388:
376:
29:
339:
613:) occurs in the same manner as the normal export of cellular mRNAs. On the other hand, unspliced and incompletely spliced mRNAs which code for the late, structural proteins are Rev-dependent. The Rev protein is expressed as an early gene from completely spliced transcripts, so the expression of late phase structural proteins cannot occur until an initial amount of Rev is produced.
335:-β (a nuclear transport factor). The Rev NLS is a highly similar sequence to that of the importin-β-binding site present within importin-α, which allows for the interaction between Rev and importin-β. The NLS overlaps with the sequence required for RNA-binding. This prevents the NLS from counteracting the export of RRE-containing mRNA transcripts.
371:
Within this purine-rich stem-loop, IIB, are non-canonical base pairs that form as a result of the mRNA stem loop-secondary structure. These base pairs include guanine-adenine (nucleotides 47 and 73, respectively) and guanine-guanine (nucleotides 48–71, respectively). The two base pairs are separated
367:
gene. The RRE remains functional if translocated, but needs to remain in the same orientation (cannot be inverted). The RRE is retained by incompletely processed mRNA transcripts. The secondary structure of the RRE creates eight stem-loops. Rev initially binds to the purine-rich stem-loop IIB, then
213:
mechanism. Therefore, the name of the protein was modified from Art to Trs (transregulator of splicing). The most recent studies have shown that the protein has multiple functions in the regulation of HIV-1 proteins, and its name has been changed to Rev (regulator of expression of virion proteins),
383:
The ARM contains residues R35 and R39 that make base-specific contacts with residues on the RRE mRNA, specifically to bases uracil 66, guanine 67, and guanine 70, respectively. On the opposite side of these bases, residues N40 and R44 make base-specific contacts with nucleotides uracil 45, guanine
342:
The secondary structure of the IIB binding site shows non-canonical base pairs G47 (magenta)-A73 (orange) and G48-G71 (magenta). Bulging, non-paired uridine nucleotide points outward from the secondary helix (colored red). The mRNA forms a stem-loop like structure with intricate folding (PDB
254:. The ARM is a highly specific sequence which allows for the multimerization of Rev proteins, prior to RNA binding. A single base substitution alters Rev's ability to form a tetramer. The arginine-rich domain of Rev interacts with the rev-binding element (RBE), which is part of the
570:
encoded. Rev also decreases the quantity of completely spliced viral messages expressed by exporting pre-mRNA before it can be spliced. This results in decreased expression of the regulatory proteins, Rev and Tat. Since Rev is continuously shuttled between the
2277:
Plavec I, Agarwal M, Ho KE, Pineda M, Auten J, Baker J, et al. (February 1997). "High transdominant RevM10 protein levels are required to inhibit HIV-1 replication in cell lines and primary T cells: implication for gene therapy of AIDS".
33:
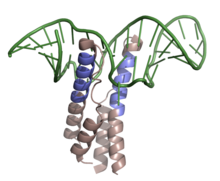
Shown above is the RNP complex formed by HIV-1 REV binding to the RRE upon mRNA of the env gene. Highlighted in slate is the ARM of the RNA binding domain, while colored in green is the mRNA with secondary stem-loop-like structure (PDB
306:
HIV-1 regulatory proteins (including Rev) are translated from completely processed mRNA transcripts, while structural proteins are translated from incompletely spliced transcripts. Completely spliced transcripts are exported from the
627:
B (LMB) binds to CRM1 which prevents the formation of the complex required for export(CRM1/NES/RanGTP/RRE) and ultimately reduces the production of incompletely spliced RNAs. Therefore, structural proteins, which are necessary for
208:
mRNA. The unknown protein functioned by removing repression of regulatory sequences and was named Art (anti-repression transactivator). Later studies suggested that the protein was involved in regulation of the
379:
Shown are residues Arg35 and Arg39 (colored by element with IUPAC standards) that make specific contacts with residues uracil 66 (red), guanine 67, and guanine 70 (magenta) during RNA binding (PDB 4PMI).
391:
Shown are residues N40 and R44 (colored by element with IUPAC standards) making specific contacts with residues uracil 45 (red), guanine 46 (magenta), guanine 47 (magenta), and adenine 73 (orange) (PDB
1828:
Kubota S, Siomi H, Satoh T, Endo S, Maki M, Hatanaka M (August 1989). "Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein".
1419:
Kubota S, Siomi H, Satoh T, Endo S, Maki M, Hatanaka M (August 1989). "Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein".
1912:
Feinberg MB, Jarrett RF, Aldovini A, Gallo RC, Wong-Staal F (September 1986). "HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA".
752:
Feinberg MB, Jarrett RF, Aldovini A, Gallo RC, Wong-Staal F (September 1986). "HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA".
1239:"Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals"
384:
46, guanine 47, and adenine 73. In addition to these stabilizing contacts, additional Arg residues within the ARM, as well as T34, make nonspecific contacts with bases on the mRNA.
2207:
DeJong ES, Chang CE, Gilson MK, Marino JP (July 2003). "Proflavine acts as a Rev inhibitor by targeting the high-affinity Rev binding site of the Rev responsive element of HIV-1".
499:
are exported, as opposed to the mechanism for export of cellular mRNAs. Rev is able to facilitate export of pre-mRNA transcripts that would otherwise typically remain in the
621:
Since Rev is absolutely necessary for HIV-1 replication and it is expressed early on in infection, it has been suggested that Rev is a good target for antiviral therapies.
2586:
2581:
4008:
701:
Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W (May 1986). "A second post-transcriptional trans-activator gene required for HTLV-III replication".
129:
404:
mRNA in the cytoplasm. The RRE also facilitates multimerization of the Rev proteins, which is required for Rev binding and function. The Rev protein binds unspliced
2638:
387:
375:
230:. Rev's sequence contains two specific domains which contribute to its nuclear import and export. The protein typically performs its function as a tetramer.
2242:
Xiao G, Kumar A, Li K, Rigl CT, Bajic M, Davis TM, et al. (May 2001). "Inhibition of the HIV-1 rev-RRE complex formation by unfused aromatic cations".
2137:
Xiao G, Kumar A, Li K, Rigl CT, Bajic M, Davis TM, et al. (May 2001). "Inhibition of the HIV-1 rev-RRE complex formation by unfused aromatic cations".
1065:
Surendran R, Herman P, Cheng Z, Daly TJ, Ching Lee J (March 2004). "HIV Rev self-assembly is linked to a molten-globule to compact structural transition".
887:
Surendran R, Herman P, Cheng Z, Daly TJ, Ching Lee J (March 2004). "HIV Rev self-assembly is linked to a molten-globule to compact structural transition".
3166:
3161:
3156:
3151:
3113:
3108:
563:
loop, which regulates Rev production. There is a decrease in Rev production when Rev protein levels are higher than is necessary for the given amount of
1957:"The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region"
1456:"The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region"
315:
by the same mechanism as cellular mRNA. However, Rev is needed to export incompletely spliced mRNAs in order to produce the viral structural proteins.
3488:
3146:
3141:
2617:
2612:
2607:
2331:
1689:"Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif"
1008:"Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif"
1503:
Nakatani K, Horie S, Goto Y, Kobori A, Hagihara S (August 2006). "Evaluation of mismatch-binding ligands as inhibitors for Rev-RRE interaction".
3869:
1100:
Auer, Manfred; Gremlich, Hans Ulrich; Seifert, Jan Marcus; Daly, Thomas J.; Parslow, Tristram G.; Casari, Georg; Gstach, Hubert (1994-03-15).
551:
expression is lower in the absence of Rev and higher in the presence of Rev. Rev negatively regulates the expression of the regulatory genes (
3632:
77:
852:
Cochrane A, Kramer R, Ruben S, Levine J, Rosen CA (July 1989). "The human immunodeficiency virus rev protein is a nuclear phosphoprotein".
647:
are small molecules that can prevent Rev from binding to the RRE sequence. If Rev is incapable of binding to the RRE on the pre-mRNA, the
511:
Rev acts post-transcriptionally to positively regulate the expression of structural genes and to negatively regulate the expression of
2444:
3602:
3536:
2763:
270:
motif, which allows the REV protein to stably bind to the RRE RNA to form the ribonucleoprotein complex. The domain also contains a
480:. Once the pre-mRNAs are in the cytoplasm, Rev dissociates, revealing the NLS. Exposure of the NLS allows for Rev interaction with
3230:
2898:
267:
192:. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
1540:"A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA"
583:
transcripts. Maintenance of the proper balance between early and late viral gene quantities leads to an overall increase in
149:
1865:"The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs"
3749:
460:
into Ran-GTP, causing the importation complex to disassemble. Upon disassembly, Rev's NES forms a new complex with CRM1 (
3223:
3218:
2324:
3851:
3786:
3458:
3434:
3241:
3126:
3032:
2958:
2883:
2778:
2662:
2597:
2517:
2483:
2434:
445:
324:
271:
177:
2527:
2454:
2419:
3409:
3213:
2414:
3908:
3796:
3722:
3660:
3612:
3564:
3516:
3468:
3347:
3196:
3091:
2852:
2758:
2738:
2721:
2571:
2459:
2394:
255:
294:-rich. Binding of Rev to viral RNAs containing the RRE allows for mRNA export out of the nucleus and into the
137:
28:
1372:"A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus"
2317:
2561:
3971:
3807:
3692:
2532:
468:-GTP at the RRE sequence within incompletely spliced transcripts. Following assembly of the complex, the
3941:
3832:
3712:
3584:
3444:
449:
283:
133:
3743:
3681:
3368:
2812:
2803:
2725:
2672:
2493:
2384:
1700:
1019:
808:
710:
681:
635:
It has been shown that various organic compounds have the ability to target the Rev/RRE interaction.
90:
3414:
2808:
2798:
2717:
1955:
Hadzopoulou-Cladaras M, Felber BK, Cladaras C, Athanassopoulos A, Tse A, Pavlakis GN (March 1989).
1454:
Hadzopoulou-Cladaras M, Felber BK, Cladaras C, Athanassopoulos A, Tse A, Pavlakis GN (March 1989).
251:
242:
region of Rev contains an arginine-rich sequence. The arginine-rich motif (ARM) is located between
3936:
3813:
2119:
1937:
1894:
1317:
834:
777:
734:
372:
by a non-stacked and bulging uridine that points outwards, away from the ARM-RNA interactions.
4003:
3388:
3136:
3075:
2840:
2295:
2259:
2224:
2189:
2154:
2111:
2070:
2021:
1986:
1929:
1886:
1845:
1810:
1769:
1728:
1669:
1620:
1569:
1520:
1485:
1436:
1401:
1352:
1309:
1268:
1219:
1170:
1129:
1121:
1082:
1047:
988:
953:
904:
869:
826:
769:
726:
678:
Rev protein for the RRE binding site, and therefore decrease Rev's normal cellular functions.
560:
124:
3650:
3256:
2993:
2988:
2983:
2978:
2973:
2968:
2947:
2287:
2251:
2216:
2181:
2146:
2101:
2060:
2052:
2013:
1976:
1968:
1921:
1876:
1837:
1800:
1759:
1718:
1708:
1659:
1651:
1610:
1600:
1559:
1551:
1512:
1475:
1467:
1428:
1391:
1383:
1344:
1299:
1258:
1250:
1209:
1201:
1160:
1113:
1074:
1037:
1027:
980:
943:
935:
896:
861:
816:
761:
718:
610:
512:
527:
Rev-mediated export from the nucleus increases cytoplasmic levels of the structural mRNAs (
338:
116:
3880:
3337:
3327:
3080:
2041:"Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein"
1704:
1190:"Mutational definition of the human immunodeficiency virus type 1 Rev activation domain"
1023:
812:
714:
3954:
3837:
3382:
3207:
3102:
2708:
2628:
1615:
1588:
1564:
1539:
1387:
1165:
1148:
939:
327:(NLS), allows Rev to enter the nucleus. Entry requires binding between a Rev multimer,
2255:
2150:
2106:
2089:
2065:
2040:
1981:
1956:
1805:
1788:
1664:
1639:
1480:
1455:
1396:
1371:
1304:
1287:
1263:
1238:
1214:
1189:
948:
923:
3997:
3903:
3554:
3015:
2652:
2369:
2344:
2340:
1925:
1881:
1864:
1841:
1723:
1688:
1638:
Hammarskjöld ML, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D (May 1989).
1432:
1042:
1007:
865:
765:
667:
658:
Other therapies target the Rev protein itself, since it is an essential component of
465:
457:
328:
223:
82:
2123:
2056:
1972:
1941:
1898:
1655:
1471:
1321:
1254:
1205:
838:
781:
58:
3623:
3575:
3527:
3479:
3400:
3185:
3021:
2698:
2507:
2374:
2348:
1640:"Regulation of human immunodeficiency virus env expression by the rev gene product"
1348:
1101:
738:
572:
500:
485:
473:
453:
441:
308:
210:
185:
112:
2309:
1555:
352:
The rev response element (RRE) is a 240 base-pair sequence located in the second
70:
3963:
3506:
2826:
2789:
243:
1693:
Proceedings of the
National Academy of Sciences of the United States of America
1012:
Proceedings of the
National Academy of Sciences of the United States of America
3845:
3777:
3374:
3180:
3011:
3007:
1589:"HIV Rev Assembly on the Rev Response Element (RRE): A Structural Perspective"
1516:
1078:
900:
663:
644:
624:
287:
239:
227:
173:
2185:
1125:
266:
gene. The alpha-helical secondary structure specifically can be considered a
3886:
3827:
3821:
3065:
2942:
2932:
2920:
2693:
2357:
1713:
1032:
675:
652:
576:
477:
437:
312:
295:
2263:
2228:
2193:
2158:
2025:
2017:
1764:
1747:
1624:
1573:
1524:
1086:
992:
984:
908:
188:, where it is involved in the export of unspliced and incompletely spliced
2299:
2291:
2115:
2074:
1990:
1933:
1890:
1849:
1814:
1773:
1732:
1673:
1489:
1440:
1405:
1356:
1313:
1272:
1223:
1174:
1133:
1051:
957:
873:
830:
795:
Gallo R, Wong-Staal F, Montagnier L, Haseltine WA, Yoshida M (June 1988).
773:
730:
86:
2473:
636:
481:
461:
332:
65:
2172:
Baba M (2004). "Inhibitors of HIV-1 gene expression and transcription".
1117:
1748:"Effect of Rev on the intranuclear localization of HIV-1 unspliced RNA"
671:
491:
Rev-directed export of viral RNAs is similar to the mechanism by which
291:
165:
2220:
3733:
3671:
3419:
3358:
3251:
2749:
2405:
1863:
Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R (August 1995).
1605:
821:
796:
722:
640:
629:
584:
567:
469:
360:
353:
259:
144:
2090:"CRM1 is an export receptor for leucine-rich nuclear export signals"
1288:"CRM1 is an export receptor for leucine-rich nuclear export signals"
1149:"Transport of macromolecules between the nucleus and the cytoplasm"
3304:
3294:
3289:
3279:
3274:
3269:
3264:
2871:
2866:
2861:
659:
598:
595:
564:
503:, suggesting that the Rev NES is dominant over nuclear retention.
492:
386:
374:
357:
337:
169:
3856:
3687:
3299:
3284:
2713:
2449:
606:
580:
496:
189:
106:
53:
2313:
200:
A novel protein was found to be involved in the translation of
3917:
3913:
3896:
3042:
2893:
1335:
Pollard VW, Malim MH (October 1998). "The HIV-1 Rev protein".
648:
602:
428:
transcripts at the RRE, facilitating export to the cytoplasm.
1746:
Favaro JP, Borg KT, Arrigo SJ, Schmidt MG (September 1998).
2088:
Fornerod M, Ohno M, Yoshida M, Mattaj IW (September 1997).
1370:
Görlich D, Henklein P, Laskey RA, Hartmann E (April 1996).
1286:
Fornerod M, Ohno M, Yoshida M, Mattaj IW (September 1997).
655:, also resulting in lack of necessary structural proteins.
605:
or from intron-containing RNA. The export of fully spliced
323:
The arginine-rich domain of the Rev protein, containing a
184:
gene, which allows the Rev protein to be localized to the
1687:
Zapp ML, Hope TJ, Parslow TG, Green MR (September 1991).
1006:
Zapp ML, Hope TJ, Parslow TG, Green MR (September 1991).
684:
was also identified as a Rev-export inhibitory congener.
662:
infection. M10 is a mutated form of Rev and has a single
1188:
Malim MH, McCarn DF, Tiley LS, Cullen BR (August 1991).
674:). If delivered to cells, Rev M10 will compete with the
922:
Kjems J, Calnan BJ, Frankel AD, Sharp PA (March 1992).
579:, small amounts of the protein are able to impact many
298:
by a mechanism different than that of cellular mRNAs.
1538:
Daugherty MD, D'Orso I, Frankel AD (September 2008).
924:"Specific binding of a basic peptide from HIV-1 Rev"
400:-acting, and is necessary to achieve high levels of
3962:
3953:
3930:
3867:
3794:
3785:
3776:
3732:
3720:
3711:
3670:
3658:
3649:
3622:
3610:
3601:
3574:
3562:
3553:
3526:
3514:
3505:
3478:
3466:
3457:
3432:
3399:
3357:
3345:
3336:
3321:
3239:
3194:
3179:
3124:
3089:
3074:
3059:
3030:
3006:
2956:
2941:
2928:
2919:
2881:
2850:
2839:
2788:
2776:
2748:
2736:
2707:
2689:
2660:
2651:
2626:
2595:
2569:
2560:
2549:
2515:
2506:
2481:
2472:
2432:
2404:
2392:
2383:
2365:
2356:
2039:Felber BK, Drysdale CM, Pavlakis GN (August 1990).
2004:Hope TJ (May 1999). "The ins and outs of HIV Rev".
1830:
1421:
971:Hope TJ (May 1999). "The ins and outs of HIV Rev".
143:
123:
105:
100:
76:
64:
52:
44:
39:
21:
591:Transition from early to late phase HIV-1 genes
1237:Meyer BE, Meinkoth JL, Malim MH (April 1996).
2325:
601:are expressed from either completely spliced
515:. Rev positively regulates the expression of
278:Rev-activation domain (Nuclear export signal)
214:which more generally describes its function.
8:
444:. The shuttling of Rev is regulated by its
3959:
3950:
3791:
3782:
3729:
3717:
3667:
3655:
3619:
3607:
3571:
3559:
3523:
3511:
3475:
3463:
3354:
3342:
3333:
3191:
3086:
3071:
3027:
2953:
2938:
2925:
2847:
2785:
2745:
2733:
2704:
2657:
2566:
2557:
2512:
2478:
2401:
2389:
2380:
2362:
2332:
2318:
2310:
1587:Rausch, Jason; Grice, Stuart (June 2015).
368:binds to a secondary site in stem-loop I.
97:
27:
3489:Parainfluenza hemagglutinin-neuraminidase
2105:
2064:
1980:
1880:
1804:
1763:
1722:
1712:
1663:
1614:
1604:
1563:
1479:
1395:
1303:
1262:
1213:
1164:
1041:
1031:
947:
820:
436:Rev is continuously shuttled between the
4009:Viral regulatory and accessory proteins
2006:Archives of Biochemistry and Biophysics
973:Archives of Biochemistry and Biophysics
693:
617:Rev as a target for antiviral therapies
484:-β in order to shuttle Rev back to the
472:-containing RNAs are exported from the
168:that is essential to the regulation of
18:
3633:Respiratory syncytial virus G protein
2174:Current Topics in Medicinal Chemistry
1102:"Helix-Loop-Helix Motif in HIV-1 Rev"
412:transcripts and incompletely spliced
7:
2244:Bioorganic & Medicinal Chemistry
2139:Bioorganic & Medicinal Chemistry
1505:Bioorganic & Medicinal Chemistry
286:is located in residues 71–82 of the
1147:Izaurralde E, Adam S (April 1998).
1388:10.1002/j.1460-2075.1996.tb00530.x
940:10.1002/j.1460-2075.1992.tb05152.x
14:
3537:Mumps hemagglutinin-neuraminidase
507:Regulation of HIV gene expression
2899:Hepatitis B virus DNA polymerase
363:, immediately downstream of the
250:gene and forms an alpha-helical
226:protein that is composed of 116
2730:(non-enveloped circular ds-DNA)
2057:10.1128/JVI.64.8.3734-3741.1990
1973:10.1128/JVI.63.3.1265-1274.1989
1656:10.1128/JVI.63.5.1959-1966.1989
1472:10.1128/JVI.63.3.1265-1274.1989
1255:10.1128/JVI.70.4.2350-2359.1996
1206:10.1128/JVI.65.8.4248-4254.1991
432:Genomic export from the nucleus
319:Rev localization to the nucleus
1349:10.1146/annurev.micro.52.1.491
1:
3750:Vesiculovirus matrix proteins
2256:10.1016/s0968-0896(00)00344-8
2151:10.1016/s0968-0896(00)00344-8
2107:10.1016/s0092-8674(00)80371-2
1806:10.1016/s1074-5521(97)90124-1
1337:Annual Review of Microbiology
1305:10.1016/s0092-8674(00)80371-2
101:Available protein structures:
22:REV protein bound to RRE mRNA
1926:10.1016/0092-8674(86)90062-0
1882:10.1016/0092-8674(95)90436-0
1842:10.1016/0006-291x(89)90767-5
1556:10.1016/j.molcel.2008.07.016
1433:10.1016/0006-291x(89)90767-5
866:10.1016/0042-6822(89)90535-7
797:"HIV/HTLV gene nomenclature"
766:10.1016/0092-8674(86)90062-0
651:will not be exported to the
632:assembly, are not produced.
3787:Structure and genome of HIV
2844:(circular partially ds-DNA)
446:nuclear localization signal
325:nuclear localization signal
272:nuclear localization signal
178:nuclear localization signal
4025:
2420:Herpesvirus glycoprotein B
2415:HHV capsid portal protein
1517:10.1016/j.bmc.2006.03.038
1079:10.1016/j.bpc.2003.10.013
901:10.1016/j.bpc.2003.10.013
452:. Once Rev is inside the
348:Binding of Rev to the RRE
96:
26:
2186:10.2174/1568026043388466
256:HIV Rev response element
176:) protein expression. A
16:HIV-1 regulating protein
3410:Influenza hemagglutinin
1793:Chemistry & Biology
1714:10.1073/pnas.88.17.7734
1033:10.1073/pnas.88.17.7734
3972:Gag-onc fusion protein
2018:10.1006/abbi.1999.1207
1765:10.1006/viro.1998.9312
985:10.1006/abbi.1999.1207
393:
380:
344:
3942:murine leukemia virus
3833:Reverse transcriptase
3713:Indiana vesiculovirus
3585:Measles hemagglutinin
2292:10.1038/sj.gt.3300369
1067:Biophysical Chemistry
889:Biophysical Chemistry
450:nuclear export signal
390:
378:
341:
284:nuclear export signal
164:is a transactivating
2673:Early 35 kDa protein
1787:Hope TJ (May 1997).
396:The RRE sequence is
258:(RRE) located in an
2045:Journal of Virology
1961:Journal of Virology
1705:1991PNAS...88.7734Z
1644:Journal of Virology
1460:Journal of Virology
1243:Journal of Virology
1194:Journal of Virology
1118:10.1021/bi00176a031
1024:1991PNAS...88.7734Z
813:1988Natur.333..504G
715:1986Natur.321..412S
252:secondary structure
234:Arginine-rich motif
3937:Rous sarcoma virus
1789:"Viral RNA export"
394:
381:
345:
262:downstream of the
180:is encoded in the
3991:
3990:
3987:
3986:
3983:
3982:
3979:
3978:
3926:
3925:
3772:
3771:
3768:
3767:
3764:
3763:
3760:
3759:
3707:
3706:
3703:
3702:
3645:
3644:
3641:
3640:
3597:
3596:
3593:
3592:
3549:
3548:
3545:
3544:
3501:
3500:
3497:
3496:
3453:
3452:
3428:
3427:
3317:
3316:
3313:
3312:
3175:
3174:
3055:
3054:
3051:
3050:
3002:
3001:
2915:
2914:
2911:
2910:
2907:
2906:
2835:
2834:
2822:
2821:
2772:
2771:
2685:
2684:
2681:
2680:
2647:
2646:
2545:
2544:
2541:
2540:
2502:
2501:
2468:
2467:
2428:
2427:
2221:10.1021/bi034252z
1112:(10): 2988–2996.
639:B, diphenylfuran
561:negative feedback
159:
158:
155:
154:
150:structure summary
4016:
3960:
3951:
3792:
3783:
3730:
3718:
3668:
3656:
3651:Zaire ebolavirus
3620:
3608:
3572:
3560:
3524:
3512:
3476:
3464:
3355:
3343:
3334:
3257:3C-like protease
3192:
3087:
3072:
3028:
2954:
2948:Duplornaviricota
2939:
2926:
2848:
2786:
2746:
2734:
2705:
2658:
2567:
2558:
2513:
2479:
2402:
2390:
2381:
2363:
2334:
2327:
2320:
2311:
2304:
2303:
2274:
2268:
2267:
2239:
2233:
2232:
2204:
2198:
2197:
2169:
2163:
2162:
2134:
2128:
2127:
2109:
2085:
2079:
2078:
2068:
2036:
2030:
2029:
2001:
1995:
1994:
1984:
1952:
1946:
1945:
1909:
1903:
1902:
1884:
1860:
1854:
1853:
1825:
1819:
1818:
1808:
1784:
1778:
1777:
1767:
1743:
1737:
1736:
1726:
1716:
1684:
1678:
1677:
1667:
1635:
1629:
1628:
1618:
1608:
1606:10.3390/v7062760
1599:(6): 3053–3075.
1584:
1578:
1577:
1567:
1535:
1529:
1528:
1500:
1494:
1493:
1483:
1451:
1445:
1444:
1416:
1410:
1409:
1399:
1376:The EMBO Journal
1367:
1361:
1360:
1332:
1326:
1325:
1307:
1283:
1277:
1276:
1266:
1234:
1228:
1227:
1217:
1185:
1179:
1178:
1168:
1144:
1138:
1137:
1097:
1091:
1090:
1062:
1056:
1055:
1045:
1035:
1003:
997:
996:
968:
962:
961:
951:
928:The EMBO Journal
919:
913:
912:
884:
878:
877:
849:
843:
842:
824:
822:10.1038/333504a0
792:
786:
785:
749:
743:
742:
723:10.1038/321412a0
698:
611:regulatory genes
559:) by creating a
513:regulatory genes
268:helix-loop-helix
98:
31:
19:
4024:
4023:
4019:
4018:
4017:
4015:
4014:
4013:
3994:
3993:
3992:
3975:
3922:
3881:transactivators
3863:
3756:
3699:
3637:
3589:
3541:
3493:
3449:
3424:
3395:
3338:Influenza virus
3328:Negarnaviricota
3325:
3323:
3309:
3235:
3183:
3171:
3120:
3081:Kitrinoviricota
3078:
3063:
3061:
3047:
3019:
2998:
2945:
2930:
2903:
2877:
2843:
2831:
2818:
2768:
2729:
2711:
2697:
2691:
2677:
2643:
2622:
2591:
2553:
2552:(Duplodnaviria,
2551:
2550:circular ds-DNA
2537:
2498:
2464:
2424:
2373:
2367:
2352:
2338:
2308:
2307:
2276:
2275:
2271:
2250:(5): 1097–113.
2241:
2240:
2236:
2215:(26): 8035–46.
2206:
2205:
2201:
2171:
2170:
2166:
2145:(5): 1097–113.
2136:
2135:
2131:
2087:
2086:
2082:
2038:
2037:
2033:
2003:
2002:
1998:
1954:
1953:
1949:
1911:
1910:
1906:
1862:
1861:
1857:
1827:
1826:
1822:
1786:
1785:
1781:
1745:
1744:
1740:
1686:
1685:
1681:
1637:
1636:
1632:
1586:
1585:
1581:
1537:
1536:
1532:
1502:
1501:
1497:
1453:
1452:
1448:
1418:
1417:
1413:
1369:
1368:
1364:
1334:
1333:
1329:
1285:
1284:
1280:
1236:
1235:
1231:
1187:
1186:
1182:
1146:
1145:
1141:
1099:
1098:
1094:
1073:(1–3): 101–19.
1064:
1063:
1059:
1005:
1004:
1000:
970:
969:
965:
921:
920:
916:
895:(1–3): 101–19.
886:
885:
881:
851:
850:
846:
794:
793:
789:
751:
750:
746:
709:(6068): 412–7.
700:
699:
695:
690:
682:Dihydrovaltrate
619:
593:
509:
434:
350:
321:
304:
280:
236:
220:
198:
35:
17:
12:
11:
5:
4022:
4020:
4012:
4011:
4006:
3996:
3995:
3989:
3988:
3985:
3984:
3981:
3980:
3977:
3976:
3974:
3969:
3967:
3957:
3955:Fusion protein
3948:
3947:
3946:
3945:
3944:
3939:
3928:
3927:
3924:
3923:
3921:
3920:
3911:
3906:
3901:
3900:
3899:
3894:
3889:
3876:
3874:
3865:
3864:
3862:
3861:
3860:
3859:
3854:
3842:
3841:
3840:
3838:HIV-1 protease
3835:
3830:
3818:
3817:
3816:
3803:
3801:
3789:
3780:
3774:
3773:
3770:
3769:
3766:
3765:
3762:
3761:
3758:
3757:
3755:
3754:
3753:
3752:
3744:matrix protein
3739:
3737:
3727:
3715:
3709:
3708:
3705:
3704:
3701:
3700:
3698:
3697:
3696:
3695:
3690:
3682:matrix protein
3677:
3675:
3665:
3653:
3647:
3646:
3643:
3642:
3639:
3638:
3636:
3635:
3629:
3627:
3617:
3605:
3599:
3598:
3595:
3594:
3591:
3590:
3588:
3587:
3581:
3579:
3569:
3557:
3551:
3550:
3547:
3546:
3543:
3542:
3540:
3539:
3533:
3531:
3521:
3509:
3503:
3502:
3499:
3498:
3495:
3494:
3492:
3491:
3485:
3483:
3473:
3461:
3455:
3454:
3451:
3450:
3448:
3447:
3441:
3439:
3430:
3429:
3426:
3425:
3423:
3422:
3417:
3412:
3406:
3404:
3397:
3396:
3394:
3393:
3392:
3391:
3383:viral envelope
3379:
3378:
3377:
3369:matrix protein
3364:
3362:
3352:
3340:
3331:
3324:negative-sense
3319:
3318:
3315:
3314:
3311:
3310:
3308:
3307:
3302:
3297:
3292:
3287:
3282:
3277:
3272:
3267:
3262:
3261:
3260:
3248:
3246:
3237:
3236:
3234:
3233:
3228:
3227:
3226:
3221:
3216:
3208:viral envelope
3203:
3201:
3189:
3177:
3176:
3173:
3172:
3170:
3169:
3164:
3159:
3154:
3149:
3144:
3139:
3133:
3131:
3122:
3121:
3119:
3118:
3117:
3116:
3111:
3103:viral envelope
3098:
3096:
3084:
3069:
3062:positive-sense
3057:
3056:
3053:
3052:
3049:
3048:
3046:
3045:
3039:
3037:
3025:
3004:
3003:
3000:
2999:
2997:
2996:
2991:
2986:
2981:
2976:
2971:
2965:
2963:
2951:
2936:
2923:
2917:
2916:
2913:
2912:
2909:
2908:
2905:
2904:
2902:
2901:
2896:
2890:
2888:
2879:
2878:
2876:
2875:
2874:
2864:
2859:
2857:
2845:
2837:
2836:
2833:
2832:
2830:
2829:
2823:
2820:
2819:
2817:
2816:
2806:
2801:
2795:
2793:
2783:
2774:
2773:
2770:
2769:
2767:
2766:
2761:
2755:
2753:
2743:
2731:
2709:Polyomaviridae
2702:
2687:
2686:
2683:
2682:
2679:
2678:
2676:
2675:
2669:
2667:
2655:
2649:
2648:
2645:
2644:
2642:
2641:
2635:
2633:
2624:
2623:
2621:
2620:
2615:
2610:
2604:
2602:
2593:
2592:
2590:
2589:
2584:
2578:
2576:
2564:
2555:
2554:Varidnaviria?)
2547:
2546:
2543:
2542:
2539:
2538:
2536:
2535:
2530:
2524:
2522:
2510:
2504:
2503:
2500:
2499:
2497:
2496:
2490:
2488:
2476:
2470:
2469:
2466:
2465:
2463:
2462:
2457:
2452:
2447:
2441:
2439:
2430:
2429:
2426:
2425:
2423:
2422:
2417:
2411:
2409:
2399:
2387:
2385:Herpes simplex
2378:
2360:
2354:
2353:
2341:Viral proteins
2339:
2337:
2336:
2329:
2322:
2314:
2306:
2305:
2269:
2234:
2199:
2164:
2129:
2100:(6): 1051–60.
2080:
2051:(8): 3734–41.
2031:
1996:
1967:(3): 1265–74.
1947:
1904:
1855:
1820:
1779:
1738:
1699:(17): 7734–8.
1679:
1650:(5): 1959–66.
1630:
1579:
1544:Molecular Cell
1530:
1511:(15): 5384–8.
1495:
1466:(3): 1265–74.
1446:
1411:
1362:
1343:(1): 491–532.
1327:
1298:(6): 1051–60.
1278:
1229:
1200:(8): 4248–54.
1180:
1139:
1092:
1057:
1018:(17): 7734–8.
998:
963:
934:(3): 1119–29.
914:
879:
844:
787:
744:
692:
691:
689:
686:
666:substitution (
618:
615:
592:
589:
508:
505:
458:phosphorylated
433:
430:
349:
346:
320:
317:
303:
300:
290:region and is
279:
276:
235:
232:
219:
216:
197:
194:
157:
156:
153:
152:
147:
141:
140:
127:
121:
120:
110:
103:
102:
94:
93:
80:
74:
73:
68:
62:
61:
56:
50:
49:
46:
42:
41:
37:
36:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
4021:
4010:
4007:
4005:
4002:
4001:
3999:
3973:
3970:
3968:
3965:
3961:
3958:
3956:
3952:
3949:
3943:
3940:
3938:
3935:
3934:
3933:
3932:
3929:
3919:
3915:
3912:
3910:
3907:
3905:
3902:
3898:
3895:
3893:
3890:
3888:
3885:
3884:
3883:
3882:
3878:
3877:
3875:
3873:
3871:
3866:
3858:
3855:
3853:
3850:
3849:
3848:
3847:
3843:
3839:
3836:
3834:
3831:
3829:
3826:
3825:
3824:
3823:
3819:
3815:
3812:
3811:
3810:
3809:
3805:
3804:
3802:
3800:
3798:
3793:
3790:
3788:
3784:
3781:
3779:
3775:
3751:
3748:
3747:
3746:
3745:
3741:
3740:
3738:
3735:
3731:
3728:
3726:
3724:
3719:
3716:
3714:
3710:
3694:
3691:
3689:
3686:
3685:
3684:
3683:
3679:
3678:
3676:
3673:
3669:
3666:
3664:
3662:
3657:
3654:
3652:
3648:
3634:
3631:
3630:
3628:
3625:
3621:
3618:
3616:
3614:
3609:
3606:
3604:
3600:
3586:
3583:
3582:
3580:
3577:
3573:
3570:
3568:
3566:
3561:
3558:
3556:
3552:
3538:
3535:
3534:
3532:
3529:
3525:
3522:
3520:
3518:
3513:
3510:
3508:
3504:
3490:
3487:
3486:
3484:
3481:
3477:
3474:
3472:
3470:
3465:
3462:
3460:
3459:Parainfluenza
3456:
3446:
3443:
3442:
3440:
3438:
3436:
3431:
3421:
3418:
3416:
3415:Neuraminidase
3413:
3411:
3408:
3407:
3405:
3402:
3398:
3390:
3387:
3386:
3385:
3384:
3380:
3376:
3373:
3372:
3371:
3370:
3366:
3365:
3363:
3360:
3356:
3353:
3351:
3349:
3344:
3341:
3339:
3335:
3332:
3329:
3320:
3306:
3303:
3301:
3298:
3296:
3293:
3291:
3288:
3286:
3283:
3281:
3278:
3276:
3273:
3271:
3268:
3266:
3263:
3258:
3255:
3254:
3253:
3250:
3249:
3247:
3245:
3243:
3238:
3232:
3229:
3225:
3222:
3220:
3217:
3215:
3212:
3211:
3210:
3209:
3205:
3204:
3202:
3200:
3198:
3193:
3190:
3187:
3182:
3178:
3168:
3165:
3163:
3160:
3158:
3155:
3153:
3150:
3148:
3145:
3143:
3140:
3138:
3135:
3134:
3132:
3130:
3128:
3123:
3115:
3112:
3110:
3107:
3106:
3105:
3104:
3100:
3099:
3097:
3095:
3093:
3088:
3085:
3082:
3077:
3073:
3070:
3067:
3058:
3044:
3041:
3040:
3038:
3036:
3034:
3029:
3026:
3023:
3017:
3013:
3009:
3005:
2995:
2992:
2990:
2987:
2985:
2982:
2980:
2977:
2975:
2972:
2970:
2967:
2966:
2964:
2962:
2960:
2955:
2952:
2949:
2944:
2940:
2937:
2934:
2927:
2924:
2922:
2918:
2900:
2897:
2895:
2892:
2891:
2889:
2887:
2885:
2880:
2873:
2870:
2869:
2868:
2865:
2863:
2860:
2858:
2856:
2854:
2849:
2846:
2842:
2838:
2828:
2825:
2824:
2814:
2810:
2807:
2805:
2802:
2800:
2797:
2796:
2794:
2791:
2787:
2784:
2782:
2780:
2775:
2765:
2762:
2760:
2757:
2756:
2754:
2751:
2747:
2744:
2742:
2740:
2735:
2732:
2727:
2723:
2719:
2715:
2710:
2706:
2703:
2700:
2695:
2688:
2674:
2671:
2670:
2668:
2666:
2664:
2659:
2656:
2654:
2653:Baculoviridae
2650:
2640:
2637:
2636:
2634:
2632:
2630:
2625:
2619:
2616:
2614:
2611:
2609:
2606:
2605:
2603:
2601:
2599:
2594:
2588:
2585:
2583:
2580:
2579:
2577:
2575:
2573:
2568:
2565:
2563:
2559:
2556:
2548:
2534:
2531:
2529:
2526:
2525:
2523:
2521:
2519:
2514:
2511:
2509:
2505:
2495:
2492:
2491:
2489:
2487:
2485:
2480:
2477:
2475:
2471:
2461:
2458:
2456:
2453:
2451:
2448:
2446:
2443:
2442:
2440:
2438:
2436:
2431:
2421:
2418:
2416:
2413:
2412:
2410:
2407:
2403:
2400:
2398:
2396:
2391:
2388:
2386:
2382:
2379:
2376:
2371:
2370:Duplodnaviria
2366:linear ds-DNA
2364:
2361:
2359:
2355:
2350:
2346:
2342:
2335:
2330:
2328:
2323:
2321:
2316:
2315:
2312:
2301:
2297:
2293:
2289:
2286:(2): 128–39.
2285:
2281:
2273:
2270:
2265:
2261:
2257:
2253:
2249:
2245:
2238:
2235:
2230:
2226:
2222:
2218:
2214:
2210:
2203:
2200:
2195:
2191:
2187:
2183:
2180:(9): 871–82.
2179:
2175:
2168:
2165:
2160:
2156:
2152:
2148:
2144:
2140:
2133:
2130:
2125:
2121:
2117:
2113:
2108:
2103:
2099:
2095:
2091:
2084:
2081:
2076:
2072:
2067:
2062:
2058:
2054:
2050:
2046:
2042:
2035:
2032:
2027:
2023:
2019:
2015:
2012:(2): 186–91.
2011:
2007:
2000:
1997:
1992:
1988:
1983:
1978:
1974:
1970:
1966:
1962:
1958:
1951:
1948:
1943:
1939:
1935:
1931:
1927:
1923:
1920:(6): 807–17.
1919:
1915:
1908:
1905:
1900:
1896:
1892:
1888:
1883:
1878:
1875:(3): 475–83.
1874:
1870:
1866:
1859:
1856:
1851:
1847:
1843:
1839:
1836:(3): 963–70.
1835:
1831:
1824:
1821:
1816:
1812:
1807:
1802:
1799:(5): 335–44.
1798:
1794:
1790:
1783:
1780:
1775:
1771:
1766:
1761:
1758:(2): 286–96.
1757:
1753:
1749:
1742:
1739:
1734:
1730:
1725:
1720:
1715:
1710:
1706:
1702:
1698:
1694:
1690:
1683:
1680:
1675:
1671:
1666:
1661:
1657:
1653:
1649:
1645:
1641:
1634:
1631:
1626:
1622:
1617:
1612:
1607:
1602:
1598:
1594:
1590:
1583:
1580:
1575:
1571:
1566:
1561:
1557:
1553:
1550:(6): 824–34.
1549:
1545:
1541:
1534:
1531:
1526:
1522:
1518:
1514:
1510:
1506:
1499:
1496:
1491:
1487:
1482:
1477:
1473:
1469:
1465:
1461:
1457:
1450:
1447:
1442:
1438:
1434:
1430:
1427:(3): 963–70.
1426:
1422:
1415:
1412:
1407:
1403:
1398:
1393:
1389:
1385:
1382:(8): 1810–7.
1381:
1377:
1373:
1366:
1363:
1358:
1354:
1350:
1346:
1342:
1338:
1331:
1328:
1323:
1319:
1315:
1311:
1306:
1301:
1297:
1293:
1289:
1282:
1279:
1274:
1270:
1265:
1260:
1256:
1252:
1249:(4): 2350–9.
1248:
1244:
1240:
1233:
1230:
1225:
1221:
1216:
1211:
1207:
1203:
1199:
1195:
1191:
1184:
1181:
1176:
1172:
1167:
1162:
1159:(4): 351–64.
1158:
1154:
1150:
1143:
1140:
1135:
1131:
1127:
1123:
1119:
1115:
1111:
1107:
1103:
1096:
1093:
1088:
1084:
1080:
1076:
1072:
1068:
1061:
1058:
1053:
1049:
1044:
1039:
1034:
1029:
1025:
1021:
1017:
1013:
1009:
1002:
999:
994:
990:
986:
982:
979:(2): 186–91.
978:
974:
967:
964:
959:
955:
950:
945:
941:
937:
933:
929:
925:
918:
915:
910:
906:
902:
898:
894:
890:
883:
880:
875:
871:
867:
863:
859:
855:
848:
845:
840:
836:
832:
828:
823:
818:
814:
810:
807:(6173): 504.
806:
802:
798:
791:
788:
783:
779:
775:
771:
767:
763:
760:(6): 807–17.
759:
755:
748:
745:
740:
736:
732:
728:
724:
720:
716:
712:
708:
704:
697:
694:
687:
685:
683:
679:
677:
673:
669:
668:Aspartic acid
665:
661:
656:
654:
650:
646:
642:
638:
633:
631:
626:
622:
616:
614:
612:
608:
604:
600:
597:
590:
588:
586:
582:
578:
574:
569:
566:
562:
558:
554:
550:
546:
542:
538:
534:
530:
526:
522:
518:
514:
506:
504:
502:
498:
494:
489:
487:
483:
479:
475:
471:
467:
463:
459:
456:, Ran-GDP is
455:
451:
447:
443:
439:
431:
429:
427:
423:
419:
415:
411:
407:
403:
399:
389:
385:
377:
373:
369:
366:
362:
359:
355:
347:
340:
336:
334:
330:
326:
318:
316:
314:
310:
301:
299:
297:
293:
289:
285:
277:
275:
273:
269:
265:
261:
257:
253:
249:
246:38–49 of the
245:
241:
233:
231:
229:
225:
222:Rev is a 13-k
217:
215:
212:
207:
203:
195:
193:
191:
187:
183:
179:
175:
171:
167:
163:
151:
148:
146:
142:
139:
135:
131:
128:
126:
122:
118:
114:
111:
108:
104:
99:
95:
92:
88:
84:
81:
79:
75:
72:
69:
67:
63:
60:
57:
55:
51:
47:
43:
38:
30:
25:
20:
3891:
3879:
3868:
3844:
3820:
3806:
3795:
3742:
3721:
3680:
3659:
3624:glycoprotein
3611:
3576:glycoprotein
3563:
3528:glycoprotein
3515:
3480:glycoprotein
3467:
3433:
3401:glycoprotein
3381:
3367:
3346:
3240:
3231:Nucleocapsid
3206:
3195:
3186:Pisuviricota
3125:
3101:
3090:
3031:
3022:Pisuviricota
2957:
2882:
2851:
2777:
2737:
2699:Monodnaviria
2661:
2627:
2596:
2570:
2562:Epstein–Barr
2516:
2508:Adenoviridae
2482:
2433:
2393:
2375:Varidnaviria
2283:
2280:Gene Therapy
2279:
2272:
2247:
2243:
2237:
2212:
2209:Biochemistry
2208:
2202:
2177:
2173:
2167:
2142:
2138:
2132:
2097:
2093:
2083:
2048:
2044:
2034:
2009:
2005:
1999:
1964:
1960:
1950:
1917:
1913:
1907:
1872:
1868:
1858:
1833:
1829:
1823:
1796:
1792:
1782:
1755:
1751:
1741:
1696:
1692:
1682:
1647:
1643:
1633:
1596:
1592:
1582:
1547:
1543:
1533:
1508:
1504:
1498:
1463:
1459:
1449:
1424:
1420:
1414:
1379:
1375:
1365:
1340:
1336:
1330:
1295:
1291:
1281:
1246:
1242:
1232:
1197:
1193:
1183:
1156:
1152:
1142:
1109:
1106:Biochemistry
1105:
1095:
1070:
1066:
1060:
1015:
1011:
1001:
976:
972:
966:
931:
927:
917:
892:
888:
882:
860:(1): 264–6.
857:
853:
847:
804:
800:
790:
757:
753:
747:
706:
702:
696:
680:
657:
634:
623:
620:
594:
587:production.
556:
552:
548:
544:
540:
536:
532:
528:
524:
520:
516:
510:
490:
435:
425:
421:
417:
413:
409:
405:
401:
397:
395:
382:
370:
364:
351:
322:
305:
281:
263:
247:
237:
221:
211:RNA splicing
205:
201:
199:
181:
161:
160:
3964:oncoprotein
3076:Hepatitis C
2841:Hepatitis B
2827:Agnoprotein
2790:oncoprotein
2764:VP2 and VP3
495:and the 5s
244:amino acids
228:amino acids
172:(and other
40:Identifiers
3998:Categories
3389:M2 protein
3375:M1 protein
3181:SARS-CoV-2
688:References
664:amino acid
645:proflavine
625:Leptomycin
331:-GDP, and
288:C-terminal
240:N-terminal
174:lentiviral
113:structures
3931:Multiple
3828:Integrase
3066:Riboviria
2943:Rotavirus
2933:Riboviria
2694:Riboviria
1126:0006-2960
676:wild-type
653:cytoplasm
577:cytoplasm
478:cytoplasm
476:into the
438:cytoplasm
313:cytoplasm
296:cytoplasm
218:Structure
71:IPR000625
4004:HIV/AIDS
3224:Membrane
3219:Envelope
2813:SV40 Tag
2474:Vaccinia
2264:11377168
2229:12834355
2194:15134546
2159:11377168
2124:15119502
2026:10328811
1942:37813869
1899:15414237
1752:Virology
1625:26075509
1574:18922466
1525:16603366
1322:15119502
1087:15043924
993:10328811
909:15043924
854:Virology
839:33832642
782:37813869
637:Neomycin
609:(early,
482:importin
464:-1) and
462:exportin
448:and its
333:importin
302:Function
130:RCSB PDB
66:InterPro
3555:Measles
3008:Rhinov.
2455:ICP34.5
2300:9081703
2116:9323133
2075:2196381
1991:2783738
1934:3638988
1891:7543368
1850:2788417
1815:9195877
1774:9791020
1733:1715576
1701:Bibcode
1674:2704072
1616:4488727
1593:Viruses
1565:2651398
1490:2783738
1441:2788417
1406:8617226
1357:9891806
1314:9323133
1273:8642662
1224:2072452
1175:9630243
1166:1369623
1134:7510518
1052:1715576
1020:Bibcode
958:1547776
874:2741343
831:2836736
809:Bibcode
774:3638988
739:4306352
731:3012355
711:Bibcode
672:Leucine
573:nucleus
501:nucleus
486:nucleus
474:nucleus
454:nucleus
442:nucleus
356:of the
311:to the
309:nucleus
292:leucine
196:History
186:nucleus
166:protein
59:PF00424
3734:capsid
3672:capsid
3420:HA-tag
3359:capsid
3322:ss-RNA
3252:ORF1ab
3060:ss-RNA
3020:etc. (
2929:ds-RNA
2750:capsid
2618:EBNA-3
2613:EBNA-2
2608:EBNA-1
2406:capsid
2298:
2262:
2227:
2192:
2157:
2122:
2114:
2073:
2066:249668
2063:
2024:
1989:
1982:247823
1979:
1940:
1932:
1897:
1889:
1848:
1813:
1772:
1731:
1721:
1672:
1665:250609
1662:
1623:
1613:
1572:
1562:
1523:
1488:
1481:247823
1478:
1439:
1404:
1397:450097
1394:
1355:
1320:
1312:
1271:
1264:190077
1261:
1222:
1215:248862
1212:
1173:
1163:
1132:
1124:
1085:
1050:
1040:
991:
956:
949:556554
946:
907:
872:
837:
829:
801:Nature
780:
772:
737:
729:
703:Nature
643:, and
641:cation
630:virion
585:virion
568:genome
535:, and
523:, and
493:snRNAs
470:intron
392:4PMI).
361:genome
354:intron
343:4PMI).
282:Rev's
260:intron
145:PDBsum
119:
109:
91:SUPFAM
45:Symbol
34:4PMI).
3870:VRAPs
3852:gp120
3507:Mumps
3305:ORF9b
3295:ORF7b
3290:ORF7a
3280:ORF3d
3275:ORF3c
3270:ORF3b
3265:ORF3a
3259:(NS5)
3214:Spike
3016:Hep A
3012:Polio
2872:HBeAg
2867:HBcAg
2862:HBsAg
2726:HaPyV
2722:MCPyV
2690:other
2629:ncRNA
2587:LMP-2
2582:LMP-1
2460:ICP47
2445:vmw65
2345:early
2120:S2CID
1938:S2CID
1895:S2CID
1724:52377
1318:S2CID
1043:52377
835:S2CID
778:S2CID
735:S2CID
660:HIV-1
607:mRNAs
599:genes
596:HIV-1
565:HIV-1
497:rRNAs
358:HIV-1
190:mRNAs
170:HIV-1
87:SCOPe
78:SCOP2
3857:gp41
3797:VSPs
3723:VSPs
3693:VP24
3688:VP40
3661:VSPs
3613:VSPs
3565:VSPs
3517:VSPs
3469:VSPs
3435:VNPs
3348:VSPs
3300:ORF8
3285:ORF6
3242:VNPs
3197:VSPs
3167:NS5B
3162:NS5A
3157:NS4B
3152:NS4A
3127:VNPs
3092:VSPs
3033:VNPs
2994:NSP6
2989:NSP5
2984:NSP4
2979:NSP3
2974:NSP2
2969:NSP1
2959:VNPs
2884:VNPs
2853:VSPs
2809:LTag
2804:MTag
2799:STag
2779:VNPs
2739:VSPs
2718:MPyV
2714:SV40
2663:VNPs
2639:EBER
2598:VNPs
2572:VSPs
2518:VNPs
2494:B13R
2484:VNPs
2450:ICP8
2435:VNPs
2395:VSPs
2349:late
2347:and
2296:PMID
2260:PMID
2225:PMID
2190:PMID
2155:PMID
2112:PMID
2094:Cell
2071:PMID
2022:PMID
1987:PMID
1930:PMID
1914:Cell
1887:PMID
1869:Cell
1846:PMID
1811:PMID
1770:PMID
1729:PMID
1670:PMID
1621:PMID
1570:PMID
1521:PMID
1486:PMID
1437:PMID
1402:PMID
1353:PMID
1310:PMID
1292:Cell
1269:PMID
1220:PMID
1171:PMID
1130:PMID
1122:ISSN
1083:PMID
1048:PMID
989:PMID
954:PMID
905:PMID
870:PMID
827:PMID
770:PMID
754:Cell
727:PMID
581:mRNA
575:and
555:and
547:and
525:env.
440:and
424:and
408:and
238:The
204:and
138:PDBj
134:PDBe
117:ECOD
107:Pfam
83:484d
54:Pfam
3918:Vpx
3916:or
3914:Vpu
3909:Vif
3904:Nef
3897:Vpr
3892:Rev
3887:Tat
3846:env
3822:pol
3814:p24
3808:gag
3603:RSV
3445:NS1
3147:NS3
3142:NS2
3043:VPg
2921:RNA
2894:HBx
2759:VP1
2533:E1B
2528:E1A
2358:DNA
2288:doi
2252:doi
2217:doi
2182:doi
2147:doi
2102:doi
2061:PMC
2053:doi
2014:doi
2010:365
1977:PMC
1969:doi
1922:doi
1877:doi
1838:doi
1834:162
1801:doi
1760:doi
1756:249
1719:PMC
1709:doi
1660:PMC
1652:doi
1611:PMC
1601:doi
1560:PMC
1552:doi
1513:doi
1476:PMC
1468:doi
1429:doi
1425:162
1392:PMC
1384:doi
1345:doi
1300:doi
1259:PMC
1251:doi
1210:PMC
1202:doi
1161:PMC
1153:RNA
1114:doi
1075:doi
1071:108
1038:PMC
1028:doi
981:doi
977:365
944:PMC
936:doi
897:doi
893:108
862:doi
858:171
817:doi
805:333
762:doi
719:doi
707:321
670:to
649:RNA
603:RNA
557:tat
553:rev
549:env
545:pol
541:Gag
539:).
537:env
533:pol
529:gag
521:pol
517:gag
466:Ran
426:vpu
422:vpr
418:vif
414:env
410:pol
406:gag
402:env
398:cis
365:env
329:Ran
264:env
248:rev
206:env
202:gag
182:rev
162:Rev
125:PDB
48:REV
4000::
3778:RT
3137:P7
3114:E2
3109:E1
3014:,
3010:,
2724:,
2720:,
2716:,
2294:.
2282:.
2258:.
2246:.
2223:.
2213:42
2211:.
2188:.
2176:.
2153:.
2141:.
2118:.
2110:.
2098:90
2096:.
2092:.
2069:.
2059:.
2049:64
2047:.
2043:.
2020:.
2008:.
1985:.
1975:.
1965:63
1963:.
1959:.
1936:.
1928:.
1918:46
1916:.
1893:.
1885:.
1873:82
1871:.
1867:.
1844:.
1832:.
1809:.
1795:.
1791:.
1768:.
1754:.
1750:.
1727:.
1717:.
1707:.
1697:88
1695:.
1691:.
1668:.
1658:.
1648:63
1646:.
1642:.
1619:.
1609:.
1595:.
1591:.
1568:.
1558:.
1548:31
1546:.
1542:.
1519:.
1509:14
1507:.
1484:.
1474:.
1464:63
1462:.
1458:.
1435:.
1423:.
1400:.
1390:.
1380:15
1378:.
1374:.
1351:.
1341:52
1339:.
1316:.
1308:.
1296:90
1294:.
1290:.
1267:.
1257:.
1247:70
1245:.
1241:.
1218:.
1208:.
1198:65
1196:.
1192:.
1169:.
1155:.
1151:.
1128:.
1120:.
1110:33
1108:.
1104:.
1081:.
1069:.
1046:.
1036:.
1026:.
1016:88
1014:.
1010:.
987:.
975:.
952:.
942:.
932:11
930:.
926:.
903:.
891:.
868:.
856:.
833:.
825:.
815:.
803:.
799:.
776:.
768:.
758:46
756:.
733:.
725:.
717:.
705:.
543:,
531:,
519:,
488:.
420:,
416:,
274:.
224:Da
136:;
132:;
115:/
89:/
85:/
3966::
3872::
3799::
3736::
3725::
3674::
3663::
3626::
3615::
3578::
3567::
3530::
3519::
3482::
3471::
3437::
3403::
3361::
3350::
3330:)
3326:(
3244::
3199::
3188:)
3184:(
3129::
3094::
3083:)
3079:(
3068:)
3064:(
3035::
3024:)
3018:,
2961::
2950:)
2946:(
2935:)
2931:(
2886::
2855::
2815:)
2811:(
2792::
2781::
2752::
2741::
2728:)
2712:(
2701:)
2696:,
2692:(
2665::
2631::
2600::
2574::
2520::
2486::
2437::
2408::
2397::
2377:)
2372:,
2368:(
2351:)
2343:(
2333:e
2326:t
2319:v
2302:.
2290::
2284:4
2266:.
2254::
2248:9
2231:.
2219::
2196:.
2184::
2178:4
2161:.
2149::
2143:9
2126:.
2104::
2077:.
2055::
2028:.
2016::
1993:.
1971::
1944:.
1924::
1901:.
1879::
1852:.
1840::
1817:.
1803::
1797:4
1776:.
1762::
1735:.
1711::
1703::
1676:.
1654::
1627:.
1603::
1597:7
1576:.
1554::
1527:.
1515::
1492:.
1470::
1443:.
1431::
1408:.
1386::
1359:.
1347::
1324:.
1302::
1275:.
1253::
1226:.
1204::
1177:.
1157:4
1136:.
1116::
1089:.
1077::
1054:.
1030::
1022::
995:.
983::
960:.
938::
911:.
899::
876:.
864::
841:.
819::
811::
784:.
764::
741:.
721::
713::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.