364:
282:
to release of Shine–Dalgarno and the AUG start codon. RNA Thermometers can also be found in some plant symbiotes or pathogens, symbiotes and pathogens use the RNA thermometers to regulate the plant's gene expression. A well studied symbiotic bacteria is the
Rhizobiaceae family. In majority of the rhizobial species, ROSE elements (cis-acting) were visible controlling heat-shock genes.
291:
278:
but once a change occurs in temperature, it melts and activates protein production. C. reinhardtii’s RNA thermometer research is the entryway to observing the chloroplast of photosynthetic organisms for gene regulation and how it can be used for agriculture at some point in the future since it helps plants get accustomed to external temperature.
22:
222:, found in the 5’-UTR of the psaA mRNA. Its function was different especially because it was considered absent, it has a hairpin-type secondary structure that protects the Shine–Dalgarno sequence when temperature is low, but once a change occurs in temperature, it melts and activates protein production.
210:
Biological reactions and organism are sensitive to temperature for cell function. RNA thermometers are an efficient way to respond to temperature because as they allow cells to monitor and sense changes to maintain the cell alive and stable. DNA, RNA, or protein-induced mechanisms avoid small changes
281:
ROSE elements, are a bacterial RNA thermometer class that regulates the activation of genes that have small heat shock proteins. It melts at a moderate level parallel to the increase of the temperature surrounding its environment. Once it fully melts at a high temperature of ~42 °C, it proceeds
277:
The first RNA thermometer discovered in chloroplast of
Chlamydomonas reinhardtii, found in the 5’-UTR of the psaA mRNA. Its function was different especially because it was considered absent, it has a hairpin-type secondary structure that protects the Shine–Dalgarno sequence when temperature is low,
214:
Bacteria use RNA thermometers to enter and survive in their hosts by mounting themselves to their host and causing fluctuations in their temperature. The bacteria can respond quickly against heat-shock and cold-shock conditions since RNA thermometers control gene expression at a translational level.
323:
Detailed structural analysis of the ROSE RNA thermometer revealed that the mismatched bases are actually engaged in nonstandard basepairing that preserves the helical structure of the RNA (see figure). The unusual basepairs consist of G-G, U-U, and UC-U pairs. Since these noncanonical base pairs
561:
due to their wide-scale distribution in distantly-related organisms. It has been proposed that, in the RNA world, RNA thermosensors would have been responsible for temperature-dependent regulation of other RNA molecules. RNA thermometers in modern organisms may be
391:
UTR of messenger RNA, upstream of a protein-coding gene. Here they are able to occlude the ribosome binding site (RBS) and prevent translation of the mRNA into protein. As temperature increases, the hairpin structure can 'melt' and expose the RBS or
226:
RNA thermometer research is the entryway to observing the chloroplast of photosynthetic organisms for gene regulation and how it can be used for agriculture at some point in the future since it helps plants get accustomed to external temperature.
57:. Its unique characteristic it is that it does not need proteins or metabolites to function, but only reacts to temperature changes. RNA thermometers often regulate genes required during either a
199:
searches have been employed to uncover several novel candidate RNA thermometers. Traditional sequence-based searches are inefficient, however, as the secondary structure of the element is much more
1099:
Altuvia S, Kornitzer D, Teff D, Oppenheim AB (November 1989). "Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation".
613:
603:
131:
1943:
Mega R, Manzoku M, Shinkai A, Nakagawa N, Kuramitsu S, Masui R (August 2010). "Very rapid induction of a cold shock protein by temperature downshift in
Thermus thermophilus".
320:
with a small number of mismatched base pairs which reduce the stability of the structure, thereby allowing easier unfolding in response to a temperature increase.
466:
Though typically associated with heat-induced protein expression, RNA thermometers can also regulate cold-shock proteins. For example, the expression of two 7
1317:
Waldminghaus T, Gaubig LC, Narberhaus F (November 2007). "Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers".
404:, typically found 8 nucleotides downstream of the Shine-Dalgarno sequence, signals the beginning of a protein-coding gene which is then translated to a
1186:
Altuvia S, Kornitzer D, Kobi S, Oppenheim AB (April 1991). "Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda".
170:
life cycle in λ phage, with high concentrations of cIII promoting lysogeny. Further study of this upstream RNA region identified two alternative
324:
are relatively unstable, increased temperature causes local melting of the RNA structure in this region, exposing the Shine-Dalgarno sequence.
1363:
Sharma P, Mondal K, Kumar S, Tamang S, Najar IN, Das S, et al. (October 2022). "RNA thermometers in bacteria: Role in thermoregulation".
1465:
839:
2249:
178:
concentration and temperature. This RNA thermometer is now thought to encourage entry to a lytic cycle under heat stress in order for the
1595:
Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F (December 2005). "RNA thermometers are common in alpha- and gamma-proteobacteria".
904:"Translational regulation of Hsp90 mRNA. AUG-proximal 5'-untranslated region elements essential for preferential heat shock translation"
997:"Role for cis-acting RNA sequences in the temperature-dependent expression of the multiadhesive lig proteins in Leptospira interrogans"
736:
Raza A, Siddique KH, Hu Z (February 2024). "Chloroplast gene control: unlocking RNA thermometer mechanisms in photosynthetic systems".
543:
388:
112:
84:
in response to temperature fluctuations. This structural transition can then expose or occlude important regions of RNA such as a
1492:
Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E (March 2006). "RNA-mediated response to heat shock in mammalian cells".
342:
650:
645:
492:
451:
ion concentration has also been shown to affect the stability of FourU. The most well-studied RNA thermometer is found in the
447:
opposite the Shine-Dalgarno sequence becomes unpaired and allows the mRNA to enter the ribosome for translation to occur.
271:
575:
123:
150:
The first temperature-sensitive RNA element was reported in 1989. Prior to this research, mutations upstream from the
236:
162:
were found to affect the level of translation of the cIII protein. This protein is involved in selection of either a
542:, with different sequences acting as biocatalysts, regulators and sensors. The hypothesis then proposes that modern
655:
313:
634:
517:
459:. This thermosensor upregulates heat shock proteins under high temperatures through σ, a specialised heat-shock
327:
Some RNA thermometers are significantly more complex than a single hairpin, as in the case of a region found in
628:
510:
151:
1267:"Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor"
2244:
1736:"Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli"
393:
376:
30:
302:
RNA thermometers are structurally simple and can be made from short RNA sequences; the smallest is just 44
505:
413:
558:
204:
89:
85:
328:
316:. Generally these RNA elements range in length from 60 to 110 nucleotides and they typically contain a
2037:
1897:
1501:
533:
476:
175:
104:
427:
660:
435:
171:
135:
77:
62:
1620:
1525:
1342:
1139:"Translational regulatory signals within the coding region of the bacteriophage lambda cIII gene"
538:
The RNA world hypothesis states that RNA was once both the carrier of hereditary information and
240:
200:
81:
1693:
Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, et al. (January 2010).
1409:"Translational control of bacterial heat shock and virulence genes by temperature-sensing mRNAs"
2221:
2172:
2151:"Multiple layers of control govern expression of the Escherichia coli ibpAB heat-shock operon"
2131:
2096:
2001:
1960:
1925:
1866:
1817:
1765:
1716:
1672:
1612:
1577:
1517:
1471:
1461:
1430:
1380:
1334:
1296:
1247:
1203:
1168:
1116:
1078:
1026:
977:
925:
881:
835:
805:
749:
709:
547:
481:
341:
RNA thermometers have been designed with just a simple single-hairpin structure. However, the
338:
139:
119:
26:
2211:
2203:
2162:
2123:
2086:
2078:
2045:
2028:
1991:
1952:
1915:
1905:
1856:
1848:
1807:
1799:
1755:
1747:
1706:
1662:
1654:
1604:
1567:
1559:
1509:
1453:
1420:
1372:
1326:
1286:
1278:
1237:
1195:
1158:
1150:
1108:
1068:
1060:
1016:
1008:
967:
959:
915:
871:
795:
785:
741:
701:
624:
607:
467:
295:
191:
1695:"The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA"
946:
Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F (December 2001).
167:
54:
1978:
Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P (September 2002).
2041:
1901:
1505:
2091:
2066:
2019:
1920:
1885:
1861:
1836:
1812:
1787:
1760:
1735:
1667:
1642:
1572:
1547:
1073:
1048:
1021:
996:
800:
773:
346:
196:
174:; experimental study found the structures to be interchangeable, and dependent on both
50:
2216:
2191:
1996:
1980:"An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes"
1979:
1452:. Contributions to Microbiology. Vol. 16. Basel: S. Karger AG. pp. 150–160.
1291:
1266:
1163:
1138:
2238:
2207:
2192:"Replicon-specific regulation of small heat shock genes in Agrobacterium tumefaciens"
1199:
1112:
972:
947:
876:
859:
563:
308:
244:
179:
66:
2067:"Ribozymes, riboswitches and beyond: regulation of gene expression without proteins"
1624:
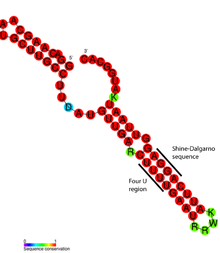
1346:
2114:
Bocobza SE, Aharoni A (October 2008). "Switching the light on plant riboswitches".
1529:
705:
460:
420:
186:
155:
108:
100:
2127:
1154:
745:
1910:
1711:
1694:
1658:
1376:
588:, increasing the translation rate of the heat shock protein at high temperatures.
433:
A specific example of an RNA thermometer motif is the FourU thermometer found in
270:(HSF1) and induces protective proteins when cell temperature exceeds 37 °C (
185:
The term "RNA thermometer" was not coined until 1999, when it was applied to the
551:
471:
401:
163:
46:
1956:
642:
mRNA, and repress heat shock protein translation at physiological temperatures.
1330:
692:
Narberhaus F, Waldminghaus T, Chowdhury S (January 2006). "RNA thermometers".
584:
363:
332:
303:
255:
70:
58:
1563:
1047:
Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F (April 2011).
963:
948:"A mRNA-based thermosensor controls expression of rhizobial heat shock genes"
790:
566:
which could hint at a previously more widespread importance in an RNA world.
500:
448:
444:
440:
317:
2225:
2176:
2167:
2150:
2135:
2100:
2005:
1964:
1929:
1870:
1821:
1769:
1720:
1676:
1616:
1581:
1521:
1475:
1434:
1384:
1338:
1300:
1251:
1082:
1030:
981:
929:
920:
903:
885:
809:
753:
713:
247:
and inherent difficulties of detecting short, unconserved RNA sequences in
1837:"Modulation of the stability of the Salmonella fourU-type RNA thermometer"
1425:
1408:
1265:
Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T (March 1999).
1207:
1172:
1120:
1064:
290:
1852:
1751:
1282:
1242:
1225:
774:"RNA structure mediated thermoregulation: What can we learn from plants?"
488:
409:
345:
of such short RNA thermometers can be sensitive to mutation, as a single
248:
1803:
1608:
1513:
1012:
487:
RNA thermometers sensitive to temperatures of 37 °C can be used by
65:
response, but have been implicated in other regulatory roles such as in
405:
351:
1457:
832:
The RNA world: the nature of modern RNA suggests a prebiotic RNA world
2050:
2023:
595:
539:
524:, and fluorescence was observed at 37 °C but not at 30 °C.
259:
2082:
1835:
Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H (October 2011).
520:
gene; the gene fusion was then transcribed from the T7 promoter in
579:
362:
289:
263:
20:
1365:
Biochimica et
Biophysica Acta (BBA) - Gene Regulatory Mechanisms
1448:
Johansson J (2009). "RNA thermosensors in bacterial pathogens".
858:
Waldminghaus T, Heidrich N, Brantl S, Narberhaus F (July 2007).
267:
243:—though it has been suggested this fact may be due, in part, to
159:
93:
21:
1548:"Molecular basis for temperature sensing by an RNA thermometer"
430:
where it is thought to be involved in the starvation response.
266:. The candidate thermosensor heat shock RNA-1 (HSR1) activates
397:
380:
602:
is predicted to contain two co-operative RNA thermometers: a
306:
and is found in the mRNA of a heat-shock protein, hsp17, in
2190:
Balsiger S, Ragaz C, Baron C, Narberhaus F (October 2004).
1546:
Chowdhury S, Maris C, Allain FH, Narberhaus F (June 2006).
1049:"Translation on demand by a simple RNA-based thermosensor"
400:), which then assembles other translation machinery. The
2149:
Gaubig LC, Waldminghaus T, Narberhaus F (January 2011).
1788:"Thermodynamics of RNA melting, one base pair at a time"
1886:"Is thermosensing property of RNA thermometers unique?"
834:. Plainview, N.Y: Cold Spring Harbor Laboratory Press.
491:
to activate infection-specific genes. For example, the
218:
The first RNA thermometer discovered in chloroplast of
76:
In general, RNA thermometers operate by changing their
860:"FourU: a novel type of RNA thermometer in Salmonella"
1312:
1310:
439:. When exposed to temperatures above 45 °C, the
557:
RNA thermometers and riboswitches are thought to be
470:
proteins are regulated by an RNA thermometer in the
1945:
1884:Shah P, Gilchrist MA (July 2010). Spirin AS (ed.).
853:
851:
1688:
1686:
1402:
1400:
1398:
1396:
1394:
995:Matsunaga J, Schlax PJ, Haake DA (November 2013).
396:to permit binding of the small ribosomal subunit (
111:present in cells, and was replaced by the current
107:. This theory proposes that RNA was once the sole
1487:
1485:
772:Thomas SE, Balcerowicz M, Chung BY (2022-08-17).
687:
685:
683:
681:
679:
677:
675:
1781:
1779:
1541:
1539:
550:replaced the majority of RNA's roles with other
258:, a potential RNA thermometer has been found in
1636:
1634:
1132:
1130:
1094:
1092:
1042:
1040:
480:and a similar mechanism has been identified in
274:), thus preventing the cells from overheating.
235:Most known RNA thermometers are located in the
182:to rapidly replicate and escape the host cell.
1734:Neupert J, Karcher D, Bock R (November 2008).
1358:
1356:
499:, encoding a key transcriptional regulator of
1786:Nikolova EN, Al-Hashimi HM (September 2010).
1219:
1217:
941:
939:
825:
823:
821:
819:
767:
765:
763:
379:becomes exposed, allowing the binding of the
8:
897:
895:
731:
729:
727:
725:
723:
638:respectively. They exist in the 5′ UTR of
294:3D representation of the structure of the
2215:
2166:
2090:
2049:
1995:
1919:
1909:
1860:
1811:
1759:
1710:
1666:
1571:
1424:
1290:
1241:
1162:
1072:
1020:
971:
919:
875:
799:
789:
528:Implications for the RNA world hypothesis
103:, are used as examples in support of the
830:Atkins JF, Gesteland RF, Cech T (2006).
211:because by sensing any external changes
671:
2065:Serganov A, Patel DJ (October 2007).
1137:Altuvia S, Oppenheim AB (July 1986).
118:Examples of RNA thermometers include
7:
902:Ahmed R, Duncan RF (November 2004).
908:The Journal of Biological Chemistry
371:) unwinds at a higher temperature (
16:Temperature-dependent RNA structure
426:RNA thermometer has been found in
387:RNA thermometers are found in the
14:
509:, was demonstrated by fusing the
268:heat-shock transcription factor 1
2208:10.1128/JB.186.20.6824-6829.2004
877:10.1111/j.1365-2958.2007.05794.x
349:can render the hairpin inactive
335:, as well as multiple hairpins.
239:(UTR) of messenger RNA encoding
92:rate of a nearby protein-coding
1450:Bacterial Sensing and Signaling
1319:Molecular Genetics and Genomics
646:Cyanobacterial RNA thermometers
544:DNA, RNA and protein-based life
419:mechanism, a lone example of a
1643:"RNA switches out in the cold"
706:10.1111/j.1574-6976.2005.004.x
331:which is thought to contain a
254:Though predominantly found in
1:
2128:10.1016/j.tplants.2008.07.004
1997:10.1016/S0092-8674(02)00905-4
1155:10.1128/jb.167.1.415-419.1986
746:10.1016/j.tplants.2024.01.005
99:RNA thermometers, along with
1911:10.1371/journal.pone.0011308
1712:10.1016/j.molcel.2009.11.033
1659:10.1016/j.molcel.2009.12.032
1377:10.1016/j.bbagrm.2022.194871
1200:10.1016/0022-2836(91)90261-4
1188:Journal of Molecular Biology
1113:10.1016/0022-2836(89)90329-X
1101:Journal of Molecular Biology
576:Hsp90 cis-regulatory element
2250:Cis-regulatory RNA elements
1641:Breaker RR (January 2010).
2266:
1957:10.1016/j.bbrc.2010.07.065
778:Frontiers in Plant Science
656:Neisseria RNA thermometers
651:Intergenic RNA thermometer
531:
189:RNA element identified in
88:, which then affects the
1331:10.1007/s00438-007-0272-7
694:FEMS Microbiology Reviews
635:Agrobacterium tumefaciens
518:green fluorescent protein
220:Chlamydomonas reinhardtii
53:molecule which regulates
2071:Nature Reviews. Genetics
1564:10.1038/sj.emboj.7601128
791:10.3389/fpls.2022.938570
629:Bradyrhizobium japonicum
152:transcription start site
2196:Journal of Bacteriology
2116:Trends in Plant Science
1271:Genes & Development
1230:Genes & Development
1143:Journal of Bacteriology
1001:Journal of Bacteriology
738:Trends in Plant Science
394:Shine-Dalgarno sequence
377:Shine-Dalgarno sequence
31:Shine-Dalgarno sequence
2168:10.1099/mic.0.043802-0
1841:Nucleic Acids Research
1740:Nucleic Acids Research
1224:Storz G (March 1999).
1053:Nucleic Acids Research
964:10.1093/nar/29.23.4800
952:Nucleic Acids Research
921:10.1074/jbc.M404681200
864:Molecular Microbiology
559:evolutionarily ancient
506:Listeria monocytogenes
412:. In addition to this
384:
299:
237:5′ untranslated region
34:
1426:10.4161/rna.7.1.10501
1407:Narberhaus F (2010).
366:
293:
205:nucleic acid sequence
86:ribosome binding site
24:
1597:Biological Chemistry
1283:10.1101/gad.13.6.655
1243:10.1101/gad.13.6.633
1226:"An RNA thermometer"
540:enzymatically active
534:RNA world hypothesis
477:Thermus thermophilus
172:secondary structures
105:RNA world hypothesis
29:RNA motif, with the
2042:1986Natur.319..618G
1902:2010PLoSO...511308S
1804:10.1261/rna.2235010
1609:10.1515/BC.2005.145
1514:10.1038/nature04518
1506:2006Natur.440..556S
1065:10.1093/nar/gkq1252
1013:10.1128/jb.00663-13
914:(48): 49919–49930.
661:Lig RNA thermometer
436:Salmonella enterica
383:ribosomal subunit.
375:). The highlighted
343:secondary structure
241:heat shock proteins
136:Lig RNA thermometer
128:-regulatory element
113:DNA → RNA → protein
78:secondary structure
1853:10.1093/nar/gkr314
1752:10.1093/nar/gkn545
385:
367:A stable hairpin (
300:
82:tertiary structure
35:
2202:(20): 6824–6829.
2022:(February 1986).
1847:(18): 8258–8270.
1603:(12): 1279–1286.
1558:(11): 2487–2497.
1500:(7083): 556–560.
1467:978-3-8055-9132-4
1458:10.1159/000219378
1007:(22): 5092–5101.
958:(23): 4800–4807.
841:978-0-87969-739-6
564:molecular fossils
482:Enterobacteriales
195:. More recently,
140:Hsp17 thermometer
27:FourU thermometer
2257:
2230:
2229:
2219:
2187:
2181:
2180:
2170:
2146:
2140:
2139:
2111:
2105:
2104:
2094:
2062:
2056:
2055:
2053:
2051:10.1038/319618a0
2016:
2010:
2009:
1999:
1975:
1969:
1968:
1940:
1934:
1933:
1923:
1913:
1881:
1875:
1874:
1864:
1832:
1826:
1825:
1815:
1798:(9): 1687–1691.
1783:
1774:
1773:
1763:
1731:
1725:
1724:
1714:
1690:
1681:
1680:
1670:
1638:
1629:
1628:
1592:
1586:
1585:
1575:
1552:The EMBO Journal
1543:
1534:
1533:
1489:
1480:
1479:
1445:
1439:
1438:
1428:
1404:
1389:
1388:
1360:
1351:
1350:
1314:
1305:
1304:
1294:
1262:
1256:
1255:
1245:
1221:
1212:
1211:
1183:
1177:
1176:
1166:
1134:
1125:
1124:
1096:
1087:
1086:
1076:
1059:(7): 2855–2868.
1044:
1035:
1034:
1024:
992:
986:
985:
975:
943:
934:
933:
923:
899:
890:
889:
879:
855:
846:
845:
827:
814:
813:
803:
793:
769:
758:
757:
733:
718:
717:
689:
625:hyphomicrobiales
608:IbpB thermometer
457:Escherichia coli
298:RNA thermometer.
272:body temperature
224:C. reinhardtii’s
192:Escherichia coli
156:lambda (λ) phage
43:RNA thermosensor
2265:
2264:
2260:
2259:
2258:
2256:
2255:
2254:
2235:
2234:
2233:
2189:
2188:
2184:
2161:(Pt 1): 66–76.
2148:
2147:
2143:
2122:(10): 526–533.
2113:
2112:
2108:
2083:10.1038/nrg2172
2077:(10): 776–790.
2064:
2063:
2059:
2024:"The RNA World"
2018:
2017:
2013:
1977:
1976:
1972:
1942:
1941:
1937:
1883:
1882:
1878:
1834:
1833:
1829:
1785:
1784:
1777:
1733:
1732:
1728:
1692:
1691:
1684:
1640:
1639:
1632:
1594:
1593:
1589:
1545:
1544:
1537:
1491:
1490:
1483:
1468:
1447:
1446:
1442:
1406:
1405:
1392:
1362:
1361:
1354:
1316:
1315:
1308:
1264:
1263:
1259:
1223:
1222:
1215:
1185:
1184:
1180:
1136:
1135:
1128:
1098:
1097:
1090:
1046:
1045:
1038:
994:
993:
989:
945:
944:
937:
901:
900:
893:
857:
856:
849:
842:
829:
828:
817:
771:
770:
761:
735:
734:
721:
691:
690:
673:
669:
621:
617:
572:
536:
530:
408:product by the
361:
288:
233:
148:
55:gene expression
39:RNA thermometer
17:
12:
11:
5:
2263:
2261:
2253:
2252:
2247:
2245:Non-coding RNA
2237:
2236:
2232:
2231:
2182:
2141:
2106:
2057:
2011:
1990:(5): 551–561.
1970:
1951:(3): 336–340.
1935:
1876:
1827:
1775:
1726:
1699:Molecular Cell
1682:
1647:Molecular Cell
1630:
1587:
1535:
1481:
1466:
1440:
1390:
1352:
1325:(5): 555–564.
1306:
1277:(6): 655–665.
1257:
1236:(6): 633–636.
1213:
1194:(4): 723–733.
1178:
1149:(1): 415–419.
1126:
1107:(2): 265–280.
1088:
1036:
987:
935:
891:
870:(2): 413–424.
847:
840:
815:
759:
719:
670:
668:
665:
664:
663:
658:
653:
648:
643:
619:
615:
611:
589:
571:
570:Other examples
568:
532:Main article:
529:
526:
360:
357:
287:
284:
232:
229:
197:bioinformatics
147:
144:
51:non-coding RNA
15:
13:
10:
9:
6:
4:
3:
2:
2262:
2251:
2248:
2246:
2243:
2242:
2240:
2227:
2223:
2218:
2213:
2209:
2205:
2201:
2197:
2193:
2186:
2183:
2178:
2174:
2169:
2164:
2160:
2156:
2152:
2145:
2142:
2137:
2133:
2129:
2125:
2121:
2117:
2110:
2107:
2102:
2098:
2093:
2088:
2084:
2080:
2076:
2072:
2068:
2061:
2058:
2052:
2047:
2043:
2039:
2036:(6055): 618.
2035:
2031:
2030:
2025:
2021:
2015:
2012:
2007:
2003:
1998:
1993:
1989:
1985:
1981:
1974:
1971:
1966:
1962:
1958:
1954:
1950:
1946:
1939:
1936:
1931:
1927:
1922:
1917:
1912:
1907:
1903:
1899:
1896:(7): e11308.
1895:
1891:
1887:
1880:
1877:
1872:
1868:
1863:
1858:
1854:
1850:
1846:
1842:
1838:
1831:
1828:
1823:
1819:
1814:
1809:
1805:
1801:
1797:
1793:
1789:
1782:
1780:
1776:
1771:
1767:
1762:
1757:
1753:
1749:
1745:
1741:
1737:
1730:
1727:
1722:
1718:
1713:
1708:
1704:
1700:
1696:
1689:
1687:
1683:
1678:
1674:
1669:
1664:
1660:
1656:
1652:
1648:
1644:
1637:
1635:
1631:
1626:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1591:
1588:
1583:
1579:
1574:
1569:
1565:
1561:
1557:
1553:
1549:
1542:
1540:
1536:
1531:
1527:
1523:
1519:
1515:
1511:
1507:
1503:
1499:
1495:
1488:
1486:
1482:
1477:
1473:
1469:
1463:
1459:
1455:
1451:
1444:
1441:
1436:
1432:
1427:
1422:
1418:
1414:
1410:
1403:
1401:
1399:
1397:
1395:
1391:
1386:
1382:
1378:
1374:
1371:(7): 194871.
1370:
1366:
1359:
1357:
1353:
1348:
1344:
1340:
1336:
1332:
1328:
1324:
1320:
1313:
1311:
1307:
1302:
1298:
1293:
1288:
1284:
1280:
1276:
1272:
1268:
1261:
1258:
1253:
1249:
1244:
1239:
1235:
1231:
1227:
1220:
1218:
1214:
1209:
1205:
1201:
1197:
1193:
1189:
1182:
1179:
1174:
1170:
1165:
1160:
1156:
1152:
1148:
1144:
1140:
1133:
1131:
1127:
1122:
1118:
1114:
1110:
1106:
1102:
1095:
1093:
1089:
1084:
1080:
1075:
1070:
1066:
1062:
1058:
1054:
1050:
1043:
1041:
1037:
1032:
1028:
1023:
1018:
1014:
1010:
1006:
1002:
998:
991:
988:
983:
979:
974:
969:
965:
961:
957:
953:
949:
942:
940:
936:
931:
927:
922:
917:
913:
909:
905:
898:
896:
892:
887:
883:
878:
873:
869:
865:
861:
854:
852:
848:
843:
837:
833:
826:
824:
822:
820:
816:
811:
807:
802:
797:
792:
787:
783:
779:
775:
768:
766:
764:
760:
755:
751:
747:
743:
739:
732:
730:
728:
726:
724:
720:
715:
711:
707:
703:
699:
695:
688:
686:
684:
682:
680:
678:
676:
672:
666:
662:
659:
657:
654:
652:
649:
647:
644:
641:
637:
636:
631:
630:
626:
623:are found in
622:
612:
609:
605:
601:
597:
594:
590:
587:
586:
581:
577:
574:
573:
569:
567:
565:
560:
555:
553:
549:
545:
541:
535:
527:
525:
523:
519:
515:
514:
508:
507:
502:
498:
494:
490:
485:
483:
479:
478:
473:
469:
464:
462:
458:
454:
450:
446:
442:
438:
437:
431:
429:
425:
423:
418:
416:
411:
407:
403:
399:
395:
390:
382:
378:
374:
370:
365:
358:
356:
354:
353:
348:
344:
340:
336:
334:
330:
325:
321:
319:
315:
311:
310:
309:Synechocystis
305:
297:
292:
285:
283:
279:
275:
273:
269:
265:
261:
257:
252:
250:
246:
245:sampling bias
242:
238:
230:
228:
225:
221:
216:
212:
208:
206:
202:
198:
194:
193:
188:
183:
181:
180:bacteriophage
177:
176:magnesium ion
173:
169:
165:
161:
157:
153:
145:
143:
141:
137:
133:
129:
127:
121:
116:
114:
110:
106:
102:
97:
95:
91:
87:
83:
79:
74:
72:
68:
67:pathogenicity
64:
60:
56:
52:
48:
44:
40:
32:
28:
23:
19:
2199:
2195:
2185:
2158:
2155:Microbiology
2154:
2144:
2119:
2115:
2109:
2074:
2070:
2060:
2033:
2027:
2014:
1987:
1983:
1973:
1948:
1944:
1938:
1893:
1889:
1879:
1844:
1840:
1830:
1795:
1791:
1746:(19): e124.
1743:
1739:
1729:
1705:(1): 21–33.
1702:
1698:
1650:
1646:
1600:
1596:
1590:
1555:
1551:
1497:
1493:
1449:
1443:
1419:(1): 84–89.
1416:
1412:
1368:
1364:
1322:
1318:
1274:
1270:
1260:
1233:
1229:
1191:
1187:
1181:
1146:
1142:
1104:
1100:
1056:
1052:
1004:
1000:
990:
955:
951:
911:
907:
867:
863:
831:
781:
777:
737:
697:
693:
639:
633:
627:
604:ROSE element
599:
592:
583:
556:
552:biomolecules
546:evolved and
537:
521:
512:
504:
496:
493:upregulation
486:
475:
472:thermophilic
465:
461:sigma factor
456:
452:
434:
432:
421:
414:
386:
372:
368:
350:
337:
326:
322:
307:
301:
280:
276:
253:
249:genomic data
234:
231:Distribution
223:
219:
217:
213:
209:
190:
184:
149:
132:ROSE element
125:
117:
109:nucleic acid
101:riboswitches
98:
75:
42:
38:
36:
33:highlighted.
18:
1413:RNA Biology
700:(1): 3–16.
402:start codon
347:base change
304:nucleotides
256:prokaryotes
90:translation
49:-sensitive
47:temperature
2239:Categories
1653:(1): 1–2.
784:: 938570.
667:References
585:Drosophila
578:regulates
511:5′ DNA of
474:bacterium
445:base-pairs
333:pseudoknot
262:including
138:, and the
71:starvation
63:cold shock
59:heat shock
2020:Gilbert W
548:selection
503:genes in
501:virulence
489:pathogens
441:stem-loop
428:RpoS mRNA
359:Mechanism
339:Synthetic
329:CspA mRNA
286:Structure
203:than the
201:conserved
168:lysogenic
146:Discovery
2226:15466035
2177:20864473
2136:18778966
2101:17846637
2006:12230973
1965:20655297
1930:20625392
1890:PLOS ONE
1871:21727085
1822:20660079
1770:18753148
1721:20129052
1677:20129048
1625:84557068
1617:16336122
1582:16710302
1522:16554823
1476:19494584
1435:20009504
1385:36041664
1347:24747327
1339:17647020
1301:10090722
1252:10090718
1083:21131278
1031:24013626
982:11726689
930:15347681
886:17630972
810:36092413
754:38311501
714:16438677
618:and ROSE
606:and the
455:gene in
410:ribosome
314:PCC 6803
312:species
115:system.
2092:4689321
2038:Bibcode
1921:2896394
1898:Bibcode
1862:3185406
1813:2924531
1761:2577334
1668:5315359
1573:1478195
1530:4311262
1502:Bibcode
1208:1827163
1173:2941413
1121:2532257
1074:3074152
1022:3811586
801:9450479
600:E. coli
522:E. coli
516:to the
424:-acting
417:-acting
406:peptide
352:in vivo
318:hairpin
260:mammals
45:) is a
2224:
2217:522190
2214:
2175:
2134:
2099:
2089:
2029:Nature
2004:
1963:
1928:
1918:
1869:
1859:
1820:
1810:
1768:
1758:
1719:
1675:
1665:
1623:
1615:
1580:
1570:
1528:
1520:
1494:Nature
1474:
1464:
1433:
1383:
1345:
1337:
1299:
1292:316556
1289:
1250:
1206:
1171:
1164:212897
1161:
1119:
1081:
1071:
1029:
1019:
980:
970:
928:
884:
838:
808:
798:
752:
712:
596:operon
264:humans
134:, the
130:, the
124:Hsp90
122:, the
1621:S2CID
1526:S2CID
1343:S2CID
973:96696
593:ibpAB
580:hsp90
443:that
422:trans
373:right
164:lytic
158:cIII
154:in a
120:FourU
2222:PMID
2173:PMID
2132:PMID
2097:PMID
2002:PMID
1984:Cell
1961:PMID
1926:PMID
1867:PMID
1818:PMID
1766:PMID
1717:PMID
1673:PMID
1613:PMID
1578:PMID
1518:PMID
1472:PMID
1462:ISBN
1431:PMID
1381:PMID
1369:1865
1335:PMID
1297:PMID
1248:PMID
1204:PMID
1169:PMID
1117:PMID
1079:PMID
1027:PMID
978:PMID
926:PMID
882:PMID
836:ISBN
806:PMID
750:PMID
710:PMID
640:HspA
632:and
614:ROSE
591:The
513:prfA
497:prfA
453:rpoH
369:left
296:ROSE
187:rpoH
160:mRNA
94:gene
80:and
69:and
41:(or
25:The
2212:PMC
2204:doi
2200:186
2163:doi
2159:157
2124:doi
2087:PMC
2079:doi
2046:doi
2034:319
1992:doi
1988:110
1953:doi
1949:399
1916:PMC
1906:doi
1857:PMC
1849:doi
1808:PMC
1800:doi
1792:RNA
1756:PMC
1748:doi
1707:doi
1663:PMC
1655:doi
1605:doi
1601:386
1568:PMC
1560:doi
1510:doi
1498:440
1454:doi
1421:doi
1373:doi
1327:doi
1323:278
1287:PMC
1279:doi
1238:doi
1196:doi
1192:218
1159:PMC
1151:doi
1147:167
1109:doi
1105:210
1069:PMC
1061:doi
1017:PMC
1009:doi
1005:195
968:PMC
960:doi
916:doi
912:279
872:doi
796:PMC
786:doi
742:doi
702:doi
620:AT2
598:of
582:in
495:of
468:kDa
415:cis
398:30S
381:30S
166:or
126:cis
61:or
37:An
2241::
2220:.
2210:.
2198:.
2194:.
2171:.
2157:.
2153:.
2130:.
2120:13
2118:.
2095:.
2085:.
2073:.
2069:.
2044:.
2032:.
2026:.
2000:.
1986:.
1982:.
1959:.
1947:.
1924:.
1914:.
1904:.
1892:.
1888:.
1865:.
1855:.
1845:39
1843:.
1839:.
1816:.
1806:.
1796:16
1794:.
1790:.
1778:^
1764:.
1754:.
1744:36
1742:.
1738:.
1715:.
1703:37
1701:.
1697:.
1685:^
1671:.
1661:.
1651:37
1649:.
1645:.
1633:^
1619:.
1611:.
1599:.
1576:.
1566:.
1556:25
1554:.
1550:.
1538:^
1524:.
1516:.
1508:.
1496:.
1484:^
1470:.
1460:.
1429:.
1415:.
1411:.
1393:^
1379:.
1367:.
1355:^
1341:.
1333:.
1321:.
1309:^
1295:.
1285:.
1275:13
1273:.
1269:.
1246:.
1234:13
1232:.
1228:.
1216:^
1202:.
1190:.
1167:.
1157:.
1145:.
1141:.
1129:^
1115:.
1103:.
1091:^
1077:.
1067:.
1057:39
1055:.
1051:.
1039:^
1025:.
1015:.
1003:.
999:.
976:.
966:.
956:29
954:.
950:.
938:^
924:.
910:.
906:.
894:^
880:.
868:65
866:.
862:.
850:^
818:^
804:.
794:.
782:13
780:.
776:.
762:^
748:.
740:.
722:^
708:.
698:30
696:.
674:^
554:.
484:.
463:.
449:Mg
389:5′
355:.
251:.
207:.
142:.
96:.
73:.
2228:.
2206::
2179:.
2165::
2138:.
2126::
2103:.
2081::
2075:8
2054:.
2048::
2040::
2008:.
1994::
1967:.
1955::
1932:.
1908::
1900::
1894:5
1873:.
1851::
1824:.
1802::
1772:.
1750::
1723:.
1709::
1679:.
1657::
1627:.
1607::
1584:.
1562::
1532:.
1512::
1504::
1478:.
1456::
1437:.
1423::
1417:7
1387:.
1375::
1349:.
1329::
1303:.
1281::
1254:.
1240::
1210:.
1198::
1175:.
1153::
1123:.
1111::
1085:.
1063::
1033:.
1011::
984:.
962::
932:.
918::
888:.
874::
844:.
812:.
788::
756:.
744::
716:.
704::
616:1
610:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.