992:
870:), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factor proteins (in 2018 Lambert et al. indicated there were about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern the level of transcription of the target gene.
425:
124:
780:
1003:(DNA-PKcs, KU70, KU80 and DNA LIGASE IV) (see figure). These enzymes repair the double-strand breaks within about 15 minutes to 2 hours. The double-strand breaks in the promoter are thus associated with TOP2B and at least these four repair enzymes. These proteins are present simultaneously on a single promoter nucleosome (there are about 147 nucleotides in the DNA sequence wrapped around a single nucleosome) located near the transcription start site of their target gene.
896:
636:
355:
937:. These MBD proteins have both a methyl-CpG-binding domain and a transcriptional repression domain. They bind to methylated DNA and guide or direct protein complexes with chromatin remodeling and/or histone modifying activity to methylated CpG islands. MBD proteins generally repress local chromatin by means such as catalyzing the introduction of repressive histone marks or creating an overall repressive chromatin environment through
36:
1006:
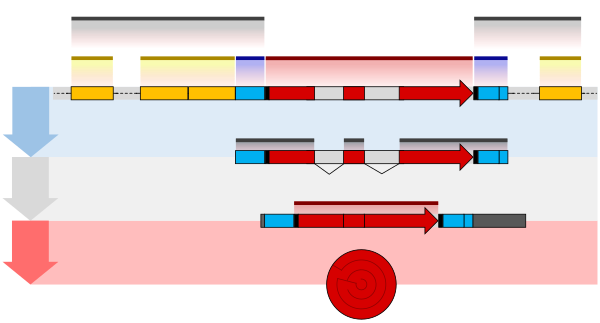
The double-strand break introduced by TOP2B apparently frees the part of the promoter at an RNA polymerase–bound transcription start site to physically move to its associated enhancer. This allows the enhancer, with its bound transcription factors and mediator proteins, to directly interact with the
958:
While only small amounts of EGR1 protein are detectable in cells that are un-stimulated, EGR1 translation into protein at one hour after stimulation is markedly elevated. Expression of EGR1 in various types of cells can be stimulated by growth factors, neurotransmitters, hormones, stress and injury.
954:
is a transcription factor important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences. There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located
947:
are proteins that bind to specific DNA sequences in order to regulate the expression of a given gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. There are approximately 1,400 different transcription factors encoded in the human genome and they
857:
are sequences of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene expression programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. In a study of brain cortical neurons, 24,937 loops
882:
may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of a transcription factor bound to an enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating a
967:
the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their
815:
shown by a small red star on a transcription factor on the enhancer) the enhancer is activated and can now activate its target promoter. The active enhancer is transcribed on each strand of DNA in opposite directions by bound RNAP IIs. Mediator (a complex consisting of about 26 proteins in an
1010:
Similarly, topoisomerase I (TOP1) enzymes appear to be located at many enhancers, and those enhancers become activated when TOP1 introduces a single-strand break. TOP1 causes single-strand breaks in particular enhancer DNA regulatory sequences when signaled by a specific enhancer-binding
877:
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the
858:
were found, bringing enhancers to promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and coordinate with each other to control expression of their common target gene.
751:(mRNA) molecules, where they are used to control mRNA biogenesis or translation. A variety of biological molecules may bind to the RNA to accomplish this regulation, including proteins (e.g., translational repressors and splicing factors), other RNA molecules (e.g.,
930:. About 59% of promoter sequences have a CpG island while only about 6% of enhancer sequences have a CpG island. CpG islands constitute regulatory sequences, since if CpG islands are methylated in the promoter of a gene this can reduce or silence gene expression.
926:). About 28 million CpG dinucleotides occur in the human genome. In most tissues of mammals, on average, 70% to 80% of CpG cytosines are methylated (forming 5-methyl-CpG, or 5-mCpG). Methylated cytosines within CpG sequences often occur in groups, called
991:
874:(a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (RNAP II) enzyme bound to the promoter.
948:
constitute about 6% of all human protein coding genes. About 94% of transcription factor binding sites that are associated with signal-responsive genes occur in enhancers while only about 6% of such sites occur in promoters.
861:
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of
1011:
transcription factor. Topoisomerase I breaks are associated with different DNA repair factors than those surrounding TOP2B breaks. In the case of TOP1, the breaks are associated most immediately with DNA repair enzymes
728:), while activators facilitate formation of a productive complex. Furthermore, DNA motifs have been shown to be predictive of epigenomic modifications, suggesting that transcription factors play a role in regulating the
806:
of the gene. The loop is stabilized by one architectural protein anchored to the enhancer and one anchored to the promoter and these proteins are joined to form a dimer (red zigzags). Specific regulatory
933:
DNA methylation regulates gene expression through interaction with methyl binding domain (MBD) proteins, such as MeCP2, MBD1 and MBD2. These MBD proteins bind most strongly to highly methylated
827:
that are located in DNA regions distant from the promoters of genes can have very large effects on gene expression, with some genes undergoing up to 100-fold increased expression due to such a
2625:
Jang WG, Kim EJ, Park KG, Park YB, Choi HS, Kim HJ, et al. (January 2007). "Glucocorticoid receptor mediated repression of human insulin gene expression is regulated by PGC-1alpha".
988:
is carried out in a mouse, this conditioning causes hundreds of gene-associated DSBs in the medial prefrontal cortex and hippocampus, which are important for learning and memory.
2438:
Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG (June 2006). "A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription".
922:
marker found predominantly on cytosines within CpG dinucleotides, which consist of a cytosine is followed by a guanine reading in the 5′ to 3′ direction along the DNA strand (
771:
A regulatory DNA sequence does not regulate unless it is activated. Different regulatory sequences are activated and then implement their regulation by different mechanisms.
811:
bind to DNA sequence motifs on the enhancer. General transcription factors bind to the promoter. When a transcription factor is activated by a signal (here indicated as
959:
In the brain, when neurons are activated, EGR1 proteins are upregulated, and they bind to (recruit) pre-existing TET1 enzymes, which are highly expressed in neurons.
2695:
424:
123:
963:
can catalyze demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can
980:(TOP2B) for activation. The induction of particular double-strand breaks is specific with respect to the inducing signal. When neurons are activated
2063:
Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM (April 2009). "A census of human transcription factors: function, expression and evolution".
976:
About 600 regulatory sequences in promoters and about 800 regulatory sequences in enhancers appear to depend on double-strand breaks initiated by
1020:
955:
in promoters and half in enhancers. The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.
1207:
995:
Regulatory sequence in a promoter at a transcription start site with a paused RNA polymerase and a TOP2B-induced double-strand break
388:
288:
2545:"A comparative analysis of relative occurrence of transcription factor binding sites in vertebrate genomes and gene promoter areas"
2226:"Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations"
1298:
Whitaker JW, Zhao Chen, Wei Wang. (2014) Predicting the Human
Epigenome from DNA Motifs. Nature Methods. doi:10.1038/nmeth.3065
820:
Expression of genes in mammals can be upregulated when signals are transmitted to the promoters associated with the genes.
2862:
2681:
1189:
1032:
100:
816:
interacting structure) communicates regulatory signals from the enhancer DNA-bound transcription factors to the promoter.
67:
63:
2688:
2791:
1125:
1000:
1796:"Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems"
1698:"The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription"
2867:
1411:
Spitz F, Furlong EE (September 2012). "Transcription factors: from enhancer binding to developmental control".
705:
672:
568:
384:
239:
2273:"Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks"
1457:
Schoenfelder S, Fraser P (August 2019). "Long-range enhancer-promoter contacts in gene expression control".
1202:
803:
779:
2837:
2832:
2827:
2816:
2806:
2801:
2796:
2781:
1172:
1167:
1162:
1151:
1141:
1136:
1131:
1115:
871:
821:
617:
327:
2821:
2811:
2786:
1156:
1146:
1120:
944:
848:
740:
709:
680:
656:
603:
596:
589:
582:
561:
404:
372:
306:
232:
1747:"MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300"
1313:"Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression"
1794:
Carullo NV, Phillips Iii RA, Simon RC, Soto SA, Hinds JE, Salisbury AJ, et al. (September 2020).
2704:
2447:
2389:
2126:
879:
844:
808:
713:
1696:
Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EE (January 2018).
1227:
899:
A methyl group is added on the carbon at the number 5 position of the ring to form 5-methylcytosine
854:
840:
836:
787:
725:
676:
668:
660:
519:
491:
484:
477:
469:
461:
454:
380:
376:
211:
204:
197:
189:
161:
154:
146:
2471:
2253:
2088:
1892:
Jabbari K, Bernardi G (May 2004). "Cytosine methylation and CpG, TpG (CpA) and TpA frequencies".
1482:
1436:
1222:
1194:
1049:
664:
512:
505:
297:
175:
895:
2378:"Profiling DNA break sites and transcriptional changes in response to contextual fear learning"
2224:
Dellino GI, Palluzzi F, Chiariello AM, Piccioni R, Bianco S, Furia L, et al. (June 2019).
2115:"EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity"
2642:
2607:
2566:
2525:
2463:
2417:
2358:
2304:
2245:
2206:
2152:
2080:
2045:
1999:
1958:
1909:
1874:
1825:
1776:
1727:
1678:
1626:
1572:
1531:
1474:
1428:
1393:
1342:
1281:
985:
964:
847:
and tethering elements. Among this constellation of sequences, enhancers and their associated
799:
48:
1500:
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. (December 2017).
2634:
2597:
2556:
2515:
2507:
2455:
2407:
2397:
2348:
2340:
2294:
2284:
2237:
2196:
2186:
2142:
2134:
2072:
2035:
1989:
1948:
1940:
1901:
1864:
1856:
1815:
1807:
1766:
1758:
1717:
1709:
1668:
1660:
1616:
1606:
1562:
1521:
1513:
1466:
1420:
1383:
1373:
1332:
1324:
1271:
1217:
1007:
RNA polymerase that had been paused at the transcription start site to start transcription.
903:
2763:
2753:
2748:
1096:
1086:
1081:
1070:
812:
756:
96:
1311:
Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, et al. (June 2020).
2451:
2393:
2271:
Singh S, Szlachta K, Manukyan A, Raimer HM, Dinda M, Bekiranov S, Wang YH (March 2020).
2130:
1549:
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. (February 2018).
999:
Such TOP2B-induced double-strand breaks are accompanied by at least four enzymes of the
2520:
2495:
2412:
2377:
2353:
2328:
2299:
2272:
2201:
2175:"Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation"
2174:
2147:
2114:
1953:
1928:
1869:
1844:
1820:
1795:
1722:
1697:
1673:
1648:
1621:
1594:
1526:
1501:
1388:
1361:
1337:
1312:
1184:
1035:
often contain regulatory regions, and so they are often the subject of these analyses.
721:
1771:
1746:
747:, or transcriptional termination. Regulatory sequences are frequently associated with
2856:
2738:
2475:
2327:
Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. (June 2015).
2257:
1486:
1440:
1064:
795:
748:
2561:
2544:
2329:"Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes"
1929:"Pervasive and CpG-dependent promoter-like characteristics of transcribed enhancers"
984:, just 22 TOP2B-induced double-strand breaks occur in their genomes. However, when
2773:
2225:
2092:
1595:"Positional specificity of different transcription factor classes within enhancers"
1108:
1016:
760:
744:
92:
1745:
Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M (January 2003).
17:
2402:
786:. An active enhancer regulatory sequence of DNA is enabled to interact with the
2728:
2173:
Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, et al. (2009).
1054:
960:
919:
907:
641:
540:
533:
320:
218:
56:
2638:
2511:
2344:
2138:
1905:
1599:
Proceedings of the
National Academy of Sciences of the United States of America
1567:
1550:
1517:
2602:
2585:
2241:
1593:
Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES (July 2018).
1470:
1378:
1328:
938:
724:
from forming a productive complex with the transcriptional initiation region (
717:
554:
547:
360:
225:
52:
2289:
2191:
1285:
2723:
2496:"Ligand-dependent enhancer activation regulated by topoisomerase-I activity"
2459:
1845:"DNA methylation in human epigenomes depends on local topology of CpG sites"
1611:
1276:
1259:
1044:
923:
729:
2646:
2611:
2570:
2529:
2494:
Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, et al. (January 2015).
2467:
2421:
2362:
2308:
2249:
2210:
2156:
2084:
2049:
2003:
1962:
1913:
1878:
1829:
1780:
1762:
1731:
1713:
1682:
1630:
1576:
1535:
1478:
1432:
1397:
1346:
712:) that activate or inhibit transcription. Transcription factors may act as
400:
396:
1944:
1811:
2733:
2718:
1860:
1235:
1059:
1039:
934:
927:
915:
752:
708:). It is accomplished through the sequence-specific binding of proteins (
526:
498:
313:
182:
168:
1362:"The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1"
2673:
2113:
Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, et al. (August 2019).
1994:
1977:
1031:
Genomes can be analyzed systematically to identify regulatory regions.
624:
343:
27:
Segment of nucleic acid that affects the expression of associated genes
2040:
2023:
1212:
1012:
644:
433:
392:
281:
274:
2076:
1664:
1424:
395:(light grey) and add a 5' cap and poly-A tail (dark grey). The mRNA
2758:
1091:
1076:
977:
778:
2841:
951:
863:
791:
701:
648:
635:
610:
364:
354:
334:
267:
260:
253:
2677:
2668:
2024:"Methyl-CpG-binding domain proteins: readers of the epigenome"
911:
884:
867:
736:
704:
expression normally happens at the level of RNA biosynthesis (
697:
575:
246:
29:
2543:
Stepanova M, Tiazhelova T, Skoblov M, Baranova A (May 2005).
1649:"The Mediator complex: a central integrator of transcription"
739:, regulation may occur at the level of protein biosynthesis (
103:
is an essential feature of all living organisms and viruses.
1927:
Steinhaus R, Gonzalez T, Seelow D, Robinson PN (June 2020).
1360:
Verheul TC, van Hijfte L, Perenthaler E, Barakat TS (2020).
34:
2663:
1502:"YY1 Is a Structural Regulator of Enhancer-Promoter Loops"
851:
have a leading role in the regulation of gene expression.
95:
molecule which is capable of increasing or decreasing the
1843:
Lövkvist C, Dodd IB, Sneppen K, Haerter JO (June 2016).
794:
by formation of a chromosome loop. This can initiate
1001:
non-homologous end joining (NHEJ) DNA repair pathway
415:
114:
2772:
2711:
2627:
371:controls when and where expression occurs for the
655:controls when expression occurs for the multiple
1978:"DNA methylation patterns and epigenetic memory"
1253:
1251:
883:promoter to initiate transcription of messenger
2669:ReMap - database of transcriptional regulators
2664:ORegAnno - Open Regulatory Annotation Database
1588:
1586:
1452:
1450:
720:, or both. Repressors often act by preventing
2689:
2584:Melloul D, Marshak S, Cerasi E (March 2002).
2322:
2320:
2318:
972:Activation by double- or single-strand breaks
8:
2022:Du Q, Luu PL, Stirzaker C, Clark SJ (2015).
1642:
1640:
2168:
2166:
1366:Frontiers in Cell and Developmental Biology
1260:"Eukaryotic and prokaryotic gene structure"
2696:
2682:
2674:
2586:"Regulation of insulin gene transcription"
2601:
2560:
2519:
2411:
2401:
2352:
2298:
2288:
2200:
2190:
2146:
2039:
1993:
1952:
1868:
1819:
1770:
1721:
1672:
1620:
1610:
1566:
1525:
1387:
1377:
1336:
1275:
941:remodeling and chromatin reorganization.
1306:
1304:
990:
894:
891:CpG island methylation and demethylation
2108:
2106:
2104:
2102:
1247:
802:(RNAP II) bound to the promoter at the
2489:
2487:
2485:
1653:Nature Reviews. Molecular Cell Biology
790:DNA regulatory sequence of its target
784:Regulation of transcription in mammals
775:Enhancer activation and implementation
604:Protein coding region
597:Protein coding region
576:DNA
282:Intron
275:Intron
268:Exon
261:Exon
254:Exon
247:DNA
99:of specific genes within an organism.
2433:
2431:
2376:Stott RT, Kritsky O, Tsai LH (2021).
2017:
2015:
2013:
403:untranslated regions (blue) regulate
387:of the gene into a pre-mRNA which is
7:
1258:Shafee, Thomas; Lowe, Rohan (2017).
321:Poly-A tail
314:5'cap
307:Protein coding region
1647:Allen BL, Taatjes DJ (March 2015).
1208:Open Regulatory Annotation Database
675:of the gene into an mRNA. The mRNA
625:Protein
611:mRNA
344:Protein
25:
1551:"The Human Transcription Factors"
683:into the final protein products.
634:
423:
353:
122:
407:into the final protein product.
1033:Conserved non-coding sequences
835:-regulatory sequences include
671:regions (yellow) regulate the
383:regions (yellow) regulate the
1:
2562:10.1093/bioinformatics/bti307
1190:Regulation of gene expression
1107:Regulatory sequences for the
849:transcription factor proteins
767:Activation and implementation
101:Regulation of gene expression
2403:10.1371/journal.pone.0249691
986:contextual fear conditioning
831:-regulatory sequence. These
527:3'UTR
499:5'UTR
448:Regulatory sequence
441:Regulatory sequence
300:mRNA
183:3'UTR
176:Open reading frame
169:5'UTR
140:Regulatory sequence
133:Regulatory sequence
2792:negative regulatory element
1126:negative regulatory element
2884:
2639:10.1016/j.bbrc.2006.11.074
2512:10.1016/j.cell.2014.12.023
2345:10.1016/j.cell.2015.05.032
2139:10.1038/s41467-019-11905-3
1906:10.1016/j.gene.2004.02.043
1568:10.1016/j.cell.2018.01.029
1518:10.1016/j.cell.2017.11.008
918:(see figure). 5-mC is an
68:the image placement policy
47:may require adjustment of
2603:10.1007/s00125-001-0728-y
2242:10.1038/s41588-019-0421-z
1471:10.1038/s41576-019-0128-0
1379:10.3389/fcell.2020.592164
1329:10.1038/s41593-020-0634-6
825:-regulatory DNA sequences
2290:10.1074/jbc.RA119.011665
2192:10.1186/gb-2009-10-4-r41
2065:Nature Reviews. Genetics
1459:Nature Reviews. Genetics
1413:Nature Reviews. Genetics
804:transcription start site
70:for further information.
2460:10.1126/science.1127196
1982:Genes & Development
1976:Bird A (January 2002).
1702:Genes & Development
1612:10.1073/pnas.1804663115
1264:WikiJournal of Medicine
1203:Gene regulatory network
1933:Nucleic Acids Research
1849:Nucleic Acids Research
1800:Nucleic Acids Research
1714:10.1101/gad.308619.117
996:
900:
887:from its target gene.
872:Mediator (coactivator)
817:
657:protein coding regions
45:This article's images
39:
2822:CAAT enhancer binding
2812:cAMP response element
2119:Nature Communications
1277:10.15347/wjm/2017.002
1157:CAAT enhancer binding
1147:cAMP response element
994:
945:Transcription factors
898:
809:transcription factors
782:
710:transcription factors
373:protein coding region
38:
2863:Regulatory sequences
2705:Regulatory sequences
1763:10.1093/emboj/cdg028
1512:(7): 1573–1588.e28.
880:transcription factor
798:(mRNA) synthesis by
677:untranslated regions
434:Polycistronic operon
289:Post-transcriptional
64:the picture tutorial
2452:2006Sci...312.1798J
2394:2021PLoSO..1649691S
2131:2019NatCo..10.3892S
1945:10.1093/nar/gkaa223
1812:10.1093/nar/gkaa671
1605:(30): E7222–E7230.
1317:Nature Neuroscience
653:Regulatory sequence
640:The structure of a
369:Regulatory sequence
359:The structure of a
337:mRNA
89:regulatory sequence
2446:(5781): 1798–802.
1995:10.1101/gad.947102
1861:10.1093/nar/gkw124
1050:Operator (biology)
997:
901:
818:
647:of protein-coding
91:is a segment of a
40:
18:Regulatory regions
2850:
2849:
2283:(12): 3990–4000.
2041:10.2217/epi.15.39
1939:(10): 5306–5317.
1806:(17): 9550–9570.
800:RNA polymerase II
759:, in the case of
743:), RNA cleavage,
694:
693:
690:
689:
414:
413:
85:
84:
16:(Redirected from
2875:
2698:
2691:
2684:
2675:
2651:
2650:
2622:
2616:
2615:
2605:
2581:
2575:
2574:
2564:
2540:
2534:
2533:
2523:
2491:
2480:
2479:
2435:
2426:
2425:
2415:
2405:
2373:
2367:
2366:
2356:
2324:
2313:
2312:
2302:
2292:
2268:
2262:
2261:
2236:(6): 1011–1023.
2221:
2215:
2214:
2204:
2194:
2170:
2161:
2160:
2150:
2110:
2097:
2096:
2060:
2054:
2053:
2043:
2019:
2008:
2007:
1997:
1973:
1967:
1966:
1956:
1924:
1918:
1917:
1889:
1883:
1882:
1872:
1840:
1834:
1833:
1823:
1791:
1785:
1784:
1774:
1751:The EMBO Journal
1742:
1736:
1735:
1725:
1693:
1687:
1686:
1676:
1644:
1635:
1634:
1624:
1614:
1590:
1581:
1580:
1570:
1546:
1540:
1539:
1529:
1497:
1491:
1490:
1454:
1445:
1444:
1408:
1402:
1401:
1391:
1381:
1357:
1351:
1350:
1340:
1308:
1299:
1296:
1290:
1289:
1279:
1255:
1218:DNA binding site
978:topoisomerase 2β
904:5-Methylcytosine
700:, regulation of
679:(blue) regulate
638:
637:
627:
620:
613:
606:
599:
592:
585:
578:
571:
564:
557:
550:
543:
536:
529:
522:
515:
508:
501:
494:
487:
480:
472:
464:
457:
450:
443:
436:
427:
416:
357:
356:
346:
339:
330:
323:
316:
309:
302:
293:
284:
277:
270:
263:
256:
249:
242:
235:
228:
221:
214:
207:
200:
192:
185:
178:
171:
164:
157:
149:
142:
135:
126:
115:
111:
110:
80:
77:
71:
37:
30:
21:
2883:
2882:
2878:
2877:
2876:
2874:
2873:
2872:
2868:Gene expression
2853:
2852:
2851:
2846:
2768:
2707:
2702:
2660:
2655:
2654:
2624:
2623:
2619:
2583:
2582:
2578:
2542:
2541:
2537:
2493:
2492:
2483:
2437:
2436:
2429:
2388:(7): e0249691.
2375:
2374:
2370:
2339:(7): 1592–605.
2326:
2325:
2316:
2270:
2269:
2265:
2230:Nature Genetics
2223:
2222:
2218:
2172:
2171:
2164:
2112:
2111:
2100:
2077:10.1038/nrg2538
2062:
2061:
2057:
2021:
2020:
2011:
1975:
1974:
1970:
1926:
1925:
1921:
1891:
1890:
1886:
1855:(11): 5123–32.
1842:
1841:
1837:
1793:
1792:
1788:
1744:
1743:
1739:
1695:
1694:
1690:
1665:10.1038/nrm3951
1646:
1645:
1638:
1592:
1591:
1584:
1548:
1547:
1543:
1499:
1498:
1494:
1456:
1455:
1448:
1425:10.1038/nrg3207
1410:
1409:
1405:
1359:
1358:
1354:
1310:
1309:
1302:
1297:
1293:
1257:
1256:
1249:
1244:
1198:-acting element
1181:
1105:
1071:Polyadenylation
1029:
974:
893:
813:phosphorylation
777:
769:
757:small molecules
686:
685:
684:
639:
631:
630:
629:
628:
623:
621:
616:
614:
609:
607:
602:
600:
595:
593:
588:
586:
581:
579:
574:
572:
567:
565:
560:
558:
553:
551:
546:
544:
539:
537:
532:
530:
525:
523:
518:
516:
511:
509:
504:
502:
497:
495:
490:
488:
483:
481:
475:
473:
467:
465:
460:
458:
453:
451:
446:
444:
439:
437:
432:
428:
410:
409:
408:
363:protein-coding
358:
350:
349:
348:
347:
342:
340:
336:
333:
331:
326:
324:
319:
317:
312:
310:
305:
303:
299:
296:
294:
290:
287:
285:
280:
278:
273:
271:
266:
264:
259:
257:
252:
250:
245:
243:
238:
236:
231:
229:
224:
222:
217:
215:
210:
208:
203:
201:
195:
193:
188:
186:
181:
179:
174:
172:
167:
165:
160:
158:
152:
150:
145:
143:
138:
136:
131:
127:
109:
81:
75:
72:
61:
49:image placement
41:
35:
28:
23:
22:
15:
12:
11:
5:
2881:
2879:
2871:
2870:
2865:
2855:
2854:
2848:
2847:
2845:
2844:
2835:
2830:
2825:
2819:
2814:
2809:
2804:
2799:
2794:
2789:
2784:
2778:
2776:
2770:
2769:
2767:
2766:
2761:
2756:
2751:
2746:
2741:
2736:
2731:
2726:
2721:
2715:
2713:
2709:
2708:
2703:
2701:
2700:
2693:
2686:
2678:
2672:
2671:
2666:
2659:
2658:External links
2656:
2653:
2652:
2617:
2576:
2555:(9): 1789–96.
2549:Bioinformatics
2535:
2481:
2427:
2368:
2314:
2263:
2216:
2179:Genome Biology
2162:
2098:
2055:
2034:(6): 1051–73.
2009:
1968:
1919:
1884:
1835:
1786:
1737:
1688:
1636:
1582:
1561:(4): 650–665.
1541:
1492:
1465:(8): 437–455.
1446:
1403:
1352:
1323:(6): 707–717.
1300:
1291:
1246:
1245:
1243:
1240:
1239:
1238:
1233:
1231:-acting factor
1225:
1220:
1215:
1210:
1205:
1200:
1192:
1187:
1185:Regulator gene
1180:
1177:
1176:
1175:
1170:
1165:
1160:
1154:
1149:
1144:
1139:
1134:
1129:
1123:
1118:
1104:
1101:
1100:
1099:
1094:
1089:
1084:
1079:
1074:
1068:
1062:
1057:
1052:
1047:
1042:
1028:
1025:
973:
970:
892:
889:
776:
773:
768:
765:
722:RNA polymerase
692:
691:
688:
687:
633:
632:
622:
615:
608:
601:
594:
587:
580:
573:
566:
559:
552:
545:
538:
531:
524:
517:
510:
503:
496:
489:
482:
474:
466:
459:
452:
445:
438:
431:
430:
429:
422:
421:
420:
419:
412:
411:
352:
351:
341:
332:
325:
318:
311:
304:
295:
286:
279:
272:
265:
258:
251:
244:
237:
230:
223:
216:
209:
202:
194:
187:
180:
173:
166:
159:
151:
144:
137:
130:
129:
128:
121:
120:
119:
118:
108:
105:
83:
82:
44:
42:
33:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2880:
2869:
2866:
2864:
2861:
2860:
2858:
2843:
2839:
2836:
2834:
2831:
2829:
2826:
2823:
2820:
2818:
2815:
2813:
2810:
2808:
2805:
2803:
2800:
2798:
2795:
2793:
2790:
2788:
2785:
2783:
2780:
2779:
2777:
2775:
2771:
2765:
2762:
2760:
2757:
2755:
2752:
2750:
2747:
2745:
2742:
2740:
2739:SECIS element
2737:
2735:
2732:
2730:
2727:
2725:
2722:
2720:
2717:
2716:
2714:
2710:
2706:
2699:
2694:
2692:
2687:
2685:
2680:
2679:
2676:
2670:
2667:
2665:
2662:
2661:
2657:
2648:
2644:
2640:
2636:
2633:(3): 716–21.
2632:
2628:
2621:
2618:
2613:
2609:
2604:
2599:
2596:(3): 309–26.
2595:
2591:
2587:
2580:
2577:
2572:
2568:
2563:
2558:
2554:
2550:
2546:
2539:
2536:
2531:
2527:
2522:
2517:
2513:
2509:
2506:(3): 367–80.
2505:
2501:
2497:
2490:
2488:
2486:
2482:
2477:
2473:
2469:
2465:
2461:
2457:
2453:
2449:
2445:
2441:
2434:
2432:
2428:
2423:
2419:
2414:
2409:
2404:
2399:
2395:
2391:
2387:
2383:
2379:
2372:
2369:
2364:
2360:
2355:
2350:
2346:
2342:
2338:
2334:
2330:
2323:
2321:
2319:
2315:
2310:
2306:
2301:
2296:
2291:
2286:
2282:
2278:
2274:
2267:
2264:
2259:
2255:
2251:
2247:
2243:
2239:
2235:
2231:
2227:
2220:
2217:
2212:
2208:
2203:
2198:
2193:
2188:
2184:
2180:
2176:
2169:
2167:
2163:
2158:
2154:
2149:
2144:
2140:
2136:
2132:
2128:
2124:
2120:
2116:
2109:
2107:
2105:
2103:
2099:
2094:
2090:
2086:
2082:
2078:
2074:
2071:(4): 252–63.
2070:
2066:
2059:
2056:
2051:
2047:
2042:
2037:
2033:
2029:
2025:
2018:
2016:
2014:
2010:
2005:
2001:
1996:
1991:
1987:
1983:
1979:
1972:
1969:
1964:
1960:
1955:
1950:
1946:
1942:
1938:
1934:
1930:
1923:
1920:
1915:
1911:
1907:
1903:
1899:
1895:
1888:
1885:
1880:
1876:
1871:
1866:
1862:
1858:
1854:
1850:
1846:
1839:
1836:
1831:
1827:
1822:
1817:
1813:
1809:
1805:
1801:
1797:
1790:
1787:
1782:
1778:
1773:
1768:
1764:
1760:
1757:(2): 281–91.
1756:
1752:
1748:
1741:
1738:
1733:
1729:
1724:
1719:
1715:
1711:
1707:
1703:
1699:
1692:
1689:
1684:
1680:
1675:
1670:
1666:
1662:
1659:(3): 155–66.
1658:
1654:
1650:
1643:
1641:
1637:
1632:
1628:
1623:
1618:
1613:
1608:
1604:
1600:
1596:
1589:
1587:
1583:
1578:
1574:
1569:
1564:
1560:
1556:
1552:
1545:
1542:
1537:
1533:
1528:
1523:
1519:
1515:
1511:
1507:
1503:
1496:
1493:
1488:
1484:
1480:
1476:
1472:
1468:
1464:
1460:
1453:
1451:
1447:
1442:
1438:
1434:
1430:
1426:
1422:
1419:(9): 613–26.
1418:
1414:
1407:
1404:
1399:
1395:
1390:
1385:
1380:
1375:
1371:
1367:
1363:
1356:
1353:
1348:
1344:
1339:
1334:
1330:
1326:
1322:
1318:
1314:
1307:
1305:
1301:
1295:
1292:
1287:
1283:
1278:
1273:
1269:
1265:
1261:
1254:
1252:
1248:
1241:
1237:
1234:
1232:
1230:
1226:
1224:
1221:
1219:
1216:
1214:
1211:
1209:
1206:
1204:
1201:
1199:
1197:
1193:
1191:
1188:
1186:
1183:
1182:
1178:
1174:
1171:
1169:
1166:
1164:
1161:
1158:
1155:
1153:
1150:
1148:
1145:
1143:
1140:
1138:
1135:
1133:
1130:
1127:
1124:
1122:
1119:
1117:
1114:
1113:
1112:
1110:
1102:
1098:
1095:
1093:
1090:
1088:
1085:
1083:
1080:
1078:
1075:
1072:
1069:
1066:
1065:SECIS element
1063:
1061:
1058:
1056:
1053:
1051:
1048:
1046:
1043:
1041:
1038:
1037:
1036:
1034:
1026:
1024:
1022:
1018:
1014:
1008:
1004:
1002:
993:
989:
987:
983:
979:
971:
969:
966:
962:
956:
953:
949:
946:
942:
940:
936:
931:
929:
925:
921:
917:
913:
909:
905:
897:
890:
888:
886:
881:
875:
873:
869:
865:
859:
856:
852:
850:
846:
842:
838:
834:
830:
826:
824:
814:
810:
805:
801:
797:
796:messenger RNA
793:
789:
785:
781:
774:
772:
766:
764:
762:
758:
754:
750:
749:messenger RNA
746:
742:
738:
733:
731:
727:
723:
719:
715:
711:
707:
706:transcription
703:
699:
682:
678:
674:
673:transcription
670:
666:
662:
658:
654:
650:
646:
643:
626:
619:
612:
605:
598:
591:
590:RBS
584:
583:RBS
577:
570:
569:Transcription
563:
556:
549:
542:
535:
528:
521:
514:
507:
500:
493:
486:
479:
471:
463:
456:
449:
442:
435:
426:
418:
417:
406:
402:
398:
394:
390:
386:
385:transcription
382:
378:
374:
370:
366:
362:
345:
338:
329:
322:
315:
308:
301:
292:
283:
276:
269:
262:
255:
248:
241:
240:Transcription
234:
227:
220:
213:
206:
199:
191:
184:
177:
170:
163:
156:
148:
141:
134:
125:
117:
116:
113:
112:
106:
104:
102:
98:
94:
90:
79:
76:February 2022
69:
65:
60:
58:
54:
50:
43:
32:
31:
19:
2774:Insulin gene
2743:
2630:
2626:
2620:
2593:
2590:Diabetologia
2589:
2579:
2552:
2548:
2538:
2503:
2499:
2443:
2439:
2385:
2381:
2371:
2336:
2332:
2280:
2276:
2266:
2233:
2229:
2219:
2182:
2178:
2122:
2118:
2068:
2064:
2058:
2031:
2027:
1985:
1981:
1971:
1936:
1932:
1922:
1897:
1893:
1887:
1852:
1848:
1838:
1803:
1799:
1789:
1754:
1750:
1740:
1708:(1): 42–57.
1705:
1701:
1691:
1656:
1652:
1602:
1598:
1558:
1554:
1544:
1509:
1505:
1495:
1462:
1458:
1416:
1412:
1406:
1369:
1365:
1355:
1320:
1316:
1294:
1267:
1263:
1228:
1195:
1109:insulin gene
1106:
1103:Insulin gene
1073:signal, mRNA
1030:
1009:
1005:
998:
981:
975:
957:
950:
943:
932:
910:form of the
906:(5-mC) is a
902:
876:
860:
853:
832:
828:
822:
819:
783:
770:
761:riboswitches
745:RNA splicing
734:
695:
652:
447:
440:
368:
291:modification
139:
132:
93:nucleic acid
88:
86:
73:
46:
2729:Pribnow box
2277:J Biol Chem
2125:(1): 3892.
2028:Epigenomics
1988:(1): 6–21.
1055:Pribnow box
968:promoters.
965:demethylate
961:TET enzymes
935:CpG islands
928:CpG islands
741:translation
681:translation
642:prokaryotic
618:Translation
405:translation
328:Translation
107:Description
62:Please see
2857:Categories
2185:(4): R41.
1372:: 592164.
1242:References
939:nucleosome
920:epigenetic
908:methylated
845:insulators
718:repressors
714:activators
562:Terminator
555:Stop
548:Stop
541:Start
534:Start
391:to remove
361:eukaryotic
233:Terminator
226:Stop
219:Start
212:Core
205:Proximal
97:expression
53:formatting
2724:CCAAT box
2476:206508330
2258:159041612
1900:: 143–9.
1487:152283312
1441:205485256
1286:2002-4436
1045:CCAAT box
924:CpG sites
855:Enhancers
841:silencers
837:enhancers
730:epigenome
2734:TATA box
2719:CAAT box
2647:17150186
2612:11914736
2571:15699025
2530:25619691
2468:16794079
2422:34197463
2382:PLOS ONE
2363:26052046
2309:32029477
2250:31110352
2211:19374776
2157:31467272
2085:19274049
2050:25927341
2004:11782440
1963:32338759
1914:15177689
1879:26932361
1830:32810208
1781:12514134
1732:29378788
1683:25693131
1631:29987030
1577:29425488
1536:29224777
1479:31086298
1433:22868264
1398:33102493
1347:32451484
1236:ORegAnno
1223:Promoter
1179:See also
1060:TATA box
1040:CAAT box
1027:Examples
982:in vitro
916:cytosine
788:promoter
726:promoter
669:enhancer
665:operator
661:Promoter
520:UTR
513:ORF
506:ORF
492:Promoter
485:Operator
478:silencer
470:silencer
462:Enhancer
455:Enhancer
389:modified
381:enhancer
377:Promoter
198:silencer
190:Enhancer
162:Promoter
155:silencer
147:Enhancer
2712:General
2521:4422651
2448:Bibcode
2440:Science
2413:8248687
2390:Bibcode
2354:4886855
2300:7086017
2202:2688932
2148:6715719
2127:Bibcode
2093:3207586
1954:7261191
1870:4914085
1821:7515708
1723:5828394
1674:4963239
1622:6065035
1527:5785279
1389:7554316
1338:7558717
659:(red).
393:introns
375:(red).
2645:
2610:
2569:
2528:
2518:
2474:
2466:
2420:
2410:
2361:
2351:
2307:
2297:
2256:
2248:
2209:
2199:
2155:
2145:
2091:
2083:
2048:
2002:
1961:
1951:
1912:
1877:
1867:
1828:
1818:
1779:
1772:140103
1769:
1730:
1720:
1681:
1671:
1629:
1619:
1575:
1534:
1524:
1485:
1477:
1439:
1431:
1396:
1386:
1345:
1335:
1284:
1213:Operon
1067:, mRNA
755:) and
645:operon
335:Mature
55:, and
2824:(CEB)
2764:G-box
2759:E-box
2754:C-box
2749:Z-box
2744:A-box
2472:S2CID
2254:S2CID
2089:S2CID
1483:S2CID
1437:S2CID
1270:(1).
1229:Trans
1159:(CEB)
1128:(NRE)
1111:are:
1097:G-box
1092:E-box
1087:C-box
1082:Z-box
1077:A-box
1017:RAD50
1013:MRE11
914:base
753:miRNA
649:genes
2842:ILPR
2643:PMID
2608:PMID
2567:PMID
2526:PMID
2500:Cell
2464:PMID
2418:PMID
2359:PMID
2333:Cell
2305:PMID
2246:PMID
2207:PMID
2153:PMID
2081:PMID
2046:PMID
2000:PMID
1959:PMID
1910:PMID
1894:Gene
1875:PMID
1826:PMID
1777:PMID
1728:PMID
1679:PMID
1627:PMID
1573:PMID
1555:Cell
1532:PMID
1506:Cell
1475:PMID
1429:PMID
1394:PMID
1343:PMID
1282:ISSN
1019:and
952:EGR1
864:CTCF
792:gene
702:gene
667:and
399:and
379:and
365:gene
298:Pre-
66:and
57:size
2635:doi
2631:352
2598:doi
2557:doi
2516:PMC
2508:doi
2504:160
2456:doi
2444:312
2408:PMC
2398:doi
2349:PMC
2341:doi
2337:161
2295:PMC
2285:doi
2281:295
2238:doi
2197:PMC
2187:doi
2143:PMC
2135:doi
2073:doi
2036:doi
1990:doi
1949:PMC
1941:doi
1902:doi
1898:333
1865:PMC
1857:doi
1816:PMC
1808:doi
1767:PMC
1759:doi
1718:PMC
1710:doi
1669:PMC
1661:doi
1617:PMC
1607:doi
1603:115
1563:doi
1559:172
1522:PMC
1514:doi
1510:171
1467:doi
1421:doi
1384:PMC
1374:doi
1333:PMC
1325:doi
1272:doi
1196:Cis
1021:ATR
912:DNA
885:RNA
868:YY1
866:or
833:cis
829:cis
823:Cis
737:RNA
735:In
698:DNA
696:In
2859::
2840:-
2838:G1
2833:E1
2828:C1
2817:A2
2807:A3
2802:E2
2797:C2
2782:A5
2641:.
2629:.
2606:.
2594:45
2592:.
2588:.
2565:.
2553:21
2551:.
2547:.
2524:.
2514:.
2502:.
2498:.
2484:^
2470:.
2462:.
2454:.
2442:.
2430:^
2416:.
2406:.
2396:.
2386:16
2384:.
2380:.
2357:.
2347:.
2335:.
2331:.
2317:^
2303:.
2293:.
2279:.
2275:.
2252:.
2244:.
2234:51
2232:.
2228:.
2205:.
2195:.
2183:10
2181:.
2177:.
2165:^
2151:.
2141:.
2133:.
2123:10
2121:.
2117:.
2101:^
2087:.
2079:.
2069:10
2067:.
2044:.
2030:.
2026:.
2012:^
1998:.
1986:16
1984:.
1980:.
1957:.
1947:.
1937:48
1935:.
1931:.
1908:.
1896:.
1873:.
1863:.
1853:44
1851:.
1847:.
1824:.
1814:.
1804:48
1802:.
1798:.
1775:.
1765:.
1755:22
1753:.
1749:.
1726:.
1716:.
1706:32
1704:.
1700:.
1677:.
1667:.
1657:16
1655:.
1651:.
1639:^
1625:.
1615:.
1601:.
1597:.
1585:^
1571:.
1557:.
1553:.
1530:.
1520:.
1508:.
1504:.
1481:.
1473:.
1463:20
1461:.
1449:^
1435:.
1427:.
1417:13
1415:.
1392:.
1382:.
1368:.
1364:.
1341:.
1331:.
1321:23
1319:.
1315:.
1303:^
1280:.
1266:.
1262:.
1250:^
1173:G1
1168:E1
1163:C1
1152:A2
1142:A3
1137:E2
1132:C2
1116:A5
1023:.
1015:,
843:,
839:,
763:.
732:.
716:,
663:,
651:.
401:3'
397:5'
367:.
87:A
51:,
2787:Z
2697:e
2690:t
2683:v
2649:.
2637::
2614:.
2600::
2573:.
2559::
2532:.
2510::
2478:.
2458::
2450::
2424:.
2400::
2392::
2365:.
2343::
2311:.
2287::
2260:.
2240::
2213:.
2189::
2159:.
2137::
2129::
2095:.
2075::
2052:.
2038::
2032:7
2006:.
1992::
1965:.
1943::
1916:.
1904::
1881:.
1859::
1832:.
1810::
1783:.
1761::
1734:.
1712::
1685:.
1663::
1633:.
1609::
1579:.
1565::
1538:.
1516::
1489:.
1469::
1443:.
1423::
1400:.
1376::
1370:8
1349:.
1327::
1288:.
1274::
1268:4
1121:Z
476:/
468:/
196:/
153:/
78:)
74:(
59:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.