444:
His31-Asp70 salt bridge in T4 lysozyme was buried within the protein. Entropy plays a larger role in surface salt bridges where residues that normally have the ability to move are constricted by their electrostatic interaction and hydrogen bonding. This has been shown to decrease entropy enough to nearly erase the contribution of the interaction. Surface salt bridges can be studied similarly to that of buried salt bridges, employing double mutant cycles and NMR titrations. Although cases exist where buried salt bridges contribute to stability, like anything else, exceptions do exist and buried salt bridges can display a destabilizing effect. Also, surface salt bridges, under certain conditions, can display a stabilizing effect. The stabilizing or destabilizing effect must be assessed on a case by case basis and few blanket statements are able to be made.
120:
27:
107:
organic ions display at moderate ionic strength I similar salt bridge association ΔG values around 5 to 6 kJ/mol for a 1:1 combination of anion and cation, almost independent of the nature (size, polarizability etc) of the ions. The ΔG values are additive and approximately a linear function of the charges, the interaction of e.g. a doubly charged phosphate anion with a single charged ammonium cation accounts for about 2x5 = 10 kJ/mol. The ΔG values depend on the ionic strength I of the solution, as described by the
230:
288:, or the pH where the ratio of protonated: deprotonated molecules is 1:1. Continuing with the T4 lysozyme example, a titration curve is obtained through observation of a shift in the C2 proton of histidine 31 (Figure 5). Figure 5 shows the shift in the titration curve between the wild-type and the mutant in which Asp70 is Asn. The salt bridge formed is between the deprotonated Asp70 and protonated His31. This interaction causes the shift seen in His31’s p
1623:
65:. It is a most commonly observed contribution to the stability to the entropically unfavorable folded conformation of proteins. Although non-covalent interactions are known to be relatively weak interactions, small stabilizing interactions can add up to make an important contribution to the overall stability of a conformer. Not only are salt bridges found in proteins, but they can also be found in
193:'s. The distance between the residues participating in the salt bridge is also cited as being important. The N-O distance required is less than 4 Å (400 pm). Amino acids greater than this distance apart do not qualify as forming a salt bridge. Due to the numerous ionizable side chains of amino acids found throughout a protein, the pH at which a protein is placed is crucial to its stability.
1617:
453:
222:
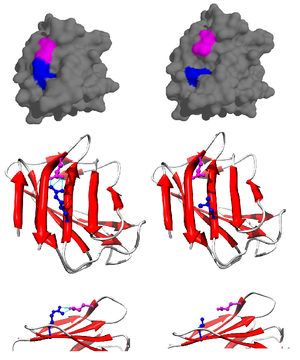
at high pH, the salt bridge’s contribution to the overall free energy of the folded protein state can be determined by performing a point-mutation, altering and, consequently, breaking the salt bridge. For example, a salt bridge was identified to exist in the T4 lysozyme between aspartic acid (Asp) at residue 70 and a histidine (His) at residue 31 (Figure 3).
211:
1629:
432:
values, and the relationship between natural logarithms and logarithms. In the T4 lysozyme example, this approach yielded a calculated contribution of about 3 kcal/mol to the overall free energy. A similar approach can be taken with the other participant in the salt bridge, such as Asp70 in the T4
221:
The contribution of a salt bridge to the overall stability to the folded state of a protein can be assessed through thermodynamic data gathered from mutagenesis studies and nuclear magnetic resonance techniques. Using a mutated pseudo-wild-type protein specifically mutated to prevent precipitation
106:
or the Fuoss equation describe ion pair association as function of the ion charges zA and zB and the dielectric constant ε of the medium; a corresponding plot of the stability ΔG vs. zAzB shows for over 200 ion pairs the expected linear correlation for a large variety of ions. Inorganic as well as
201:
Salt bridges also can form between a protein and small molecule ligands. Over 1100 unique protein-ligand complexes from the
Protein Databank were found to form salt bridges with their protein targets, indicating that salt bridges are frequent in drug-protein interaction. These contain structures
240:
Once the mutants have been established, two methods can be employed to calculate the free energy associated with a salt bridge. One method involves the observation of the melting temperature of the wild-type protein versus that of the three mutants. The denaturation can be monitored through a
443:
A word of caution when choosing the appropriate experiment involves the location of the salt bridge within the protein. The environment plays a large role in the interaction. At high ionic strengths, the salt bridge can be completely masked since an electrostatic interaction is involved. The
486:
Major contributions of supramolecular chemistry have been devoted to recognition and sensing of anions. Ion pairing is the most important driving force for anion complexation, but selectivity e.g. within the halide series has been achieved, mostly by hydrogen bonds contributions.
477:
is a field concerned with non-covalent interactions between macromolecules. Salt bridges have been used by chemists within this field in both diverse and creative ways, including sensing of anions, the synthesis of molecular capsules and double helical polymers.
280:
to calculate the free energy of the salt bridge. A titration is performed, while recording the chemical shift corresponding to the protons of the carbon adjacent to the carboxylate or ammonium group. The midpoint of the titration curve corresponds to the
531:
130:(LMNA, PDB: 1IFR). Normally, arginine 527 (blue) forms salt bridge with glutamate 537 (magenta), but R527L mutation causes loss of the complementary negative charge and structure destabilization. At the phenotype level this manifests with overlapping
266:
245:. A reduction in melting temperature indicates a reduction in stability. This is quantified through a method described by Becktel and Schellman where the free energy difference between the two is calculated through Δ
261:
of 360 cal/(mol·K) (1.5 kJ/(mol·K)) yields a free energy change of about −4 kcal/mol (−17 kJ/mol). This value corresponds to the amount of free energy contributed to the stability of the protein by the salt bridge.
1389:
Ikeda M, Tanaka Y, Hasegawa T, Furusho Y, Yashima E (May 2006). "Construction of double-stranded metallosupramolecular polymers with a controlled helicity by combination of salt bridges and metal coordination".
97:
etc; then the association constants depend on the pH. Entropic driving forces for ion pairing (in absence of significant H-bonding contributions) are also found in methanol as solvent. In nonpolar solvents
320:
is supported by the His31’s interaction with Asp70. To maintain the salt bridge, His31 will attempt to keep its proton as long as possible. When the salt bridge is disrupted, like in the mutant D70N, the
81:
is mostly driven by entropy, usually accompanied by unfavorable ΔH contributions on account of desolvation of the interacting ions upon association. Hydrogen bonds contribute to the stability of
944:
Anderson DE, Becktel WJ, Dahlquist FW (March 1990). "pH-induced denaturation of proteins: a single salt bridge contributes 3-5 kcal/mol to the free energy of folding of T4 lysozyme".
111:, at zero ionic strength one observes ΔG = 8 kJ/mol. The stabilities of the alkali-ion pairs as function of the anion charge z by can be described by a more detailed equation.
1052:
Sun DP, Sauer U, Nicholson H, Matthews BW (July 1991). "Contributions of engineered surface salt bridges to the stability of T4 lysozyme determined by directed mutagenesis".
69:. The thermodynamics of each are explored through experimental procedures to access the free energy contribution of the salt bridge to the overall free energy of the state.
780:
Daniele PG, Foti C, Gianguzza A, Prenesti E, Sammartano S (2008). "Weak alkali and alkaline earth metal complexes of low molecular weight ligands in aqueous solution".
1716:
523:
to create a double helical metallopolymer. Starting from their monomer and platinum(II) biphenyl (Figure 8), their metallopolymer self assembles through a series of
226:
with asparagine (Asn) (Figure 4) was done obtaining three new mutants: Asp70Asn His31 (Mutant 1), Asp70 His31Asn (Mutant 2), and Asp70Asn His31Asn (Double Mutant).
1017:
Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR (December 1990). "Strength and co-operativity of contributions of surface salt bridges to protein stability".
507:(Figure 6). Salt bridge interactions between the two halves cause them to self-assemble in solution (Figure 7). They are stable even when heated to 60 °C.
527:
reactions. The two halves of the monomer are anchored together through the salt bridge between the deprotonated carboxylate and the protonated nitrogens.
257:
of the pseudo-wild-type had previously been reported at pH 5.5 so the midpoint temperature difference of 11 °C at this pH multiplied by the reported Δ
202:
from different enzyme classes, including hydrolase, transferases, kinases, reductase, oxidoreductase, lyases, and G protein-coupled receptors (GPCRs).
1834:
1442:
807:
Al-Haggar M, Madej-Pilarczyk A, Kozlowski L, Bujnicki JM, Yahia S, Abdel-Hadi D, Shams A, Ahmad N, Hamed S, Puzianowska-Kuznicka M (November 2012).
119:
809:"A novel homozygous p.Arg527Leu LMNA mutation in two unrelated Egyptian families causes overlapping mandibuloacral dysplasia and progeria syndrome"
277:
1091:
464:
102:
with very high association constants are formed; in the gas phase the association energies of e.g. alkali halides reach up to 200 kJ/mol. The
694:
657:
566:
418:). Calculation of the free energy difference of the mutant and wild-type can now be done using the free energy equation, the definition of p
1224:
1191:
1166:
630:
1141:
372:
is the equilibrium constant of a reaction in equilibrium. The deprotonation of His31 is an acid equilibrium reaction with a special
1696:
1681:
1282:
33:
Example of salt bridge between amino acids glutamic acid and lysine demonstrating electrostatic interaction and hydrogen bonding
1505:
872:
1532:
1493:
1483:
62:
515:
Yashima and coworkers have used salt bridges to construct several polymers that adopt a double helix conformation much like
253:. There are some issues with this calculation and can only be used with very accurate data. In the T4 lysozyme example, Δ
1488:
108:
1865:
1435:
26:
380:
1756:
1751:
1860:
1741:
1731:
1706:
1676:
223:
58:
1522:
474:
131:
66:
46:
1783:
1686:
1658:
1428:
496:
217:
A salt bridge in T4 lysozyme between aspartic acid (Asp) at residue 70 and a histidine (His) at residue 31
1827:
1788:
500:
1822:
229:
1746:
1637:
1500:
1459:
1354:
Liu J, Lam JW, Tang BZ (November 2009). "Acetylenic polymers: syntheses, structures, and functions".
1212:
710:
Biedermann F, Schneider HJ (May 2016). "Experimental
Binding Energies in Supramolecular Complexes".
295:. In the unfolded wild-type protein, where the salt bridge is absent, His31 is reported to have a p
61:
forces in chemistry, in biological systems, in different materials and in many applications such as
1648:
1512:
1478:
174:(Figure 2). Although these are the most common, other residues with ionizable side chains such as
1812:
1567:
894:
Kurczab, Rafał; Śliwa, Paweł; Rataj, Krzysztof; Kafel, Rafał; Bojarski, Andrzej J. (2018-11-26).
242:
896:"Salt Bridge in Ligand-Protein Complexes-Systematic Theoretical and Statistical Investigations"
1798:
1587:
1547:
1537:
1407:
1371:
1336:
1305:
1263:
1220:
1187:
1162:
1137:
1114:
1069:
1034:
996:
961:
923:
915:
876:
838:
762:
727:
690:
653:
626:
599:
562:
339:
135:
495:
Molecular capsules are chemical scaffolds designed to capture and hold a guest molecule (see
1839:
1622:
1579:
1552:
1399:
1363:
1328:
1297:
1253:
1106:
1061:
1026:
988:
953:
907:
868:
828:
820:
789:
754:
719:
591:
99:
1691:
1562:
1323:
Kuberski B, Szumna A (April 2009). "A self-assembled chiral capsule with polar interior".
524:
82:
1817:
1216:
1726:
1527:
1283:"Advances in anion supramolecular chemistry: From recognition to chemical applications"
859:
Kumar S, Nussinov R (July 2002). "Close-range electrostatic interactions in proteins".
833:
808:
1030:
1854:
1775:
1735:
1668:
1597:
1470:
1451:
674:. J. Phys. Chem. Ref. Data. Vol. Monograph 9 (Fourth ed.). pp. 1–1951.
619:
147:
143:
103:
54:
50:
463:
452:
1721:
338:
can be quantified to reflect the salt bridge’s contribution to free energy. Using
328:
shifts back to a value of 6.9, much closer to that of His31 in the unfolded state.
86:
1807:
1557:
1258:
1241:
723:
530:
142:
The salt bridge most often arises from the anionic carboxylate (RCOO) of either
90:
20:
1616:
793:
504:
919:
911:
470:
Interlacing salt bridges that connect the two halves of the molecular capsule
1542:
1517:
175:
159:
94:
38:
1411:
1375:
1340:
1309:
1301:
1267:
1118:
927:
895:
880:
842:
766:
758:
731:
603:
1073:
1038:
1000:
992:
965:
824:
520:
179:
171:
78:
1065:
957:
210:
745:
Schneider HJ (2009). "Binding mechanisms in supramolecular complexes".
127:
1403:
1367:
1110:
979:
Becktel WJ, Schellman JA (November 1987). "Protein stability curves".
873:
10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X
595:
265:
1332:
186:
can also participate, depending on outside factors perturbing their p
183:
155:
499:). Szumna and coworkers developed a novel molecular capsule with a
1628:
529:
451:
264:
228:
209:
25:
1420:
272:
Titration curve between the wild-type (blue) and the mutant (red)
236:
Mutagenesis of T4 lysozyme salt bridge between Asp 70 and His 31
1424:
1240:
Busschaert N, Caltagirone C, Van Rossom W, Gale PA (May 2015).
89:, and with anions is formed by deprotonation as in the case of
516:
503:
interior. This capsule is made of two halves, like a plastic
1092:"Contribution of surface salt bridges to protein stability"
646:
1157:
Bowman-James K, Bianchi A, García-Espana E, eds. (2012).
1132:
Bianchi A, Bowman-James K, García-España E, eds. (1997).
306:
O buffers of moderate ionic strength. Figure 5 shows a p
206:
Methods for quantifying salt bridge stability in proteins
16:
Combination of hydrogen and ionic bonding in chemistry
57:(Figure 1). Ion pairing is one of the most important
1797:
1774:
1705:
1667:
1647:
1636:
1596:
1578:
1469:
1458:
582:Marcus Y, Hefter G (November 2006). "Ion pairing".
1242:"Applications of supramolecular anion recognition"
687:Principles and methods in supramolecular chemistry
618:
537:Self-assembly of a double helical metallopolymer
197:Salt bridges found in protein - ligand complexes
313:of the wild-type of 9.05. This difference in p
854:
852:
433:lysozyme example, by monitoring its shift in p
1436:
1209:Anion Recognition in Supramolecular Chemistry
126:Wild type (left) and mutated (right) form of
19:For the device used in electrochemistry, see
8:
939:
937:
900:Journal of Chemical Information and Modeling
1012:
1010:
685:Schneider HJ, Yatsimirsky AK, eds. (2000).
561:. Sausalito, CA: University Science Books.
1644:
1466:
1443:
1429:
1421:
1182:Sessler JL, Gale PA, Cho WS, eds. (2006).
1257:
1186:. Cambridge: Royal Society of Chemistry.
832:
552:
550:
1835:Polyhedral skeletal electron pair theory
1392:Journal of the American Chemical Society
462:
118:
1290:Angewandte Chemie International Edition
1085:
1083:
652:John Wiley & Sons, Inc., p. 35 ff
546:
278:nuclear magnetic resonance spectroscopy
77:In water, formation of salt bridges or
519:. In one example, they incorporated
7:
625:(2nd ed.). England: Longmans.
365:is the temperature in kelvins, and
1281:Evans NH, Beer PD (October 2014).
1134:Supramolecular chemistry of anions
1090:Strop P, Mayo SL (February 2000).
813:European Journal of Human Genetics
14:
648:(A. Ciferri and A. Perico, Eds),
559:Modern Physical Organic Chemistry
459:The "egg shell" molecular capsule
1627:
1621:
1615:
1207:Gale PA, Dehaen W, eds. (2010).
672:NIST-JANAF Thermochemical Tables
73:Salt bridges in chemical bonding
361:is the universal gas constant,
782:Coordination Chemistry Reviews
390:: His31-H ⇌ His31 + H. The p
150:and the cationic ammonium (RNH
115:Salt bridges found in proteins
1:
1031:10.1016/S0022-2836(99)80018-7
557:Dougherty, Dennis A. (2006).
1159:Anion coordination chemistry
1019:Journal of Molecular Biology
1259:10.1021/acs.chemrev.5b00099
724:10.1021/acs.chemrev.5b00583
276:The second method utilizes
1882:
1533:Metal–ligand multiple bond
621:Physical Organic Chemistry
381:acid dissociation constant
18:
1613:
794:10.1016/j.ccr.2007.08.005
440:after mutation of His31.
224:Site-directed mutagenesis
47:non-covalent interactions
1184:Anion receptor chemistry
912:10.1021/acs.jcim.8b00266
475:Supramolecular chemistry
448:Supramolecular chemistry
132:mandibuloacral dysplasia
67:supramolecular chemistry
45:is a combination of two
1325:Chemical Communications
1161:. Weinheim: Wiley-VCH.
1136:. New York: Wiley-VCH.
511:Double helical polymers
497:molecular encapsulation
63:ion pair chromatography
1302:10.1002/anie.201309937
759:10.1002/anie.200802947
538:
471:
460:
273:
237:
218:
139:
34:
993:10.1002/bip.360261104
689:. Chichester: Wiley.
533:
466:
455:
268:
232:
213:
122:
109:Debye–Hückel equation
85:with e.g. protonated
29:
1523:Coordinate (dipolar)
1211:. Springer Science.
825:10.1038/ejhg.2012.77
788:(10–11): 1093–1107.
404:by the following: p
1866:Protein engineering
1697:C–H···O interaction
1479:Electron deficiency
1217:2010arsc.book.....G
1066:10.1021/bi00243a015
958:10.1021/bi00461a025
397:is then related to
331:The difference in p
1682:Resonance-assisted
539:
491:Molecular capsules
482:Anion complexation
472:
461:
274:
243:circular dichroism
238:
219:
140:
35:
1848:
1847:
1799:Electron counting
1770:
1769:
1659:London dispersion
1611:
1610:
1588:Metal aromaticity
1404:10.1021/ja0619096
1368:10.1021/cr900149d
1111:10.1021/bi992257j
906:(11): 2224–2238.
747:Angewandte Chemie
696:978-0-471-97253-2
670:Chase MW (1998).
658:978-0-470-52927-0
617:Isaacs N (1996).
596:10.1021/cr040087x
568:978-1-891389-31-3
340:Gibbs free energy
136:progeria syndrome
100:contact ion pairs
1873:
1861:Chemical bonding
1840:Jemmis mno rules
1692:Dihydrogen bonds
1645:
1631:
1625:
1619:
1553:Hyperconjugation
1467:
1445:
1438:
1431:
1422:
1416:
1415:
1386:
1380:
1379:
1362:(11): 5799–867.
1356:Chemical Reviews
1351:
1345:
1344:
1333:10.1039/b820990a
1320:
1314:
1313:
1296:(44): 11716–54.
1287:
1278:
1272:
1271:
1261:
1252:(15): 8038–155.
1246:Chemical Reviews
1237:
1231:
1230:
1204:
1198:
1197:
1179:
1173:
1172:
1154:
1148:
1147:
1129:
1123:
1122:
1096:
1087:
1078:
1077:
1049:
1043:
1042:
1014:
1005:
1004:
976:
970:
969:
941:
932:
931:
891:
885:
884:
856:
847:
846:
836:
804:
798:
797:
777:
771:
770:
742:
736:
735:
712:Chemical Reviews
707:
701:
700:
682:
676:
675:
667:
661:
643:
637:
636:
624:
614:
608:
607:
590:(11): 4585–621.
584:Chemical Reviews
579:
573:
572:
554:
425:, the observed p
51:hydrogen bonding
1881:
1880:
1876:
1875:
1874:
1872:
1871:
1870:
1851:
1850:
1849:
1844:
1793:
1766:
1709:
1701:
1663:
1650:
1640:
1632:
1626:
1620:
1607:
1592:
1574:
1462:
1454:
1449:
1419:
1388:
1387:
1383:
1353:
1352:
1348:
1327:(15): 1959–61.
1322:
1321:
1317:
1285:
1280:
1279:
1275:
1239:
1238:
1234:
1227:
1206:
1205:
1201:
1194:
1181:
1180:
1176:
1169:
1156:
1155:
1151:
1144:
1131:
1130:
1126:
1094:
1089:
1088:
1081:
1060:(29): 7142–53.
1051:
1050:
1046:
1016:
1015:
1008:
987:(11): 1859–77.
978:
977:
973:
943:
942:
935:
893:
892:
888:
858:
857:
850:
819:(11): 1134–40.
806:
805:
801:
779:
778:
774:
753:(22): 3924–77.
744:
743:
739:
718:(9): 5216–300.
709:
708:
704:
697:
684:
683:
679:
669:
668:
664:
644:
640:
633:
616:
615:
611:
581:
580:
576:
569:
556:
555:
548:
544:
525:ligand exchange
513:
493:
484:
450:
439:
431:
424:
417:
410:
403:
396:
389:
378:
371:
356:
337:
327:
319:
312:
305:
301:
294:
287:
208:
199:
192:
169:
165:
153:
117:
75:
24:
17:
12:
11:
5:
1879:
1877:
1869:
1868:
1863:
1853:
1852:
1846:
1845:
1843:
1842:
1837:
1832:
1831:
1830:
1825:
1820:
1815:
1804:
1802:
1795:
1794:
1792:
1791:
1786:
1780:
1778:
1772:
1771:
1768:
1767:
1765:
1764:
1759:
1754:
1749:
1744:
1739:
1729:
1724:
1719:
1713:
1711:
1703:
1702:
1700:
1699:
1694:
1689:
1684:
1679:
1673:
1671:
1665:
1664:
1662:
1661:
1655:
1653:
1642:
1638:Intermolecular
1634:
1633:
1614:
1612:
1609:
1608:
1606:
1605:
1602:
1600:
1594:
1593:
1591:
1590:
1584:
1582:
1576:
1575:
1573:
1572:
1571:
1570:
1565:
1555:
1550:
1545:
1540:
1535:
1530:
1525:
1520:
1515:
1510:
1509:
1508:
1498:
1497:
1496:
1491:
1486:
1475:
1473:
1464:
1460:Intramolecular
1456:
1455:
1452:Chemical bonds
1450:
1448:
1447:
1440:
1433:
1425:
1418:
1417:
1398:(21): 6806–7.
1381:
1346:
1315:
1273:
1232:
1226:978-3642264702
1225:
1199:
1193:978-0854049745
1192:
1174:
1168:978-3527323708
1167:
1149:
1142:
1124:
1079:
1044:
1025:(4): 1031–44.
1006:
971:
933:
886:
848:
799:
772:
737:
702:
695:
677:
662:
638:
632:978-0582218635
631:
609:
574:
567:
545:
543:
540:
512:
509:
492:
489:
483:
480:
449:
446:
437:
429:
422:
415:
408:
401:
394:
387:
376:
369:
354:
335:
325:
317:
310:
303:
299:
292:
285:
207:
204:
198:
195:
190:
167:
163:
151:
116:
113:
74:
71:
15:
13:
10:
9:
6:
4:
3:
2:
1878:
1867:
1864:
1862:
1859:
1858:
1856:
1841:
1838:
1836:
1833:
1829:
1826:
1824:
1821:
1819:
1816:
1814:
1813:Hückel's rule
1811:
1810:
1809:
1806:
1805:
1803:
1800:
1796:
1790:
1787:
1785:
1782:
1781:
1779:
1777:
1776:Bond cleavage
1773:
1763:
1760:
1758:
1755:
1753:
1750:
1748:
1745:
1743:
1742:Intercalation
1740:
1737:
1733:
1732:Metallophilic
1730:
1728:
1725:
1723:
1720:
1718:
1715:
1714:
1712:
1708:
1704:
1698:
1695:
1693:
1690:
1688:
1685:
1683:
1680:
1678:
1675:
1674:
1672:
1670:
1666:
1660:
1657:
1656:
1654:
1652:
1649:Van der Waals
1646:
1643:
1639:
1635:
1630:
1624:
1618:
1604:
1603:
1601:
1599:
1595:
1589:
1586:
1585:
1583:
1581:
1577:
1569:
1566:
1564:
1561:
1560:
1559:
1556:
1554:
1551:
1549:
1546:
1544:
1541:
1539:
1536:
1534:
1531:
1529:
1526:
1524:
1521:
1519:
1516:
1514:
1511:
1507:
1504:
1503:
1502:
1499:
1495:
1492:
1490:
1487:
1485:
1482:
1481:
1480:
1477:
1476:
1474:
1472:
1468:
1465:
1461:
1457:
1453:
1446:
1441:
1439:
1434:
1432:
1427:
1426:
1423:
1413:
1409:
1405:
1401:
1397:
1393:
1385:
1382:
1377:
1373:
1369:
1365:
1361:
1357:
1350:
1347:
1342:
1338:
1334:
1330:
1326:
1319:
1316:
1311:
1307:
1303:
1299:
1295:
1291:
1284:
1277:
1274:
1269:
1265:
1260:
1255:
1251:
1247:
1243:
1236:
1233:
1228:
1222:
1218:
1214:
1210:
1203:
1200:
1195:
1189:
1185:
1178:
1175:
1170:
1164:
1160:
1153:
1150:
1145:
1143:9780471186229
1139:
1135:
1128:
1125:
1120:
1116:
1112:
1108:
1105:(6): 1251–5.
1104:
1100:
1093:
1086:
1084:
1080:
1075:
1071:
1067:
1063:
1059:
1055:
1048:
1045:
1040:
1036:
1032:
1028:
1024:
1020:
1013:
1011:
1007:
1002:
998:
994:
990:
986:
982:
975:
972:
967:
963:
959:
955:
952:(9): 2403–8.
951:
947:
940:
938:
934:
929:
925:
921:
917:
913:
909:
905:
901:
897:
890:
887:
882:
878:
874:
870:
867:(7): 604–17.
866:
862:
855:
853:
849:
844:
840:
835:
830:
826:
822:
818:
814:
810:
803:
800:
795:
791:
787:
783:
776:
773:
768:
764:
760:
756:
752:
748:
741:
738:
733:
729:
725:
721:
717:
713:
706:
703:
698:
692:
688:
681:
678:
673:
666:
663:
659:
655:
651:
647:
642:
639:
634:
628:
623:
622:
613:
610:
605:
601:
597:
593:
589:
585:
578:
575:
570:
564:
560:
553:
551:
547:
541:
536:
532:
528:
526:
522:
518:
510:
508:
506:
502:
498:
490:
488:
481:
479:
476:
469:
465:
458:
454:
447:
445:
441:
436:
428:
421:
414:
407:
400:
393:
386:
382:
379:known as the
375:
368:
364:
360:
353:
349:
345:
341:
334:
329:
324:
316:
309:
298:
291:
284:
279:
271:
267:
263:
260:
256:
252:
248:
244:
235:
231:
227:
225:
216:
212:
205:
203:
196:
194:
189:
185:
181:
177:
173:
161:
157:
149:
148:glutamic acid
145:
144:aspartic acid
137:
133:
129:
125:
121:
114:
112:
110:
105:
101:
96:
92:
88:
87:ammonium ions
84:
80:
72:
70:
68:
64:
60:
56:
55:ionic bonding
52:
48:
44:
40:
32:
28:
22:
1818:Baird's rule
1761:
1538:Charge-shift
1501:Hypervalence
1395:
1391:
1384:
1359:
1355:
1349:
1324:
1318:
1293:
1289:
1276:
1249:
1245:
1235:
1208:
1202:
1183:
1177:
1158:
1152:
1133:
1127:
1102:
1099:Biochemistry
1098:
1057:
1054:Biochemistry
1053:
1047:
1022:
1018:
984:
980:
974:
949:
946:Biochemistry
945:
903:
899:
889:
864:
860:
816:
812:
802:
785:
781:
775:
750:
746:
740:
715:
711:
705:
686:
680:
671:
665:
649:
645:
641:
620:
612:
587:
583:
577:
558:
534:
514:
494:
485:
473:
467:
456:
442:
434:
426:
419:
412:
405:
398:
391:
384:
373:
366:
362:
358:
351:
347:
343:
332:
330:
322:
314:
307:
296:
289:
282:
275:
269:
258:
254:
250:
246:
239:
233:
220:
214:
200:
187:
141:
123:
76:
42:
36:
30:
1808:Aromaticity
1784:Heterolysis
1762:Salt bridge
1707:Noncovalent
1677:Low-barrier
1558:Aromaticity
1548:Conjugation
1528:Pi backbond
981:Biopolymers
861:ChemBioChem
302:of 6.8 in H
160:guanidinium
91:carboxylate
59:noncovalent
43:salt bridge
21:Salt bridge
1855:Categories
1736:aurophilic
1717:Mechanical
542:References
505:easter egg
241:change in
1828:spherical
1789:Homolysis
1752:Cation–pi
1727:Chalcogen
1687:Symmetric
1543:Hapticity
920:1549-960X
535:Figure 8.
468:Figure 7.
457:Figure 6.
357:), where
350: ln(
346: = −
270:Figure 5.
234:Figure 4.
215:Figure 3.
176:histidine
124:Figure 2.
95:phosphate
83:ion pairs
79:ion pairs
39:chemistry
31:Figure 1.
1757:Anion–pi
1747:Stacking
1669:Hydrogen
1580:Metallic
1471:Covalent
1463:(strong)
1412:16719458
1376:19678641
1341:19333456
1310:25204549
1268:25996028
1119:10684603
928:30351056
881:12324994
843:22549407
767:19415701
732:27136957
604:17091929
521:platinum
180:tyrosine
172:arginine
162:(RNHC(NH
128:lamin A
1722:Halogen
1568:bicyclo
1513:Agostic
1213:Bibcode
1074:1854726
1039:2266554
1001:3689874
966:2337607
834:3476705
411:= −log(
158:or the
154:) from
104:Bjerrum
1823:Möbius
1651:forces
1641:(weak)
1410:
1374:
1339:
1308:
1266:
1223:
1190:
1165:
1140:
1117:
1072:
1037:
999:
964:
926:
918:
879:
841:
831:
765:
730:
693:
656:
629:
602:
565:
501:chiral
184:serine
182:, and
156:lysine
1801:rules
1710:other
1598:Ionic
1506:3c–4e
1494:8c–2e
1489:4c–2e
1484:3c–2e
1286:(PDF)
1095:(PDF)
170:) of
1563:homo
1518:Bent
1408:PMID
1372:PMID
1337:PMID
1306:PMID
1264:PMID
1221:ISBN
1188:ISBN
1163:ISBN
1138:ISBN
1115:PMID
1070:PMID
1035:PMID
997:PMID
962:PMID
924:PMID
916:ISSN
877:PMID
839:PMID
763:PMID
728:PMID
691:ISBN
654:ISBN
650:2012
627:ISBN
600:PMID
563:ISBN
134:and
53:and
41:, a
1400:doi
1396:128
1364:doi
1360:109
1329:doi
1298:doi
1254:doi
1250:115
1107:doi
1062:doi
1027:doi
1023:216
989:doi
954:doi
908:doi
869:doi
829:PMC
821:doi
790:doi
786:252
755:doi
720:doi
716:116
592:doi
588:106
517:DNA
342:: Δ
146:or
37:In
1857::
1406:.
1394:.
1370:.
1358:.
1335:.
1304:.
1294:53
1292:.
1288:.
1262:.
1248:.
1244:.
1219:.
1113:.
1103:39
1101:.
1097:.
1082:^
1068:.
1058:30
1056:.
1033:.
1021:.
1009:^
995:.
985:26
983:.
960:.
950:29
948:.
936:^
922:.
914:.
904:58
902:.
898:.
875:.
863:.
851:^
837:.
827:.
817:20
815:.
811:.
784:.
761:.
751:48
749:.
726:.
714:.
598:.
586:.
549:^
383:,
377:eq
370:eq
355:eq
348:RT
178:,
93:,
49::
1738:)
1734:(
1444:e
1437:t
1430:v
1414:.
1402::
1378:.
1366::
1343:.
1331::
1312:.
1300::
1270:.
1256::
1229:.
1215::
1196:.
1171:.
1146:.
1121:.
1109::
1076:.
1064::
1041:.
1029::
1003:.
991::
968:.
956::
930:.
910::
883:.
871::
865:3
845:.
823::
796:.
792::
769:.
757::
734:.
722::
699:.
660:.
635:.
606:.
594::
571:.
438:a
435:K
430:a
427:K
423:a
420:K
416:a
413:K
409:a
406:K
402:a
399:K
395:a
392:K
388:a
385:K
374:K
367:K
363:T
359:R
352:K
344:G
336:a
333:K
326:a
323:K
321:p
318:a
315:K
311:a
308:K
304:2
300:a
297:K
293:a
290:K
286:a
283:K
281:p
259:S
255:S
251:S
249:Δ
247:T
191:a
188:K
168:2
166:)
164:2
152:3
138:.
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.