyadenylate polymerase (PAP) that is used in the nuclear process, or the cytoplasmic polymerase
443:, which is the strengthening of the signal transmission from a nerve cell to another in response to nerve impulses and is important for learning and memory formation.
5440:"The Functional Role of the 3′ Untranslated Region and Poly(A) Tail of Duck Hepatitis a Virus Type 1 in Viral Replication and Regulation of IRES-Mediated Translation"
5438:
Chen, Jun-Hao; Zhang, Rui-Hua; Lin, Shao-Li; Li, Peng-Fei; Lan, Jing-Jing; Song, Sha-Sha; Gao, Ji-Ming; Wang, Yu; Xie, Zhi-Jing; Li, Fu-Chang; Jiang, Shi-Jin (2018).
200:
structure followed by a purine-rich sequence, termed histone downstream element, that directs where the RNA is cut so that the 3′ end of the histone mRNA is formed.
98:
re-polyadenylation in the cytosol. In contrast, when polyadenylation occurs in bacteria, it promotes RNA degradation. This is also sometimes the case for eukaryotic
6213:
6162:
447:
238:
2743:"sCLIP-an integrated platform to study RNA-protein interactomes in biomedical research: identification of CSTF2tau in alternative processing of small nuclear RNAs"
5636:"Polyadenylic Acid Sequences in the Heterogeneous Nuclear RNA and Rapidly-Labeled Polyribosomal RNA of HeLa Cells: Possible Evidence for a Precursor Relationship"
5234:
Slomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G (April 2008). "Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases".
330:
DNA template and ⁵'AAUAAA' on the primary transcript), the end of transcription is signaled. The polyadenylation machinery is also physically linked to the
5064:
Slomovic S, Portnoy V, Schuster G (2008). "Chapter 24 Detection and
Characterization of Polyadenylated RNA in Eukarya, Bacteria, Archaea, and Organelles".
6155:
2141:
Stumpf G, Domdey H (November 1996). "Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor".
1295:
Hunt AG, Xu R, Addepalli B, Rao S, Forbes KP, Meeks LR, Xing D, Mo M, Zhao H, Bandyopadhyay A, Dampanaboina L, Marion A, Von Lanken C, Li QQ (May 2008).
5782:
4211:
Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE (June 2007).
2429:
Nag A, Narsinh K, Martinson HG (July 2007). "The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase".
597:. Polynucleotide phosphorylase binds to the 3′ end of RNAs and the 3′ extension provided by the poly(A) tail allows it to bind to the RNAs whose
4919:"PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts"
4733:
Slomovic S, Portnoy V, Liveanu V, Schuster G (2006). "RNA Polyadenylation in
Prokaryotes and Organelles; Different Tails Tell Different Tales".
2131:
Molecular
Biology of the Cell, Chapter 6, "From DNA to RNA". 4th edition. Alberts B, Johnson A, Lewis J, et al. New York: Garland Science; 2002.
601:
would otherwise block the 3′ end. Successive rounds of polyadenylation and degradation of the 3′ end by polynucleotide phosphorylase allows the
5271:"Direct Evidence that the Poly(A) Tail of Influenza A Virus mRNA Is Synthesized by Reiterative Copying of a U Track in the Virion RNA Template"
4835:"Kinetics of polynucleotide phosphorylase: comparison of enzymes from Streptomyces and Escherichia coli and effects of nucleoside diphosphates"
2694:"Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay"
289:
cleaves the 3′-most part of a newly produced RNA and polyadenylates the end produced by this cleavage. The cleavage is catalysed by the enzyme
4260:
Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Gröne HJ, Schirmacher P, Hentze MW, Kulozik AE (February 2011).
1881:"Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping"
5950:
5607:
5081:
1190:
4105:
Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB (November 2008).
569:. Poly(A) tails have also been found on human rRNA fragments, both the form of homopolymeric (A only) and heterpolymeric (mostly A) tails.
4050:"Transcriptome 3′ end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma"
135:(for polyadenylic acid tail) reflects the way RNA nucleotides are abbreviated, with a letter for the base the nucleotide contains (A for
6223:
5834:
5814:
4646:
248:
79:
5512:"Polynucleotide Biosynthesis: Formation of a Sequence of Adenylate Units from Adenosine Triphosphate by an Enzyme from Thymus Nuclei"
2192:
Iseli C, Stevenson BJ, de Souza SJ, Samaia HB, Camargo AA, Buetow KH, Strausberg RL, Simpson AJ, Bucher P, Jongeneel CV (July 2002).
486:
data (sequencing of only mRNAs inside ribosomes) has shown that mRNA isoforms with shorter 3’ UTRs are more likely to be translated.
6178:
5922:
1297:"Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling"
620:
While many bacteria and mitochondria have polyadenylate polymerases, they also have another type of polyadenylation, performed by
3651:"Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3'-end of genes"
1822:"Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing"
1398:
Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ (November 2002). "The human and mouse replication-dependent histone genes".
6076:
6020:
490:
376:
3326:"PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila"
6015:
478:
Many protein-coding genes have more than one polyadenylation site, so a gene can code for several mRNAs that differ in their
2569:"Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein"
2795:"A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells"
1048:
6103:
6034:
5775:
213:
5381:"Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA"
197:
5839:
669:
621:
355:
354:. Poly(A)-binding protein also can bind to, and thus recruit, several proteins that affect translation, one of these is
3863:"MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene"
6218:
585:
In many bacteria, both mRNAs and non-coding RNAs can be polyadenylated. This poly(A) tail promotes degradation by the
522:
506:
243:
4882:
Nagaike T, Suzuki T, Ueda T (April 2008). "Polyadenylation in mammalian mitochondria: insights from recent studies".
435:. These shortened poly(A) tails are often less than 20 nucleotides, and are lengthened to around 80–150 nucleotides.
6054:
5754:
5107:"RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus"
3956:"Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing"
221:
131:
RNAs are a type of large biological molecules, whose individual building blocks are called nucleotides. The name
1930:"Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition"
692:
Polyadenylate polymerases are not as ancient. They have separately evolved in both bacteria and eukaryotes from
6477:
6098:
5886:
5799:
5768:
655:
558:
310:
152:
75:
68:
6424:
6245:
6059:
5876:
5861:
649:
347:
314:
41:
565:, which maintains a tail that is around 4 nucleotides long to the 3′ end. The RNA is then degraded by the
6482:
6064:
5989:
5881:
4633:
Régnier P, Arraiano CM (March 2000). "Degradation of mRNA in bacteria: emergence of ubiquitous features".
3757:"Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites"
685:
577:
440:
318:
4164:"Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR"
2243:"Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis"
5979:
5964:
5844:
5105:
Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G (December 2005).
3708:"Processing and transcriptome expansion at the mRNA 3′ end in health and disease: finding the right end"
681:
432:
166:
60:, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of
53:
3001:"RNA stabilization by a poly(A) tail 3′-end binding pocket and other modes of poly(A)-RNA interaction"
6308:
6086:
5984:
5902:
5647:
5392:
5333:
4742:
4118:
4061:
3768:
2904:
2382:"Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination"
2150:
1833:
1446:
1060:
1005:
728:
431:. This lengthens the poly(A) tail of an mRNA with a shortened poly(A) tail, so that the mRNA will be
91:
4301:
Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ (May 2008).
165:
Messenger RNA (mRNA) is RNA that has a coding region that acts as a template for protein synthesis (
6301:
5912:
598:
3109:"Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein"
2517:"mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation"
123:
6047:
5930:
4758:
4658:
4514:
3936:
3306:
3089:
3046:
2608:
Vinciguerra P, Stutz F (June 2004). "mRNA export: an assembly line from genes to nuclear pores".
2497:
2454:
2174:
1982:"An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries"
1566:
1086:
1029:
823:
514:
483:
209:
2085:"RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes"
4483:
LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (June 2005).
4213:"Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals"
1980:
Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S (October 2006).
1643:
Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH (2006).
581:
Polyadenylation in bacteria helps polynucleotide phosphorylase degrade past secondary structure
6296:
6131:
5729:
5675:
5613:
5603:
5566:
5471:
5420:
5379:
Neeleman, Lyda; Olsthoorn, René C. L.; Linthorst, Huub J. M.; Bol, John F. (4 December 2001).
5361:
5302:
5251:
5213:
5177:
5136:
5087:
5077:
5046:
4997:
4948:
4899:
4864:
4815:
4797:
4712:
4650:
4615:
4566:
4506:
4465:
4416:
4381:
4332:
4283:
4242:
4193:
4144:
4087:
4029:
3985:
3928:
3892:
3843:
3794:
3737:
3688:
3670:
3631:
3613:
3574:
3556:
3515:
3497:
3455:
3406:
3357:
3298:
3257:
3222:
3173:
3138:
3081:
3038:
3020:
2981:
2932:
2873:
2824:
2772:
2723:
2674:
2625:
2590:
2546:
2489:
2446:
2411:
2362:
2321:
2272:
2223:
2166:
2114:
2065:
2036:"Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation"
2011:
1959:
1910:
1861:
1802:
1761:
1709:
1674:
1622:
1558:
1523:
1474:
1415:
1380:
1328:
1274:
1237:
1186:
1159:
1124:
1078:
1021:
978:
929:
875:
815:
550:
465:
278:
6376:
6371:
5156:"Mycoplasma gallisepticum as the first analyzed bacterium in which RNA is not polyadenylated"
4352:"TREND-DB-a transcriptome-wide atlas of the dynamic landscape of alternative polyadenylation"
6381:
6356:
5719:
5711:
5665:
5655:
5595:
5556:
5523:
5461:
5451:
5410:
5400:
5351:
5341:
5292:
5282:
5243:
5205:
5167:
5126:
5118:
5069:
5036:
5028:
4987:
4979:
4968:"RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R"
4938:
4930:
4891:
4854:
4846:
4805:
4789:
4750:
4702:
4694:
4642:
4605:
4597:
4556:
4548:
4496:
4455:
4447:
4408:
4371:
4363:
4322:
4314:
4273:
4232:
4224:
4183:
4175:
4134:
4126:
4077:
4069:
4019:
3975:
3967:
3920:
3882:
3874:
3833:
3825:
3784:
3776:
3727:
3719:
3678:
3662:
3621:
3605:
3564:
3546:
3505:
3489:
3445:
3437:
3396:
3388:
3347:
3337:
3288:
3249:
3212:
3204:
3165:
3128:
3120:
3073:
3028:
3012:
2971:
2963:
2922:
2912:
2863:
2855:
2814:
2806:
2762:
2754:
2713:
2705:
2664:
2656:
2617:
2580:
2536:
2528:
2481:
2438:
2401:
2393:
2352:
2311:
2303:
2262:
2254:
2213:
2205:
2158:
2104:
2096:
2055:
2047:
2001:
1993:
1949:
1941:
1900:
1892:
1851:
1841:
1792:
1751:
1743:
1701:
1664:
1656:
1612:
1604:
1550:
1513:
1505:
1464:
1454:
1407:
1370:
1362:
1318:
1308:
1264:
1227:
1219:
1151:
1116:
1068:
1013:
968:
960:
919:
911:
865:
857:
805:
732:
645:
424:
196:
mRNAs. These are the only mRNAs in eukaryotes that lack a poly(A) tail, ending instead in a
5594:. Progress in Nucleic Acid Research and Molecular Biology. Vol. 71. pp. 285–389.
6454:
6255:
6238:
6233:
6108:
5791:
5489:
3064:
Wilusz CJ, Wormington M, Peltz SW (April 2001). "The cap-to-tail guide to mRNA turnover".
677:
633:
566:
526:
351:
263:
258:
83:
61:
4399:
Reinisch KM, Wolin SL (April 2007). "Emerging themes in non-coding RNA quality control".
3208:
2952:"Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation"
1692:
Lutz CS (October 2008). "Alternative polyadenylation: a twist on mRNA 3′ end formation".
5651:
5396:
5337:
4746:
4122:
4065:
3772:
2908:
2154:
1837:
1450:
1155:
1064:
1009:
309:
When the RNA is cleaved, polyadenylation starts, catalysed by polyadenylate polymerase.
6419:
6318:
6203:
6198:
5935:
5907:
5724:
5699:
5466:
5439:
5356:
5321:
5269:
Poon, Leo L. M.; Pritlove, David C.; Fodor, Ervin; Brownlee, George G. (1 April 1999).
5131:
5106:
4992:
4967:
4859:
4834:
4810:
4777:
4610:
4585:
4561:
4536:
4460:
4435:
4434:
Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E (June 2011).
4376:
4351:
4327:
4302:
4237:
4212:
4188:
4163:
4139:
4106:
4082:
4049:
3980:
3955:
3887:
3862:
3838:
3813:
3789:
3756:
3732:
3707:
3683:
3650:
3626:
3593:
3569:
3534:
3510:
3477:
3450:
3425:
3352:
3325:
3217:
3192:
3133:
3108:
3033:
3000:
2976:
2951:
2927:
2892:
2819:
2794:
2767:
2742:
2718:
2693:
2316:
2291:
2267:
2242:
2109:
2084:
2060:
2035:
2006:
1981:
1954:
1929:
1905:
1880:
1856:
1821:
1669:
1644:
1608:
1518:
1493:
1469:
1434:
1375:
1350:
1323:
1296:
1232:
1207:
973:
948:
870:
845:
609:
538:
384:
282:
99:
5700:"3′ end mRNA processing: molecular mechanisms and implications for health and disease"
5670:
5635:
5599:
5528:
5511:
5297:
5270:
5073:
5041:
5016:
4943:
4918:
4707:
4682:
3814:"Expression and function of micro-RNAs in immune cells during normal or disease state"
3426:"3′ end mRNA processing: molecular mechanisms and implications for health and disease"
3401:
3376:
2868:
2843:
2669:
2644:
2541:
2516:
2406:
2381:
2218:
2193:
1797:
1781:"A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im"
1780:
1756:
1731:
1617:
1592:
1411:
861:
810:
793:
6471:
6136:
5823:
5415:
5380:
5172:
5155:
3924:
3050:
1570:
1120:
924:
899:
709:
613:
562:
546:
420:
400:
322:
293:
and occurs 10–30 nucleotides downstream of its binding site. This site often has the
118:
37:
6147:
5287:
4762:
4662:
4518:
3310:
2178:
1090:
1033:
6265:
6186:
6010:
5940:
5809:
5631:
5587:
5507:
3940:
3093:
2501:
2458:
915:
697:
542:
372:
274:
3861:
Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B (November 2008).
2162:
827:
306:
site associated with a polyadenylation signal can vary up to some 50 nucleotides.
5346:
5247:
4895:
4793:
4485:"RNA degradation by the exosome is promoted by a nuclear polyadenylation complex"
4278:
4261:
3609:
2567:
Siddiqui N, Mangus DA, Chang TC, Palermino JM, Shyu AB, Gehring K (August 2007).
1645:"Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis"
1179:
996:
Zhuang Y, Zhang H, Lin S (June 2013). "Polyadenylation of 18S rRNA in algae(1)".
415:
There is polyadenylation in the cytosol of some animal cell types, namely in the
6444:
6291:
6250:
6113:
6042:
2967:
724:
720:
673:
586:
331:
4501:
4484:
4451:
4073:
3293:
3276:
3253:
3193:"Virtues and limitations of the preimplantation mouse embryo as a model system"
3169:
2897:
Proceedings of the
National Academy of Sciences of the United States of America
2660:
1826:
Proceedings of the
National Academy of Sciences of the United States of America
1439:
Proceedings of the
National Academy of Sciences of the United States of America
479:
395:
127:
Chemical structure of RNA. The sequence of bases differs between RNA molecules.
6071:
4754:
4412:
3723:
3594:"Global patterns of tissue-specific alternative polyadenylation in Drosophila"
3551:
2793:
Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH (2007).
2621:
2341:"Poly(A) tail length control is caused by termination of processive synthesis"
2258:
1896:
1554:
1073:
964:
740:
701:
510:
326:
5715:
5456:
5122:
4801:
4262:"p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing"
4228:
4107:"HITS-CLIP yields genome-wide insights into brain alternative RNA processing"
3674:
3617:
3560:
3501:
3441:
3024:
1997:
5015:
Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G (September 2003).
4698:
3780:
3016:
2917:
2645:"Multiple portions of poly(A)-binding protein stimulate translation in vivo"
1846:
1459:
1313:
716:
661:
189:
49:
21:
5733:
5660:
5617:
5561:
5544:
5475:
5424:
5405:
5365:
5306:
5255:
5217:
5181:
5140:
5091:
5050:
5001:
4952:
4934:
4903:
4868:
4819:
4716:
4683:"Comparative genomics and evolution of proteins involved in RNA metabolism"
4654:
4619:
4570:
4510:
4469:
4420:
4385:
4336:
4287:
4246:
4197:
4148:
4091:
4033:
4024:
4007:
3989:
3896:
3847:
3798:
3741:
3692:
3635:
3578:
3519:
3459:
3410:
3361:
3302:
3261:
3226:
3177:
3142:
3085:
3042:
2985:
2936:
2877:
2828:
2776:
2727:
2678:
2629:
2594:
2585:
2568:
2532:
2493:
2450:
2415:
2397:
2380:
Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W (August 2002).
2357:
2340:
2325:
2276:
2227:
2118:
2069:
2015:
1963:
1914:
1865:
1806:
1765:
1713:
1678:
1562:
1527:
1478:
1419:
1384:
1332:
1278:
1241:
1163:
1082:
1025:
982:
933:
879:
819:
557:, polyadenylation is a way of marking the RNA for degradation, at least in
5749:
5679:
5570:
4367:
3932:
2550:
2366:
2170:
1626:
1128:
747:) in order to emphasize their own genes' expression over the host cell's.
277:
polyadenylation complex in the nucleus of eukaryotes works on products of
6091:
6081:
6005:
4983:
4647:
10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2
4601:
3878:
3666:
3493:
3392:
3377:"A large-scale analysis of mRNA polyadenylation of human and mouse genes"
3124:
2810:
2758:
2307:
2051:
1747:
1660:
1269:
1256:
518:
494:
416:
404:
205:
140:
57:
4850:
4130:
1945:
1509:
297:
sequence AAUAAA on the RNA, but variants of it that bind more weakly to
6411:
6391:
6386:
5827:
5032:
4552:
4436:"The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex"
4318:
4179:
3971:
3342:
2859:
1730:
Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D (July 2000).
1366:
1223:
705:
629:
625:
193:
144:
136:
52:, polyadenylation is part of the process that produces mature mRNA for
45:
5209:
3829:
2709:
1705:
1017:
6366:
6361:
6341:
6277:
6228:
3077:
2485:
2442:
2209:
2100:
1541:
Amaral PP, Mattick JS (August 2008). "Noncoding RNA in development".
756:
744:
554:
428:
380:
335:
253:
148:
5592:
A history of poly A sequences: from formation to factors to function
1494:"A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis"
1142:
Stevens A (1963). "Ribonucleic Acids-Biosynthesis and
Degradation".
2292:"Molecular dissection of mRNA poly(A) tail length control in yeast"
6434:
6429:
6351:
6346:
6336:
6331:
6326:
6286:
5849:
5760:
2290:
Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L (April 2008).
2034:
Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ (May 2008).
736:
693:
502:
498:
much less common than just shortening the 3′ untranslated region.
459:
5490:"Inhibition of host poly(A)-binding protein by virus ~ ViralZone"
5320:
Wu, Hung-Yi; Ke, Ting-Yung; Liao, Wei-Yu; Chang, Nai-Yun (2013).
4303:"Regulation of alternative polyadenylation by genomic imprinting"
3755:
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (June 2008).
3275:
Piqué M, López JM, Foissac S, Guigó R, Méndez R (February 2008).
2950:
Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF (April 2008).
1732:"Patterns of variant polyadenylation signal usage in human genes"
469:
Results of using different polyadenylation sites on the same gene
6449:
6439:
6281:
5322:"Regulation of Coronaviral Poly(A) Tail Length during Infection"
5236:
Biochimica et
Biophysica Acta (BBA) - Gene Regulatory Mechanisms
4884:
Biochimica et
Biophysica Acta (BBA) - Gene Regulatory Mechanisms
4782:
Biochimica et
Biophysica Acta (BBA) - Gene Regulatory Mechanisms
3324:
Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M (June 2008).
1593:"Assembly of a processive messenger RNA polyadenylation complex"
1107:
Sarkar N (June 1997). "Polyadenylation of mRNA in prokaryotes".
769:
455:
451:
391:
298:
290:
217:
72:
6151:
5764:
2083:
Glover-Cutter K, Kim S, Espinosa J, Bentley DL (January 2008).
1492:
Yoshikawa M, Peragine A, Park MY, Poethig RS (September 2005).
5819:
446:
Cytoplasmic polyadenylation requires the RNA-binding proteins
359:
158:
114:
3277:"A combinatorial code for CPE-mediated translational control"
2741:
Kargapolova Y, Levin M, Lackner K, Danckwardt S (June 2017).
82:
segment of the newly made pre-mRNA is first cleaved off by a
2844:"A protein interaction framework for human mRNA degradation"
2643:
Gray NK, Coller JM, Dickson KS, Wickens M (September 2000).
489:
Since alternative polyadenylation changes the length of the
684:
concentrations than other nucleotides as a result of using
493:, it can also change which binding sites are available for
4006:
Shell SA, Hesse C, Morris SM, Milcarek C (December 2005).
5068:. Methods in Enzymology. Vol. 447. pp. 501–20.
1433:
Saini HK, Griffiths-Jones S, Enright AJ (November 2007).
3706:
Ogorodnikov A, Kargapolova Y, Danckwardt S (June 2016).
2194:"Long-range heterogeneity at the 3′ ends of human mRNAs"
1879:
Yang Q, Coseno M, Gilmartin GM, Doublié S (March 2011).
624:
itself. This enzyme is found in bacteria, mitochondria,
4833:
Chang SA, Cozad M, Mackie GA, Jones GH (January 2008).
3240:
Richter JD (June 2007). "CPEB: a life in translation".
1049:"RNA turnover: unexpected consequences of being tailed"
900:"Cytoplasmic polyadenylation in development and beyond"
628:
and as a constituent of the archaeal exosome (in those
44:; in other words, it is a stretch of RNA that has only
4778:"Mitochondrial poly(A) polymerase and polyadenylation"
4162:
Hall-Pogar T, Liang S, Hague LK, Lutz CS (July 2007).
700:. Its catalytic domain is homologous to that of other
5698:
Danckwardt S, Hentze MW, Kulozik AE (February 2008).
3535:"Biased alternative polyadenylation in human tissues"
3424:
Danckwardt S, Hentze MW, Kulozik AE (February 2008).
755:
Poly(A)polymerase was first identified in 1960 as an
561:. This polyadenylation is done in the nucleus by the
1928:
Venkataraman K, Brown KM, Gilmartin GM (June 2005).
696:, which is the enzyme that completes the 3′ ends of
6410:
6317:
6264:
6185:
6124:
6033:
5998:
5972:
5963:
5921:
5895:
5869:
5860:
5798:
4681:Anantharaman V, Koonin EV, Aravind L (April 2002).
4584:Slomovic S, Laufer D, Geiger D, Schuster G (2006).
3158:
Biochemical and Biophysical Research Communications
1351:"Early evolution of histone mRNA 3′ end processing"
16:
Addition of adenylic acids to 3' end of mature mRNA
5545:"Mechanism and regulation of mRNA polyadenylation"
1178:
454:, and can involve other RNA-binding proteins like
224: – a poly(A) tail is part of the mature RNA.
4586:"Polyadenylation of ribosomal RNA in human cells"
846:"Regulation of mRNA stability in mammalian cells"
5066:RNA Turnover in Bacteria, Archaea and Organelles
3478:"Alternative polyadenylation of mRNA precursors"
1435:"Genomic analysis of human microRNA transcripts"
794:"Integrating mRNA processing with transcription"
792:Proudfoot NJ, Furger A, Dye MJ (February 2002).
5640:Proceedings of the National Academy of Sciences
5385:Proceedings of the National Academy of Sciences
4676:
4674:
4672:
4530:
4528:
2515:Coller JM, Gray NK, Wickens MP (October 1998).
1591:Bienroth S, Keller W, Wahle E (February 1993).
1208:"Trading translation with RNA-binding proteins"
5634:; Vaughan, M. H.; Nakazato, H. (1 June 1971).
4350:Marini F, Scherzinger D, Danckwardt S (2021).
3533:Zhang, Haibo; Lee, Ju Youn; Tian, Bin (2005).
2893:"MicroRNAs direct rapid deadenylation of mRNA"
2562:
2560:
1177:Lehninger AL, Nelson DL, Cox MM, eds. (1993).
346:The poly(A) tail acts as the binding site for
40:(mRNA). The poly(A) tail consists of multiple
6163:
5776:
4917:Walter M, Kilian J, Kudla J (December 2002).
4728:
4726:
3812:Tili E, Michaille JJ, Calin GA (April 2008).
1820:Yang Q, Gilmartin GM, Doublié S (June 2010).
1349:Dávila López M, Samuelsson T (January 2008).
949:"Emerging features of mRNA decay in bacteria"
743:, inhibit the cell's poly-A binding protein (
241:: cleavage/polyadenylation specificity factor
67:The process of polyadenylation begins as the
25:Typical structure of a mature eukaryotic mRNA
8:
2788:
2786:
1725:
1723:
715:Polyadenylate tails are observed in several
712:, never evolved a polyadenylate polymerase.
672:. This enzyme is part of both the bacterial
589:, which contains two RNA-degrading enzymes:
5582:
5580:
4537:"RNA-specific ribonucleotidyl transferases"
3107:Jung MY, Lorenz L, Richter JD (June 2006).
1586:
1584:
1582:
1580:
787:
785:
668:The most ancient polyadenylating enzyme is
6270:
6191:
6170:
6156:
6148:
5969:
5931:Precursor mRNA (pre-mRNA / hnRNA)
5866:
5783:
5769:
5761:
1344:
1342:
904:Microbiology and Molecular Biology Reviews
5723:
5669:
5659:
5560:
5527:
5465:
5455:
5414:
5404:
5355:
5345:
5296:
5286:
5171:
5130:
5040:
4991:
4942:
4858:
4809:
4706:
4609:
4560:
4500:
4459:
4375:
4326:
4277:
4236:
4187:
4138:
4081:
4023:
3979:
3886:
3837:
3818:International Journal of Medical Sciences
3788:
3731:
3682:
3649:Lee, Ju Youn; Ji, Zhe; Tian, Bin (2008).
3625:
3568:
3550:
3509:
3449:
3400:
3351:
3341:
3292:
3216:
3132:
3032:
2975:
2926:
2916:
2867:
2818:
2766:
2717:
2668:
2584:
2540:
2431:Nature Structural & Molecular Biology
2405:
2356:
2315:
2266:
2217:
2108:
2089:Nature Structural & Molecular Biology
2059:
2005:
1953:
1904:
1855:
1845:
1796:
1755:
1668:
1616:
1517:
1468:
1458:
1374:
1322:
1312:
1268:
1231:
1072:
972:
923:
893:
891:
889:
869:
839:
837:
809:
216:RNAs that, for example, includes the RNA
2029:
2027:
2025:
1779:Brown KM, Gilmartin GM (December 2003).
1102:
1100:
576:
464:
122:
20:
3954:Tian B, Pan Z, Lee JY (February 2007).
3375:Tian B, Hu J, Zhang H, Lutz CS (2005).
1975:
1973:
1290:
1288:
781:
5543:Colgan DF, Manley JL (November 1997).
5229:
5227:
3482:Nature Reviews. Molecular Cell Biology
2891:Wu L, Fan J, Belasco JG (March 2006).
1638:
1636:
608:In as different groups as animals and
5951:Histone acetylation and deacetylation
4776:Chang, Jeong Ho; Tong, Liang (2012).
4401:Current Opinion in Structural Biology
4001:
3999:
3471:
3469:
3066:Nature Reviews Molecular Cell Biology
1255:Mattick JS, Makunin IV (April 2006).
844:Guhaniyogi J, Brewer G (March 2001).
533:Tagging for degradation in eukaryotes
7:
6016:Ribosome-nascent chain complex (RNC)
4535:Martin G, Keller W (November 2007).
3476:Tian, Bin; Manley, James L. (2017).
3209:10.1016/j.theriogenology.2007.09.032
2842:Lehner B, Sanderson CM (July 2004).
5154:Portnoy V, Schuster G (June 2008).
4012:The Journal of Biological Chemistry
2573:The Journal of Biological Chemistry
2345:The Journal of Biological Chemistry
2241:Balbo PB, Bohm A (September 2007).
1156:10.1146/annurev.bi.32.070163.000311
212: – a seemingly large group of
4735:Critical Reviews in Plant Sciences
1609:10.1002/j.1460-2075.1993.tb05690.x
403:and human cells, most notably the
390:In animals, poly(A) ribonuclease (
313:builds the poly(A) tail by adding
36:to an RNA transcript, typically a
14:
6179:Post-transcriptional modification
2692:Meaux S, Van Hoof A (July 2006).
1206:Abaza I, Gebauer F (March 2008).
1185:(2nd ed.). New York: Worth.
256:: polyadenylate binding protein 2
5748:
5510:; Abrams, Richard (April 1960).
5173:10.1111/j.1574-6968.2008.01157.x
1263:. 15 Spec No 1 (90001): R17-29.
1121:10.1146/annurev.biochem.66.1.173
358:-4G, which in turn recruits the
285:. Here, a multi-protein complex
6021:Post-translational modification
5516:Journal of Biological Chemistry
5288:10.1128/JVI.73.4.3473-3476.1999
2610:Current Opinion in Cell Biology
659:and the salt-tolerant archaean
4966:Portnoy V, Schuster G (2006).
3242:Trends in Biochemical Sciences
3113:Molecular and Cellular Biology
916:10.1128/MMBR.63.2.446-456.1999
650:last universal common ancestor
1:
5600:10.1016/S0079-6603(02)71046-5
5529:10.1016/S0021-9258(18)69494-3
5074:10.1016/S0076-6879(08)02224-6
2163:10.1126/science.274.5292.1517
1798:10.1016/S1097-2765(03)00453-2
1412:10.1016/S0888-7543(02)96850-3
1144:Annual Review of Biochemistry
1109:Annual Review of Biochemistry
862:10.1016/S0378-1119(01)00350-X
811:10.1016/S0092-8674(02)00617-7
573:In prokaryotes and organelles
287:(see components on the right)
246:: cleavage stimulation factor
171:). The rest of the mRNA, the
5347:10.1371/journal.pone.0070548
5248:10.1016/j.bbagrm.2007.12.004
4896:10.1016/j.bbagrm.2008.02.001
4794:10.1016/j.bbagrm.2011.10.012
4279:10.1016/j.molcel.2010.12.032
3925:10.1016/0092-8674(80)90615-7
3610:10.1016/j.celrep.2012.01.001
670:polynucleotide phosphorylase
622:polynucleotide phosphorylase
591:polynucleotide phosphorylase
2968:10.1534/genetics.107.084558
1047:Anderson JT (August 2005).
507:cleavage stimulatory factor
474:Alternative polyadenylation
411:Cytoplasmic polyadenylation
88:alternative polyadenylation
6501:
4502:10.1016/j.cell.2005.04.029
4452:10.1016/j.cell.2011.05.010
4074:10.1038/s41467-018-07580-5
3294:10.1016/j.cell.2007.12.038
3254:10.1016/j.tibs.2007.04.004
3170:10.1016/j.bbrc.2005.08.250
1181:Principles of biochemistry
251:: polyadenylate polymerase
112:
6273:
6194:
5444:Frontiers in Microbiology
5160:FEMS Microbiology Letters
4755:10.1080/07352680500391337
4413:10.1016/j.sbi.2007.03.012
3724:10.1007/s00424-016-1828-3
3552:10.1186/gb-2005-6-12-r100
2622:10.1016/j.ceb.2004.03.013
2339:Wahle E (February 1995).
2259:10.1016/j.str.2007.07.010
1897:10.1016/j.str.2010.12.021
1555:10.1007/s00335-008-9136-7
1074:10.1016/j.cub.2005.08.002
965:10.1017/S1355838200001023
947:Steege DA (August 2000).
648:of life implies that the
334:, a complex that removes
321:to the RNA, cleaving off
222:X chromosome inactivation
6082:sequestration (P-bodies)
5716:10.1038/sj.emboj.7601932
5457:10.3389/fmicb.2018.02250
5123:10.1038/sj.embor.7400571
4229:10.1038/sj.emboj.7601699
3442:10.1038/sj.emboj.7601932
3191:Taft RA (January 2008).
2661:10.1093/emboj/19.17.4723
1998:10.1038/sj.emboj.7601331
1261:Human Molecular Genetics
898:Richter JD (June 1999).
735:. Some viruses, such as
665:lack this modification.
656:Mycoplasma gallisepticum
311:Polyadenylate polymerase
42:adenosine monophosphates
6246:Poly(A)-binding protein
6060:Gene regulatory network
5549:Genes & Development
4839:Journal of Bacteriology
4307:Genes & Development
3781:10.1126/science.1155390
3017:10.1126/science.abe6523
2918:10.1073/pnas.0510928103
2521:Genes & Development
2474:Mayo Clinic Proceedings
1934:Genes & Development
1847:10.1073/pnas.1000848107
1498:Genes & Development
1460:10.1073/pnas.0703890104
1314:10.1186/1471-2164-9-220
348:poly(A)-binding protein
315:adenosine monophosphate
179:Nuclear polyadenylation
6065:cis-regulatory element
5661:10.1073/pnas.68.6.1336
5562:10.1101/gad.11.21.2755
5406:10.1073/pnas.251542798
4972:Nucleic Acids Research
4687:Nucleic Acids Research
4590:Nucleic Acids Research
4356:Nucleic Acids Research
4025:10.1074/jbc.M508848200
3867:Nucleic Acids Research
3655:Nucleic Acids Research
3381:Nucleic Acids Research
2799:Nucleic Acids Research
2747:Nucleic Acids Research
2586:10.1074/jbc.M701256200
2533:10.1101/gad.12.20.3226
2358:10.1074/jbc.270.6.2800
2296:Nucleic Acids Research
2040:Nucleic Acids Research
1649:Nucleic Acids Research
689:nucleotides in them).
582:
470:
441:long-term potentiation
377:3′ untranslated region
319:adenosine triphosphate
295:polyadenylation signal
151:). RNAs are produced (
128:
26:
4699:10.1093/nar/30.7.1427
4054:Nature Communications
580:
509:(CstF), increases in
468:
439:could play a role in
266:: cleavage factor II
126:
113:Further information:
32:is the addition of a
24:
6309:Alternative splicing
6087:alternative splicing
6077:Post-transcriptional
5903:Transcription factor
5757:at Wikimedia Commons
5494:viralzone.expasy.org
4935:10.1093/emboj/cdf686
3494:10.1038/nrm.2016.116
3125:10.1128/MCB.02470-05
2398:10.1093/emboj/cdf390
1748:10.1101/gr.10.7.1001
1694:ACS Chemical Biology
998:Journal of Phycology
729:Alfalfa mosaic virus
173:untranslated regions
92:alternative splicing
6011:Transfer RNA (tRNA)
5652:1971PNAS...68.1336E
5397:2001PNAS...9814286N
5391:(25): 14286–14291.
5338:2013PLoSO...870548W
5275:Journal of Virology
4851:10.1128/JB.00327-07
4747:2006CRvPS..25...65S
4368:10.1093/nar/gkaa722
4131:10.1038/nature07488
4123:2008Natur.456..464L
4066:2018NatCo...9.5331O
3773:2008Sci...320.1643S
2909:2006PNAS..103.4034W
2155:1996Sci...274.1517S
1946:10.1101/gad.1298605
1838:2010PNAS..10710062Y
1510:10.1101/gad.1352605
1451:2007PNAS..10417719S
1065:2005CBio...15.R635A
1010:2013JPcgy..49..570Z
599:secondary structure
515:lipopolysaccharides
261:: cleavage factor I
210:long noncoding RNAs
6420:5′ cap methylation
6125:Influential people
6104:Post-translational
5923:Post-transcription
5033:10.1105/tpc.013326
4984:10.1093/nar/gkl763
4602:10.1093/nar/gkl357
4553:10.1261/rna.652807
4362:(D1): D:243–D253.
4319:10.1101/gad.473408
4180:10.1261/rna.577707
3972:10.1101/gr.5532707
3879:10.1093/nar/gkn624
3667:10.1093/nar/gkn540
3393:10.1093/nar/gki158
3343:10.1242/dev.021444
3011:(6529): eabe6523.
2860:10.1101/gr.2122004
2811:10.1093/nar/gkm830
2759:10.1093/nar/gkx152
2308:10.1093/nar/gkn080
2052:10.1093/nar/gkn158
1661:10.1093/nar/gkl919
1367:10.1261/rna.782308
1270:10.1093/hmg/ddl046
1224:10.1261/rna.848208
757:enzymatic activity
662:Haloferax volcanii
583:
471:
394:) can bind to the
381:immature egg cells
342:Downstream effects
235:Proteins involved:
129:
27:
6465:
6464:
6406:
6405:
6402:
6401:
6319:pre-mRNA factors
6145:
6144:
6029:
6028:
5959:
5958:
5835:Special transfers
5753:Media related to
5609:978-0-12-540071-8
5210:10.1021/bi8012214
5083:978-0-12-374377-0
4788:(9–10): 992–997.
3830:10.7150/ijms.5.73
3661:(17): 5581–5590.
2753:(10): 6074–6086.
2710:10.1261/rna.46306
2149:(5292): 1517–20.
1706:10.1021/cb800138w
1192:978-0-87901-500-8
1018:10.1111/jpy.12068
694:CCA-adding enzyme
676:and the archaeal
356:initiation factor
279:RNA polymerase II
271:
270:
220:, which mediates
109:Background on RNA
6490:
6271:
6204:5′ cap formation
6192:
6172:
6165:
6158:
6149:
5970:
5867:
5785:
5778:
5771:
5762:
5752:
5737:
5727:
5704:The EMBO Journal
5684:
5683:
5673:
5663:
5646:(6): 1336–1340.
5628:
5622:
5621:
5584:
5575:
5574:
5564:
5540:
5534:
5533:
5531:
5522:(4): 1142–1149.
5504:
5498:
5497:
5486:
5480:
5479:
5469:
5459:
5435:
5429:
5428:
5418:
5408:
5376:
5370:
5369:
5359:
5349:
5317:
5311:
5310:
5300:
5290:
5281:(4): 3473–3476.
5266:
5260:
5259:
5231:
5222:
5221:
5204:(50): 13158–68.
5192:
5186:
5185:
5175:
5151:
5145:
5144:
5134:
5102:
5096:
5095:
5061:
5055:
5054:
5044:
5012:
5006:
5005:
4995:
4963:
4957:
4956:
4946:
4923:The EMBO Journal
4914:
4908:
4907:
4879:
4873:
4872:
4862:
4830:
4824:
4823:
4813:
4773:
4767:
4766:
4730:
4721:
4720:
4710:
4678:
4667:
4666:
4630:
4624:
4623:
4613:
4581:
4575:
4574:
4564:
4532:
4523:
4522:
4504:
4480:
4474:
4473:
4463:
4431:
4425:
4424:
4396:
4390:
4389:
4379:
4347:
4341:
4340:
4330:
4298:
4292:
4291:
4281:
4257:
4251:
4250:
4240:
4217:The EMBO Journal
4208:
4202:
4201:
4191:
4159:
4153:
4152:
4142:
4102:
4096:
4095:
4085:
4044:
4038:
4037:
4027:
4018:(48): 39950–61.
4003:
3994:
3993:
3983:
3951:
3945:
3944:
3907:
3901:
3900:
3890:
3858:
3852:
3851:
3841:
3809:
3803:
3802:
3792:
3767:(5883): 1643–7.
3752:
3746:
3745:
3735:
3703:
3697:
3696:
3686:
3646:
3640:
3639:
3629:
3589:
3583:
3582:
3572:
3554:
3530:
3524:
3523:
3513:
3473:
3464:
3463:
3453:
3430:The EMBO Journal
3421:
3415:
3414:
3404:
3372:
3366:
3365:
3355:
3345:
3321:
3315:
3314:
3296:
3272:
3266:
3265:
3237:
3231:
3230:
3220:
3188:
3182:
3181:
3153:
3147:
3146:
3136:
3104:
3098:
3097:
3078:10.1038/35067025
3061:
3055:
3054:
3036:
2996:
2990:
2989:
2979:
2947:
2941:
2940:
2930:
2920:
2888:
2882:
2881:
2871:
2839:
2833:
2832:
2822:
2790:
2781:
2780:
2770:
2738:
2732:
2731:
2721:
2689:
2683:
2682:
2672:
2649:The EMBO Journal
2640:
2634:
2633:
2605:
2599:
2598:
2588:
2579:(34): 25067–75.
2564:
2555:
2554:
2544:
2512:
2506:
2505:
2486:10.4065/77.8.785
2469:
2463:
2462:
2443:10.1038/nsmb1253
2426:
2420:
2419:
2409:
2386:The EMBO Journal
2377:
2371:
2370:
2360:
2336:
2330:
2329:
2319:
2287:
2281:
2280:
2270:
2238:
2232:
2231:
2221:
2210:10.1101/gr.62002
2189:
2183:
2182:
2138:
2132:
2129:
2123:
2122:
2112:
2101:10.1038/nsmb1352
2080:
2074:
2073:
2063:
2031:
2020:
2019:
2009:
1986:The EMBO Journal
1977:
1968:
1967:
1957:
1925:
1919:
1918:
1908:
1876:
1870:
1869:
1859:
1849:
1817:
1811:
1810:
1800:
1776:
1770:
1769:
1759:
1727:
1718:
1717:
1689:
1683:
1682:
1672:
1640:
1631:
1630:
1620:
1597:The EMBO Journal
1588:
1575:
1574:
1543:Mammalian Genome
1538:
1532:
1531:
1521:
1489:
1483:
1482:
1472:
1462:
1445:(45): 17719–24.
1430:
1424:
1423:
1395:
1389:
1388:
1378:
1346:
1337:
1336:
1326:
1316:
1292:
1283:
1282:
1272:
1257:"Non-coding RNA"
1252:
1246:
1245:
1235:
1203:
1197:
1196:
1184:
1174:
1168:
1167:
1139:
1133:
1132:
1104:
1095:
1094:
1076:
1044:
1038:
1037:
993:
987:
986:
976:
944:
938:
937:
927:
895:
884:
883:
873:
841:
832:
831:
813:
789:
733:Duck Hepatitis A
232:
231:
208:. But, for many
6500:
6499:
6493:
6492:
6491:
6489:
6488:
6487:
6478:Gene expression
6468:
6467:
6466:
6461:
6398:
6313:
6260:
6256:Polyuridylation
6209:Polyadenylation
6181:
6176:
6146:
6141:
6120:
6055:Transcriptional
6025:
5994:
5955:
5946:Polyadenylation
5917:
5891:
5856:
5850:Protein→Protein
5801:
5794:
5792:Gene expression
5789:
5755:Polyadenylation
5745:
5740:
5697:
5693:
5691:Further reading
5688:
5687:
5630:
5629:
5625:
5610:
5586:
5585:
5578:
5555:(21): 2755–66.
5542:
5541:
5537:
5506:
5505:
5501:
5488:
5487:
5483:
5437:
5436:
5432:
5378:
5377:
5373:
5319:
5318:
5314:
5268:
5267:
5263:
5233:
5232:
5225:
5194:
5193:
5189:
5153:
5152:
5148:
5117:(12): 1188–93.
5104:
5103:
5099:
5084:
5063:
5062:
5058:
5014:
5013:
5009:
4978:(20): 5923–31.
4965:
4964:
4960:
4929:(24): 6905–14.
4916:
4915:
4911:
4881:
4880:
4876:
4832:
4831:
4827:
4775:
4774:
4770:
4732:
4731:
4724:
4680:
4679:
4670:
4632:
4631:
4627:
4596:(10): 2966–75.
4583:
4582:
4578:
4547:(11): 1834–49.
4534:
4533:
4526:
4482:
4481:
4477:
4433:
4432:
4428:
4398:
4397:
4393:
4349:
4348:
4344:
4300:
4299:
4295:
4259:
4258:
4254:
4223:(11): 2658–69.
4210:
4209:
4205:
4161:
4160:
4156:
4117:(7221): 464–9.
4104:
4103:
4099:
4046:
4045:
4041:
4005:
4004:
3997:
3960:Genome Research
3953:
3952:
3948:
3909:
3908:
3904:
3873:(19): 6318–32.
3860:
3859:
3855:
3811:
3810:
3806:
3754:
3753:
3749:
3718:(6): 993–1012.
3712:Pflügers Archiv
3705:
3704:
3700:
3648:
3647:
3643:
3591:
3590:
3586:
3532:
3531:
3527:
3475:
3474:
3467:
3423:
3422:
3418:
3374:
3373:
3369:
3336:(11): 1969–79.
3323:
3322:
3318:
3274:
3273:
3269:
3239:
3238:
3234:
3190:
3189:
3185:
3155:
3154:
3150:
3119:(11): 4277–87.
3106:
3105:
3101:
3063:
3062:
3058:
2998:
2997:
2993:
2949:
2948:
2944:
2890:
2889:
2885:
2848:Genome Research
2841:
2840:
2836:
2792:
2791:
2784:
2740:
2739:
2735:
2691:
2690:
2686:
2655:(17): 4723–33.
2642:
2641:
2637:
2607:
2606:
2602:
2566:
2565:
2558:
2527:(20): 3226–35.
2514:
2513:
2509:
2471:
2470:
2466:
2428:
2427:
2423:
2392:(15): 4125–35.
2379:
2378:
2374:
2338:
2337:
2333:
2289:
2288:
2284:
2240:
2239:
2235:
2198:Genome Research
2191:
2190:
2186:
2140:
2139:
2135:
2130:
2126:
2082:
2081:
2077:
2033:
2032:
2023:
1992:(20): 4854–64.
1979:
1978:
1971:
1940:(11): 1315–27.
1927:
1926:
1922:
1878:
1877:
1873:
1832:(22): 10062–7.
1819:
1818:
1814:
1778:
1777:
1773:
1736:Genome Research
1729:
1728:
1721:
1691:
1690:
1686:
1642:
1641:
1634:
1590:
1589:
1578:
1549:(7–8): 454–92.
1540:
1539:
1535:
1504:(18): 2164–75.
1491:
1490:
1486:
1432:
1431:
1427:
1397:
1396:
1392:
1348:
1347:
1340:
1294:
1293:
1286:
1254:
1253:
1249:
1205:
1204:
1200:
1193:
1176:
1175:
1171:
1141:
1140:
1136:
1106:
1105:
1098:
1053:Current Biology
1046:
1045:
1041:
995:
994:
990:
946:
945:
941:
897:
896:
887:
843:
842:
835:
791:
790:
783:
778:
766:
753:
642:
575:
539:non-coding RNAs
535:
527:DNA methylation
513:in response to
505:, a subunit of
484:Ribo-sequencing
476:
419:, during early
413:
379:of an mRNA. In
369:
344:
262:
257:
252:
247:
242:
236:
230:
186:
181:
121:
111:
100:non-coding RNAs
84:set of proteins
62:gene expression
30:Polyadenylation
17:
12:
11:
5:
6498:
6497:
6494:
6486:
6485:
6480:
6470:
6469:
6463:
6462:
6460:
6459:
6458:
6457:
6452:
6447:
6442:
6437:
6432:
6425:mRNA decapping
6422:
6416:
6414:
6408:
6407:
6404:
6403:
6400:
6399:
6397:
6396:
6395:
6394:
6389:
6384:
6379:
6374:
6369:
6364:
6359:
6354:
6349:
6344:
6339:
6334:
6323:
6321:
6315:
6314:
6312:
6311:
6306:
6305:
6304:
6299:
6289:
6284:
6274:
6268:
6262:
6261:
6259:
6258:
6253:
6248:
6243:
6242:
6241:
6236:
6231:
6226:
6221:
6216:
6206:
6201:
6199:Precursor mRNA
6195:
6189:
6183:
6182:
6177:
6175:
6174:
6167:
6160:
6152:
6143:
6142:
6140:
6139:
6134:
6132:François Jacob
6128:
6126:
6122:
6121:
6119:
6118:
6117:
6116:
6111:
6101:
6096:
6095:
6094:
6089:
6084:
6074:
6069:
6068:
6067:
6062:
6052:
6051:
6050:
6039:
6037:
6031:
6030:
6027:
6026:
6024:
6023:
6018:
6013:
6008:
6002:
6000:
5996:
5995:
5993:
5992:
5987:
5982:
5976:
5974:
5967:
5961:
5960:
5957:
5956:
5954:
5953:
5948:
5943:
5938:
5933:
5927:
5925:
5919:
5918:
5916:
5915:
5910:
5908:RNA polymerase
5905:
5899:
5897:
5893:
5892:
5890:
5889:
5884:
5879:
5873:
5871:
5864:
5858:
5857:
5855:
5854:
5853:
5852:
5847:
5842:
5832:
5831:
5830:
5812:
5806:
5804:
5796:
5795:
5790:
5788:
5787:
5780:
5773:
5765:
5759:
5758:
5744:
5743:External links
5741:
5739:
5738:
5694:
5692:
5689:
5686:
5685:
5623:
5608:
5576:
5535:
5499:
5481:
5430:
5371:
5312:
5261:
5223:
5187:
5146:
5097:
5082:
5056:
5027:(9): 2003–19.
5021:The Plant Cell
5007:
4958:
4909:
4874:
4825:
4768:
4722:
4693:(7): 1427–64.
4668:
4625:
4576:
4524:
4475:
4446:(6): 890–901.
4426:
4391:
4342:
4293:
4272:(3): 298–310.
4266:Molecular Cell
4252:
4203:
4174:(7): 1103–15.
4154:
4097:
4039:
3995:
3946:
3919:(2): 293–301.
3902:
3853:
3804:
3747:
3698:
3641:
3604:(3): 277–289.
3584:
3539:Genome Biology
3525:
3465:
3416:
3367:
3316:
3267:
3232:
3197:Theriogenology
3183:
3148:
3099:
3056:
2991:
2962:(4): 2017–29.
2942:
2903:(11): 4034–9.
2883:
2854:(7): 1315–23.
2834:
2782:
2733:
2704:(7): 1323–37.
2684:
2635:
2600:
2556:
2507:
2480:(8): 785–808.
2464:
2421:
2372:
2331:
2302:(7): 2418–33.
2282:
2253:(9): 1117–31.
2233:
2204:(7): 1068–74.
2184:
2133:
2124:
2075:
2046:(9): 3150–61.
2021:
1969:
1920:
1871:
1812:
1791:(6): 1467–76.
1785:Molecular Cell
1771:
1742:(7): 1001–10.
1719:
1700:(10): 609–17.
1684:
1632:
1576:
1533:
1484:
1425:
1390:
1338:
1284:
1247:
1198:
1191:
1169:
1134:
1096:
1059:(16): R635-8.
1039:
988:
959:(8): 1079–90.
939:
885:
856:(1–2): 11–23.
833:
780:
779:
777:
774:
773:
772:
765:
762:
752:
749:
641:
638:
574:
571:
534:
531:
475:
472:
412:
409:
385:egg activation
371:In eukaryotic
368:
365:
343:
340:
283:precursor mRNA
269:
268:
229:
226:
185:
182:
180:
177:
110:
107:
90:), similar to
15:
13:
10:
9:
6:
4:
3:
2:
6496:
6495:
6484:
6483:Messenger RNA
6481:
6479:
6476:
6475:
6473:
6456:
6453:
6451:
6448:
6446:
6443:
6441:
6438:
6436:
6433:
6431:
6428:
6427:
6426:
6423:
6421:
6418:
6417:
6415:
6413:
6409:
6393:
6390:
6388:
6385:
6383:
6380:
6378:
6375:
6373:
6370:
6368:
6365:
6363:
6360:
6358:
6355:
6353:
6350:
6348:
6345:
6343:
6340:
6338:
6335:
6333:
6330:
6329:
6328:
6325:
6324:
6322:
6320:
6316:
6310:
6307:
6303:
6300:
6298:
6295:
6294:
6293:
6290:
6288:
6285:
6283:
6279:
6276:
6275:
6272:
6269:
6267:
6263:
6257:
6254:
6252:
6249:
6247:
6244:
6240:
6237:
6235:
6232:
6230:
6227:
6225:
6222:
6220:
6217:
6215:
6212:
6211:
6210:
6207:
6205:
6202:
6200:
6197:
6196:
6193:
6190:
6188:
6184:
6180:
6173:
6168:
6166:
6161:
6159:
6154:
6153:
6150:
6138:
6137:Jacques Monod
6135:
6133:
6130:
6129:
6127:
6123:
6115:
6112:
6110:
6107:
6106:
6105:
6102:
6100:
6099:Translational
6097:
6093:
6090:
6088:
6085:
6083:
6080:
6079:
6078:
6075:
6073:
6070:
6066:
6063:
6061:
6058:
6057:
6056:
6053:
6049:
6046:
6045:
6044:
6041:
6040:
6038:
6036:
6032:
6022:
6019:
6017:
6014:
6012:
6009:
6007:
6004:
6003:
6001:
5997:
5991:
5988:
5986:
5983:
5981:
5978:
5977:
5975:
5971:
5968:
5966:
5962:
5952:
5949:
5947:
5944:
5942:
5939:
5937:
5934:
5932:
5929:
5928:
5926:
5924:
5920:
5914:
5911:
5909:
5906:
5904:
5901:
5900:
5898:
5894:
5888:
5885:
5883:
5880:
5878:
5875:
5874:
5872:
5868:
5865:
5863:
5862:Transcription
5859:
5851:
5848:
5846:
5843:
5841:
5838:
5837:
5836:
5833:
5829:
5825:
5821:
5818:
5817:
5816:
5815:Central dogma
5813:
5811:
5808:
5807:
5805:
5803:
5797:
5793:
5786:
5781:
5779:
5774:
5772:
5767:
5766:
5763:
5756:
5751:
5747:
5746:
5742:
5735:
5731:
5726:
5721:
5717:
5713:
5710:(3): 482–98.
5709:
5705:
5701:
5696:
5695:
5690:
5681:
5677:
5672:
5667:
5662:
5657:
5653:
5649:
5645:
5641:
5637:
5633:
5627:
5624:
5619:
5615:
5611:
5605:
5601:
5597:
5593:
5589:
5583:
5581:
5577:
5572:
5568:
5563:
5558:
5554:
5550:
5546:
5539:
5536:
5530:
5525:
5521:
5517:
5513:
5509:
5508:Edmonds, Mary
5503:
5500:
5495:
5491:
5485:
5482:
5477:
5473:
5468:
5463:
5458:
5453:
5449:
5445:
5441:
5434:
5431:
5426:
5422:
5417:
5412:
5407:
5402:
5398:
5394:
5390:
5386:
5382:
5375:
5372:
5367:
5363:
5358:
5353:
5348:
5343:
5339:
5335:
5332:(7): e70548.
5331:
5327:
5323:
5316:
5313:
5308:
5304:
5299:
5294:
5289:
5284:
5280:
5276:
5272:
5265:
5262:
5257:
5253:
5249:
5245:
5242:(4): 247–55.
5241:
5237:
5230:
5228:
5224:
5219:
5215:
5211:
5207:
5203:
5199:
5191:
5188:
5183:
5179:
5174:
5169:
5166:(1): 97–103.
5165:
5161:
5157:
5150:
5147:
5142:
5138:
5133:
5128:
5124:
5120:
5116:
5112:
5108:
5101:
5098:
5093:
5089:
5085:
5079:
5075:
5071:
5067:
5060:
5057:
5052:
5048:
5043:
5038:
5034:
5030:
5026:
5022:
5018:
5011:
5008:
5003:
4999:
4994:
4989:
4985:
4981:
4977:
4973:
4969:
4962:
4959:
4954:
4950:
4945:
4940:
4936:
4932:
4928:
4924:
4920:
4913:
4910:
4905:
4901:
4897:
4893:
4889:
4885:
4878:
4875:
4870:
4866:
4861:
4856:
4852:
4848:
4845:(1): 98–106.
4844:
4840:
4836:
4829:
4826:
4821:
4817:
4812:
4807:
4803:
4799:
4795:
4791:
4787:
4783:
4779:
4772:
4769:
4764:
4760:
4756:
4752:
4748:
4744:
4740:
4736:
4729:
4727:
4723:
4718:
4714:
4709:
4704:
4700:
4696:
4692:
4688:
4684:
4677:
4675:
4673:
4669:
4664:
4660:
4656:
4652:
4648:
4644:
4641:(3): 235–44.
4640:
4636:
4629:
4626:
4621:
4617:
4612:
4607:
4603:
4599:
4595:
4591:
4587:
4580:
4577:
4572:
4568:
4563:
4558:
4554:
4550:
4546:
4542:
4538:
4531:
4529:
4525:
4520:
4516:
4512:
4508:
4503:
4498:
4495:(5): 713–24.
4494:
4490:
4486:
4479:
4476:
4471:
4467:
4462:
4457:
4453:
4449:
4445:
4441:
4437:
4430:
4427:
4422:
4418:
4414:
4410:
4407:(2): 209–14.
4406:
4402:
4395:
4392:
4387:
4383:
4378:
4373:
4369:
4365:
4361:
4357:
4353:
4346:
4343:
4338:
4334:
4329:
4324:
4320:
4316:
4313:(9): 1141–6.
4312:
4308:
4304:
4297:
4294:
4289:
4285:
4280:
4275:
4271:
4267:
4263:
4256:
4253:
4248:
4244:
4239:
4234:
4230:
4226:
4222:
4218:
4214:
4207:
4204:
4199:
4195:
4190:
4185:
4181:
4177:
4173:
4169:
4165:
4158:
4155:
4150:
4146:
4141:
4136:
4132:
4128:
4124:
4120:
4116:
4112:
4108:
4101:
4098:
4093:
4089:
4084:
4079:
4075:
4071:
4067:
4063:
4059:
4055:
4051:
4043:
4040:
4035:
4031:
4026:
4021:
4017:
4013:
4009:
4002:
4000:
3996:
3991:
3987:
3982:
3977:
3973:
3969:
3966:(2): 156–65.
3965:
3961:
3957:
3950:
3947:
3942:
3938:
3934:
3930:
3926:
3922:
3918:
3914:
3906:
3903:
3898:
3894:
3889:
3884:
3880:
3876:
3872:
3868:
3864:
3857:
3854:
3849:
3845:
3840:
3835:
3831:
3827:
3823:
3819:
3815:
3808:
3805:
3800:
3796:
3791:
3786:
3782:
3778:
3774:
3770:
3766:
3762:
3758:
3751:
3748:
3743:
3739:
3734:
3729:
3725:
3721:
3717:
3713:
3709:
3702:
3699:
3694:
3690:
3685:
3680:
3676:
3672:
3668:
3664:
3660:
3656:
3652:
3645:
3642:
3637:
3633:
3628:
3623:
3619:
3615:
3611:
3607:
3603:
3599:
3595:
3588:
3585:
3580:
3576:
3571:
3566:
3562:
3558:
3553:
3548:
3544:
3540:
3536:
3529:
3526:
3521:
3517:
3512:
3507:
3503:
3499:
3495:
3491:
3487:
3483:
3479:
3472:
3470:
3466:
3461:
3457:
3452:
3447:
3443:
3439:
3436:(3): 482–98.
3435:
3431:
3427:
3420:
3417:
3412:
3408:
3403:
3398:
3394:
3390:
3387:(1): 201–12.
3386:
3382:
3378:
3371:
3368:
3363:
3359:
3354:
3349:
3344:
3339:
3335:
3331:
3327:
3320:
3317:
3312:
3308:
3304:
3300:
3295:
3290:
3287:(3): 434–48.
3286:
3282:
3278:
3271:
3268:
3263:
3259:
3255:
3251:
3248:(6): 279–85.
3247:
3243:
3236:
3233:
3228:
3224:
3219:
3214:
3210:
3206:
3202:
3198:
3194:
3187:
3184:
3179:
3175:
3171:
3167:
3164:(4): 1181–9.
3163:
3159:
3152:
3149:
3144:
3140:
3135:
3130:
3126:
3122:
3118:
3114:
3110:
3103:
3100:
3095:
3091:
3087:
3083:
3079:
3075:
3072:(4): 237–46.
3071:
3067:
3060:
3057:
3052:
3048:
3044:
3040:
3035:
3030:
3026:
3022:
3018:
3014:
3010:
3006:
3002:
2995:
2992:
2987:
2983:
2978:
2973:
2969:
2965:
2961:
2957:
2953:
2946:
2943:
2938:
2934:
2929:
2924:
2919:
2914:
2910:
2906:
2902:
2898:
2894:
2887:
2884:
2879:
2875:
2870:
2865:
2861:
2857:
2853:
2849:
2845:
2838:
2835:
2830:
2826:
2821:
2816:
2812:
2808:
2804:
2800:
2796:
2789:
2787:
2783:
2778:
2774:
2769:
2764:
2760:
2756:
2752:
2748:
2744:
2737:
2734:
2729:
2725:
2720:
2715:
2711:
2707:
2703:
2699:
2695:
2688:
2685:
2680:
2676:
2671:
2666:
2662:
2658:
2654:
2650:
2646:
2639:
2636:
2631:
2627:
2623:
2619:
2616:(3): 285–92.
2615:
2611:
2604:
2601:
2596:
2592:
2587:
2582:
2578:
2574:
2570:
2563:
2561:
2557:
2552:
2548:
2543:
2538:
2534:
2530:
2526:
2522:
2518:
2511:
2508:
2503:
2499:
2495:
2491:
2487:
2483:
2479:
2475:
2468:
2465:
2460:
2456:
2452:
2448:
2444:
2440:
2436:
2432:
2425:
2422:
2417:
2413:
2408:
2403:
2399:
2395:
2391:
2387:
2383:
2376:
2373:
2368:
2364:
2359:
2354:
2351:(6): 2800–8.
2350:
2346:
2342:
2335:
2332:
2327:
2323:
2318:
2313:
2309:
2305:
2301:
2297:
2293:
2286:
2283:
2278:
2274:
2269:
2264:
2260:
2256:
2252:
2248:
2244:
2237:
2234:
2229:
2225:
2220:
2215:
2211:
2207:
2203:
2199:
2195:
2188:
2185:
2180:
2176:
2172:
2168:
2164:
2160:
2156:
2152:
2148:
2144:
2137:
2134:
2128:
2125:
2120:
2116:
2111:
2106:
2102:
2098:
2094:
2090:
2086:
2079:
2076:
2071:
2067:
2062:
2057:
2053:
2049:
2045:
2041:
2037:
2030:
2028:
2026:
2022:
2017:
2013:
2008:
2003:
1999:
1995:
1991:
1987:
1983:
1976:
1974:
1970:
1965:
1961:
1956:
1951:
1947:
1943:
1939:
1935:
1931:
1924:
1921:
1916:
1912:
1907:
1902:
1898:
1894:
1891:(3): 368–77.
1890:
1886:
1882:
1875:
1872:
1867:
1863:
1858:
1853:
1848:
1843:
1839:
1835:
1831:
1827:
1823:
1816:
1813:
1808:
1804:
1799:
1794:
1790:
1786:
1782:
1775:
1772:
1767:
1763:
1758:
1753:
1749:
1745:
1741:
1737:
1733:
1726:
1724:
1720:
1715:
1711:
1707:
1703:
1699:
1695:
1688:
1685:
1680:
1676:
1671:
1666:
1662:
1658:
1655:(1): 234–46.
1654:
1650:
1646:
1639:
1637:
1633:
1628:
1624:
1619:
1614:
1610:
1606:
1603:(2): 585–94.
1602:
1598:
1594:
1587:
1585:
1583:
1581:
1577:
1572:
1568:
1564:
1560:
1556:
1552:
1548:
1544:
1537:
1534:
1529:
1525:
1520:
1515:
1511:
1507:
1503:
1499:
1495:
1488:
1485:
1480:
1476:
1471:
1466:
1461:
1456:
1452:
1448:
1444:
1440:
1436:
1429:
1426:
1421:
1417:
1413:
1409:
1406:(5): 487–98.
1405:
1401:
1394:
1391:
1386:
1382:
1377:
1372:
1368:
1364:
1360:
1356:
1352:
1345:
1343:
1339:
1334:
1330:
1325:
1320:
1315:
1310:
1306:
1302:
1298:
1291:
1289:
1285:
1280:
1276:
1271:
1266:
1262:
1258:
1251:
1248:
1243:
1239:
1234:
1229:
1225:
1221:
1217:
1213:
1209:
1202:
1199:
1194:
1188:
1183:
1182:
1173:
1170:
1165:
1161:
1157:
1153:
1149:
1145:
1138:
1135:
1130:
1126:
1122:
1118:
1115:(1): 173–97.
1114:
1110:
1103:
1101:
1097:
1092:
1088:
1084:
1080:
1075:
1070:
1066:
1062:
1058:
1054:
1050:
1043:
1040:
1035:
1031:
1027:
1023:
1019:
1015:
1011:
1007:
1003:
999:
992:
989:
984:
980:
975:
970:
966:
962:
958:
954:
950:
943:
940:
935:
931:
926:
921:
917:
913:
910:(2): 446–56.
909:
905:
901:
894:
892:
890:
886:
881:
877:
872:
867:
863:
859:
855:
851:
847:
840:
838:
834:
829:
825:
821:
817:
812:
807:
804:(4): 501–12.
803:
799:
795:
788:
786:
782:
775:
771:
768:
767:
763:
761:
758:
750:
748:
746:
742:
738:
734:
730:
726:
722:
718:
713:
711:
710:cyanobacteria
707:
703:
699:
695:
690:
687:
683:
679:
675:
671:
666:
664:
663:
658:
657:
651:
647:
639:
637:
635:
632:that have an
631:
627:
623:
618:
615:
611:
606:
604:
600:
596:
592:
588:
579:
572:
570:
568:
564:
563:TRAMP complex
560:
556:
552:
548:
544:
540:
532:
530:
528:
524:
520:
516:
512:
508:
504:
499:
496:
492:
487:
485:
481:
473:
467:
463:
461:
457:
453:
449:
444:
442:
436:
434:
430:
426:
422:
421:embryogenesis
418:
410:
408:
406:
402:
401:budding yeast
397:
393:
388:
386:
382:
378:
374:
373:somatic cells
367:Deadenylation
366:
364:
361:
357:
353:
349:
341:
339:
337:
333:
328:
324:
323:pyrophosphate
320:
316:
312:
307:
303:
300:
296:
292:
288:
284:
280:
276:
267:
265:
260:
255:
250:
245:
240:
234:
233:
227:
225:
223:
219:
215:
211:
207:
201:
199:
195:
191:
183:
178:
176:
174:
170:
169:
163:
160:
156:
155:
150:
146:
142:
138:
134:
125:
120:
119:Messenger RNA
116:
108:
106:
103:
101:
95:
93:
89:
85:
81:
77:
74:
70:
69:transcription
65:
63:
59:
55:
51:
47:
43:
39:
38:messenger RNA
35:
31:
23:
19:
6280: /
6266:RNA splicing
6208:
6114:irreversible
5999:Key elements
5945:
5896:Key elements
5810:Genetic code
5800:Introduction
5707:
5703:
5643:
5639:
5626:
5591:
5552:
5548:
5538:
5519:
5515:
5502:
5493:
5484:
5447:
5443:
5433:
5388:
5384:
5374:
5329:
5325:
5315:
5278:
5274:
5264:
5239:
5235:
5201:
5198:Biochemistry
5197:
5190:
5163:
5159:
5149:
5114:
5111:EMBO Reports
5110:
5100:
5065:
5059:
5024:
5020:
5010:
4975:
4971:
4961:
4926:
4922:
4912:
4890:(4): 266–9.
4887:
4883:
4877:
4842:
4838:
4828:
4785:
4781:
4771:
4741:(1): 65–77.
4738:
4734:
4690:
4686:
4638:
4634:
4628:
4593:
4589:
4579:
4544:
4540:
4492:
4488:
4478:
4443:
4439:
4429:
4404:
4400:
4394:
4359:
4355:
4345:
4310:
4306:
4296:
4269:
4265:
4255:
4220:
4216:
4206:
4171:
4167:
4157:
4114:
4110:
4100:
4057:
4053:
4042:
4015:
4011:
3963:
3959:
3949:
3916:
3912:
3905:
3870:
3866:
3856:
3821:
3817:
3807:
3764:
3760:
3750:
3715:
3711:
3701:
3658:
3654:
3644:
3601:
3598:Cell Reports
3597:
3587:
3545:(12): R100.
3542:
3538:
3528:
3488:(1): 18–30.
3485:
3481:
3433:
3429:
3419:
3384:
3380:
3370:
3333:
3329:
3319:
3284:
3280:
3270:
3245:
3241:
3235:
3200:
3196:
3186:
3161:
3157:
3151:
3116:
3112:
3102:
3069:
3065:
3059:
3008:
3004:
2994:
2959:
2955:
2945:
2900:
2896:
2886:
2851:
2847:
2837:
2805:(19): e132.
2802:
2798:
2750:
2746:
2736:
2701:
2697:
2687:
2652:
2648:
2638:
2613:
2609:
2603:
2576:
2572:
2524:
2520:
2510:
2477:
2473:
2467:
2437:(7): 662–9.
2434:
2430:
2424:
2389:
2385:
2375:
2348:
2344:
2334:
2299:
2295:
2285:
2250:
2246:
2236:
2201:
2197:
2187:
2146:
2142:
2136:
2127:
2092:
2088:
2078:
2043:
2039:
1989:
1985:
1937:
1933:
1923:
1888:
1884:
1874:
1829:
1825:
1815:
1788:
1784:
1774:
1739:
1735:
1697:
1693:
1687:
1652:
1648:
1600:
1596:
1546:
1542:
1536:
1501:
1497:
1487:
1442:
1438:
1428:
1403:
1399:
1393:
1358:
1354:
1304:
1301:BMC Genomics
1300:
1260:
1250:
1218:(3): 404–9.
1215:
1211:
1201:
1180:
1172:
1147:
1143:
1137:
1112:
1108:
1056:
1052:
1042:
1004:(3): 570–9.
1001:
997:
991:
956:
952:
942:
907:
903:
853:
849:
801:
797:
754:
719:, including
714:
691:
667:
660:
654:
643:
619:
614:mitochondria
610:trypanosomes
607:
602:
594:
590:
584:
541:, including
536:
500:
488:
477:
445:
437:
423:and in post-
414:
389:
370:
345:
308:
304:
294:
286:
272:
237:
202:
187:
172:
167:
164:
153:
133:poly(A) tail
132:
130:
104:
96:
87:
66:
34:poly(A) tail
33:
29:
28:
18:
6292:Spliceosome
6251:RNA editing
5965:Translation
5802:to genetics
5632:Edmonds, M.
4060:(1): 5331.
3824:(2): 73–9.
3330:Development
3203:(1): 10–6.
2095:(1): 71–8.
1361:(1): 1–10.
725:Coronavirus
721:Influenza A
717:RNA viruses
702:polymerases
674:degradosome
603:degradosome
587:degradosome
511:macrophages
429:nerve cells
338:from RNAs.
332:spliceosome
327:nucleotides
317:units from
168:translation
154:transcribed
54:translation
6472:Categories
6109:reversible
6072:lac operon
6048:imprinting
6043:Epigenetic
6035:Regulation
5990:Eukaryotic
5936:5' capping
5887:Eukaryotic
5588:Edmonds, M
776:References
741:Poliovirus
653:bacterium
433:translated
281:, such as
275:processive
214:regulatory
147:and U for
76:terminates
56:. In many
50:eukaryotes
48:bases. In
6412:Cytosolic
5980:Bacterial
5877:Bacterial
4802:0006-3002
4635:BioEssays
3675:1362-4962
3618:2211-1247
3561:1474-760X
3502:1471-0080
3051:231195473
3025:0036-8075
2247:Structure
1885:Structure
1571:206956408
1150:: 15–42.
640:Evolution
537:For many
495:microRNAs
427:sites of
407:complex.
228:Mechanism
206:microRNAs
198:stem-loop
190:cytoplasm
157:) from a
6092:microRNA
6006:Ribosome
5985:Archaeal
5941:Splicing
5913:Promoter
5882:Archaeal
5826: →
5822: →
5734:18256699
5618:12102557
5590:(2002).
5476:30319572
5450:: 2250.
5425:11717411
5366:23923003
5326:PLOS ONE
5307:10074205
5256:18177749
5218:19053279
5182:18399989
5141:16282984
5092:19161858
5051:12953107
5002:17065466
4953:12486011
4904:18312863
4869:17965156
4820:22172994
4763:86607431
4717:11917006
4663:26109164
4655:10684583
4620:16738135
4571:17872511
4519:14898055
4511:15935758
4470:21663793
4421:17395456
4386:32976578
4337:18451104
4288:21292162
4247:17464285
4198:17507659
4149:18978773
4092:30552333
4034:16207706
3990:17210931
3897:18835850
3848:18392144
3799:18566288
3742:27220521
3693:18757892
3636:22685694
3579:16356263
3520:27677860
3460:18256699
3411:15647503
3362:18434412
3311:16092673
3303:18267074
3262:17481902
3227:18023855
3178:16169522
3143:16705177
3086:11283721
3043:33414189
2986:18430932
2956:Genetics
2937:16495412
2878:15231747
2829:17933768
2777:28334977
2728:16714281
2679:10970864
2630:15145353
2595:17595167
2494:12173714
2451:17572685
2416:12145212
2326:18304944
2277:17850751
2228:12097343
2179:34840144
2119:18157150
2070:18411206
2016:17024186
1964:15937220
1915:21295486
1866:20479262
1807:14690600
1766:10899149
1714:18817380
1679:17158511
1563:18839252
1528:16131612
1479:17965236
1420:12408966
1400:Genomics
1385:17998288
1333:18479511
1279:16651366
1242:18212021
1164:14140701
1091:19003617
1083:16111937
1034:19863143
1026:27007045
983:10943888
934:10357857
880:11255003
820:11909521
764:See also
626:plastids
519:lysozyme
425:synaptic
417:germline
405:CCR4-Not
184:Function
143:, G for
141:cytosine
139:, C for
58:bacteria
6392:PRPF40B
6387:PRPF40A
6377:PRPF38B
6372:PRPF38A
6187:Nuclear
5845:RNA→DNA
5840:RNA→RNA
5828:Protein
5725:2241648
5680:5288383
5648:Bibcode
5571:9353246
5467:6167517
5393:Bibcode
5357:3726627
5334:Bibcode
5132:1369208
4993:1635327
4860:2223728
4811:3307840
4743:Bibcode
4611:1474067
4562:2040100
4461:3115544
4377:7778938
4328:2335310
4238:1888663
4189:1894925
4140:2597294
4119:Bibcode
4083:6294251
4062:Bibcode
3981:1781347
3941:7448467
3933:6771018
3888:2577349
3839:2288788
3790:2587246
3769:Bibcode
3761:Science
3733:4893057
3684:2553571
3627:3368434
3570:1414089
3511:5483950
3451:2241648
3353:9154023
3218:2239213
3134:1489097
3094:9734550
3034:9491362
3005:Science
2977:2323793
2928:1449641
2905:Bibcode
2820:2095794
2768:5449641
2719:1484436
2551:9784497
2502:2237085
2459:5777074
2367:7852352
2317:2367721
2268:2032019
2171:8929410
2151:Bibcode
2143:Science
2110:2836588
2061:2396415
2007:1618107
1955:1142555
1906:3056899
1857:2890493
1834:Bibcode
1670:1802579
1627:8440247
1519:1221887
1470:2077053
1447:Bibcode
1376:2151031
1324:2391170
1307:: 220.
1233:2248257
1129:9242905
1061:Bibcode
1006:Bibcode
974:1369983
871:3340483
751:History
706:archaea
678:exosome
646:domains
634:exosome
630:archaea
595:RNase E
567:exosome
503:CstF-64
456:Pumilio
352:exosome
336:introns
194:histone
145:guanine
137:adenine
80:3′-most
46:adenine
6382:PRPF39
6367:PRPF31
6362:PRPF19
6357:PRPF18
6342:PRPF4B
6278:Intron
5732:
5722:
5678:
5671:389184
5668:
5616:
5606:
5569:
5474:
5464:
5423:
5413:
5364:
5354:
5305:
5298:104115
5295:
5254:
5216:
5180:
5139:
5129:
5090:
5080:
5049:
5042:181327
5039:
5000:
4990:
4951:
4944:139106
4941:
4902:
4867:
4857:
4818:
4808:
4800:
4761:
4715:
4708:101826
4705:
4661:
4653:
4618:
4608:
4569:
4559:
4517:
4509:
4468:
4458:
4419:
4384:
4374:
4335:
4325:
4286:
4245:
4235:
4196:
4186:
4147:
4137:
4111:Nature
4090:
4080:
4032:
3988:
3978:
3939:
3931:
3895:
3885:
3846:
3836:
3797:
3787:
3740:
3730:
3691:
3681:
3673:
3634:
3624:
3616:
3577:
3567:
3559:
3518:
3508:
3500:
3458:
3448:
3409:
3402:546146
3399:
3360:
3350:
3309:
3301:
3260:
3225:
3215:
3176:
3141:
3131:
3092:
3084:
3049:
3041:
3031:
3023:
2984:
2974:
2935:
2925:
2876:
2869:442147
2866:
2827:
2817:
2775:
2765:
2726:
2716:
2677:
2670:302064
2667:
2628:
2593:
2549:
2542:317214
2539:
2500:
2492:
2457:
2449:
2414:
2407:126137
2404:
2365:
2324:
2314:
2275:
2265:
2226:
2219:186619
2216:
2177:
2169:
2117:
2107:
2068:
2058:
2014:
2004:
1962:
1952:
1913:
1903:
1864:
1854:
1805:
1764:
1757:310884
1754:
1712:
1677:
1667:
1625:
1618:413241
1615:
1569:
1561:
1526:
1516:
1477:
1467:
1418:
1383:
1373:
1331:
1321:
1277:
1240:
1230:
1189:
1162:
1127:
1089:
1081:
1032:
1024:
981:
971:
932:
922:
878:
868:
828:478260
826:
818:
745:PABPC1
731:, and
612:, the
555:snoRNA
553:, and
491:3' UTR
480:3′ end
396:5′ cap
149:uracil
78:. The
6435:DCP1B
6430:DCP1A
6352:PRPF8
6347:PRPF6
6337:PRPF4
6332:PRPF3
6327:PLRG1
6297:minor
6287:snRNP
5973:Types
5870:Types
5416:64674
4759:S2CID
4659:S2CID
4515:S2CID
3937:S2CID
3307:S2CID
3090:S2CID
3047:S2CID
2498:S2CID
2455:S2CID
2175:S2CID
1567:S2CID
1087:S2CID
1030:S2CID
925:98972
824:S2CID
737:HIV-1
698:tRNAs
559:yeast
551:snRNA
523:TNF-α
460:GLD-2
254:PABII
71:of a
6455:EDC4
6450:EDC3
6445:DCPS
6440:DCP2
6282:Exon
6239:CFII
6229:PAB2
6219:CstF
6214:CPSF
5730:PMID
5676:PMID
5614:PMID
5604:ISBN
5567:PMID
5472:PMID
5421:PMID
5362:PMID
5303:PMID
5252:PMID
5240:1779
5214:PMID
5178:PMID
5137:PMID
5088:PMID
5078:ISBN
5047:PMID
4998:PMID
4949:PMID
4900:PMID
4888:1779
4865:PMID
4816:PMID
4798:ISSN
4786:1819
4713:PMID
4651:PMID
4616:PMID
4567:PMID
4507:PMID
4489:Cell
4466:PMID
4440:Cell
4417:PMID
4382:PMID
4333:PMID
4284:PMID
4243:PMID
4194:PMID
4145:PMID
4088:PMID
4030:PMID
3986:PMID
3929:PMID
3913:Cell
3893:PMID
3844:PMID
3795:PMID
3738:PMID
3689:PMID
3671:ISSN
3632:PMID
3614:ISSN
3575:PMID
3557:ISSN
3516:PMID
3498:ISSN
3456:PMID
3407:PMID
3358:PMID
3299:PMID
3281:Cell
3258:PMID
3223:PMID
3174:PMID
3139:PMID
3082:PMID
3039:PMID
3021:ISSN
2982:PMID
2933:PMID
2874:PMID
2825:PMID
2773:PMID
2724:PMID
2675:PMID
2626:PMID
2591:PMID
2547:PMID
2490:PMID
2447:PMID
2412:PMID
2363:PMID
2322:PMID
2273:PMID
2224:PMID
2167:PMID
2115:PMID
2066:PMID
2012:PMID
1960:PMID
1911:PMID
1862:PMID
1803:PMID
1762:PMID
1710:PMID
1675:PMID
1623:PMID
1559:PMID
1524:PMID
1475:PMID
1416:PMID
1381:PMID
1329:PMID
1275:PMID
1238:PMID
1187:ISBN
1160:PMID
1125:PMID
1079:PMID
1022:PMID
979:PMID
930:PMID
876:PMID
850:Gene
816:PMID
798:Cell
770:SV40
739:and
708:and
593:and
547:rRNA
543:tRNA
521:and
452:CPEB
450:and
448:CPSF
392:PARN
299:CPSF
291:CPSF
273:The
264:CFII
244:CstF
239:CPSF
218:Xist
117:and
73:gene
6234:CFI
6224:PAP
5824:RNA
5820:DNA
5720:PMC
5712:doi
5666:PMC
5656:doi
5596:doi
5557:doi
5524:doi
5520:235
5462:PMC
5452:doi
5411:PMC
5401:doi
5352:PMC
5342:doi
5293:PMC
5283:doi
5244:doi
5206:doi
5168:doi
5164:283
5127:PMC
5119:doi
5070:doi
5037:PMC
5029:doi
4988:PMC
4980:doi
4939:PMC
4931:doi
4892:doi
4855:PMC
4847:doi
4843:190
4806:PMC
4790:doi
4751:doi
4703:PMC
4695:doi
4643:doi
4606:PMC
4598:doi
4557:PMC
4549:doi
4541:RNA
4497:doi
4493:121
4456:PMC
4448:doi
4444:145
4409:doi
4372:PMC
4364:doi
4323:PMC
4315:doi
4274:doi
4233:PMC
4225:doi
4184:PMC
4176:doi
4168:RNA
4135:PMC
4127:doi
4115:456
4078:PMC
4070:doi
4020:doi
4016:280
3976:PMC
3968:doi
3921:doi
3883:PMC
3875:doi
3834:PMC
3826:doi
3785:PMC
3777:doi
3765:320
3728:PMC
3720:doi
3716:468
3679:PMC
3663:doi
3622:PMC
3606:doi
3565:PMC
3547:doi
3506:PMC
3490:doi
3446:PMC
3438:doi
3397:PMC
3389:doi
3348:PMC
3338:doi
3334:135
3289:doi
3285:132
3250:doi
3213:PMC
3205:doi
3166:doi
3162:336
3129:PMC
3121:doi
3074:doi
3029:PMC
3013:doi
3009:371
2972:PMC
2964:doi
2960:178
2923:PMC
2913:doi
2901:103
2864:PMC
2856:doi
2815:PMC
2807:doi
2763:PMC
2755:doi
2714:PMC
2706:doi
2698:RNA
2665:PMC
2657:doi
2618:doi
2581:doi
2577:282
2537:PMC
2529:doi
2482:doi
2439:doi
2402:PMC
2394:doi
2353:doi
2349:270
2312:PMC
2304:doi
2263:PMC
2255:doi
2214:PMC
2206:doi
2159:doi
2147:274
2105:PMC
2097:doi
2056:PMC
2048:doi
2002:PMC
1994:doi
1950:PMC
1942:doi
1901:PMC
1893:doi
1852:PMC
1842:doi
1830:107
1793:doi
1752:PMC
1744:doi
1702:doi
1665:PMC
1657:doi
1613:PMC
1605:doi
1551:doi
1514:PMC
1506:doi
1465:PMC
1455:doi
1443:104
1408:doi
1371:PMC
1363:doi
1355:RNA
1319:PMC
1309:doi
1265:doi
1228:PMC
1220:doi
1212:RNA
1152:doi
1117:doi
1069:doi
1014:doi
969:PMC
961:doi
953:RNA
920:PMC
912:doi
866:PMC
858:doi
854:265
806:doi
802:108
686:ATP
682:ADP
360:40S
259:CFI
249:PAP
159:DNA
115:RNA
6474::
6302:U1
5728:.
5718:.
5708:27
5706:.
5702:.
5674:.
5664:.
5654:.
5644:68
5642:.
5638:.
5612:.
5602:.
5579:^
5565:.
5553:11
5551:.
5547:.
5518:.
5514:.
5492:.
5470:.
5460:.
5446:.
5442:.
5419:.
5409:.
5399:.
5389:98
5387:.
5383:.
5360:.
5350:.
5340:.
5328:.
5324:.
5301:.
5291:.
5279:73
5277:.
5273:.
5250:.
5238:.
5226:^
5212:.
5202:47
5200:.
5176:.
5162:.
5158:.
5135:.
5125:.
5113:.
5109:.
5086:.
5076:.
5045:.
5035:.
5025:15
5023:.
5019:.
4996:.
4986:.
4976:34
4974:.
4970:.
4947:.
4937:.
4927:21
4925:.
4921:.
4898:.
4886:.
4863:.
4853:.
4841:.
4837:.
4814:.
4804:.
4796:.
4784:.
4780:.
4757:.
4749:.
4739:25
4737:.
4725:^
4711:.
4701:.
4691:30
4689:.
4685:.
4671:^
4657:.
4649:.
4639:22
4637:.
4614:.
4604:.
4594:34
4592:.
4588:.
4565:.
4555:.
4545:13
4543:.
4539:.
4527:^
4513:.
4505:.
4491:.
4487:.
4464:.
4454:.
4442:.
4438:.
4415:.
4405:17
4403:.
4380:.
4370:.
4360:49
4358:.
4354:.
4331:.
4321:.
4311:22
4309:.
4305:.
4282:.
4270:41
4268:.
4264:.
4241:.
4231:.
4221:26
4219:.
4215:.
4192:.
4182:.
4172:13
4170:.
4166:.
4143:.
4133:.
4125:.
4113:.
4109:.
4086:.
4076:.
4068:.
4056:.
4052:.
4028:.
4014:.
4010:.
3998:^
3984:.
3974:.
3964:17
3962:.
3958:.
3935:.
3927:.
3917:20
3915:.
3891:.
3881:.
3871:36
3869:.
3865:.
3842:.
3832:.
3820:.
3816:.
3793:.
3783:.
3775:.
3763:.
3759:.
3736:.
3726:.
3714:.
3710:.
3687:.
3677:.
3669:.
3659:36
3657:.
3653:.
3630:.
3620:.
3612:.
3600:.
3596:.
3573:.
3563:.
3555:.
3541:.
3537:.
3514:.
3504:.
3496:.
3486:18
3484:.
3480:.
3468:^
3454:.
3444:.
3434:27
3432:.
3428:.
3405:.
3395:.
3385:33
3383:.
3379:.
3356:.
3346:.
3332:.
3328:.
3305:.
3297:.
3283:.
3279:.
3256:.
3246:32
3244:.
3221:.
3211:.
3201:69
3199:.
3195:.
3172:.
3160:.
3137:.
3127:.
3117:26
3115:.
3111:.
3088:.
3080:.
3068:.
3045:.
3037:.
3027:.
3019:.
3007:.
3003:.
2980:.
2970:.
2958:.
2954:.
2931:.
2921:.
2911:.
2899:.
2895:.
2872:.
2862:.
2852:14
2850:.
2846:.
2823:.
2813:.
2803:35
2801:.
2797:.
2785:^
2771:.
2761:.
2751:45
2749:.
2745:.
2722:.
2712:.
2702:12
2700:.
2696:.
2673:.
2663:.
2653:19
2651:.
2647:.
2624:.
2614:16
2612:.
2589:.
2575:.
2571:.
2559:^
2545:.
2535:.
2525:12
2523:.
2519:.
2496:.
2488:.
2478:77
2476:.
2453:.
2445:.
2435:14
2433:.
2410:.
2400:.
2390:21
2388:.
2384:.
2361:.
2347:.
2343:.
2320:.
2310:.
2300:36
2298:.
2294:.
2271:.
2261:.
2251:15
2249:.
2245:.
2222:.
2212:.
2202:12
2200:.
2196:.
2173:.
2165:.
2157:.
2145:.
2113:.
2103:.
2093:15
2091:.
2087:.
2064:.
2054:.
2044:36
2042:.
2038:.
2024:^
2010:.
2000:.
1990:25
1988:.
1984:.
1972:^
1958:.
1948:.
1938:19
1936:.
1932:.
1909:.
1899:.
1889:19
1887:.
1883:.
1860:.
1850:.
1840:.
1828:.
1824:.
1801:.
1789:12
1787:.
1783:.
1760:.
1750:.
1740:10
1738:.
1734:.
1722:^
1708:.
1696:.
1673:.
1663:.
1653:35
1651:.
1647:.
1635:^
1621:.
1611:.
1601:12
1599:.
1595:.
1579:^
1565:.
1557:.
1547:19
1545:.
1522:.
1512:.
1502:19
1500:.
1496:.
1473:.
1463:.
1453:.
1441:.
1437:.
1414:.
1404:80
1402:.
1379:.
1369:.
1359:14
1357:.
1353:.
1341:^
1327:.
1317:.
1303:.
1299:.
1287:^
1273:.
1259:.
1236:.
1226:.
1216:14
1214:.
1210:.
1158:.
1148:32
1146:.
1123:.
1113:66
1111:.
1099:^
1085:.
1077:.
1067:.
1057:15
1055:.
1051:.
1028:.
1020:.
1012:.
1002:49
1000:.
977:.
967:.
955:.
951:.
928:.
918:.
908:63
906:.
902:.
888:^
874:.
864:.
852:.
848:.
836:^
822:.
814:.
800:.
796:.
784:^
727:,
723:,
549:,
545:,
387:.
102:.
94:.
64:.
6171:e
6164:t
6157:v
5784:e
5777:t
5770:v
5736:.
5714::
5682:.
5658::
5650::
5620:.
5598::
5573:.
5559::
5532:.
5526::
5496:.
5478:.
5454::
5448:9
5427:.
5403::
5395::
5368:.
5344::
5336::
5330:8
5309:.
5285::
5258:.
5246::
5220:.
5208::
5184:.
5170::
5143:.
5121::
5115:6
5094:.
5072::
5053:.
5031::
5004:.
4982::
4955:.
4933::
4906:.
4894::
4871:.
4849::
4822:.
4792::
4765:.
4753::
4745::
4719:.
4697::
4665:.
4645::
4622:.
4600::
4573:.
4551::
4521:.
4499::
4472:.
4450::
4423:.
4411::
4388:.
4366::
4339:.
4317::
4290:.
4276::
4249:.
4227::
4200:.
4178::
4151:.
4129::
4121::
4094:.
4072::
4064::
4058:9
4036:.
4022::
3992:.
3970::
3943:.
3923::
3899:.
3877::
3850:.
3828::
3822:5
3801:.
3779::
3771::
3744:.
3722::
3695:.
3665::
3638:.
3608::
3602:1
3581:.
3549::
3543:6
3522:.
3492::
3462:.
3440::
3413:.
3391::
3364:.
3340::
3313:.
3291::
3264:.
3252::
3229:.
3207::
3180:.
3168::
3145:.
3123::
3096:.
3076::
3070:2
3053:.
3015::
2988:.
2966::
2939:.
2915::
2907::
2880:.
2858::
2831:.
2809::
2779:.
2757::
2730:.
2708::
2681:.
2659::
2632:.
2620::
2597:.
2583::
2553:.
2531::
2504:.
2484::
2461:.
2441::
2418:.
2396::
2369:.
2355::
2328:.
2306::
2279:.
2257::
2230:.
2208::
2181:.
2161::
2153::
2121:.
2099::
2072:.
2050::
2018:.
1996::
1966:.
1944::
1917:.
1895::
1868:.
1844::
1836::
1809:.
1795::
1768:.
1746::
1716:.
1704::
1698:3
1681:.
1659::
1629:.
1607::
1573:.
1553::
1530:.
1508::
1481:.
1457::
1449::
1422:.
1410::
1387:.
1365::
1335:.
1311::
1305:9
1281:.
1267::
1244:.
1222::
1195:.
1166:.
1154::
1131:.
1119::
1093:.
1071::
1063::
1036:.
1016::
1008::
985:.
963::
957:6
936:.
914::
882:.
860::
830:.
808::
462:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.