1606:
1681:
344:
321:
218:
243:
1893:
these disorders have been found to be in vivo and in vitro substrates of tTG. Although tTG is up regulated in the areas of the brain affected by
Huntington's disease, a recent study showed that increasing levels of tTG do not affect the onset and/or progression of the disease in mice. Recent studies show that tTG may not be involved in AD as studies show it is associated with erythrocyte lysis and is a consequence of the disease rather than a cause.
2029:
602:
595:
609:
350:
249:
4354:
1664:-1 has been shown to activate extracellular tTG by reducing the disulfide bond. Another disuplhide bond can form in tTG, between the residues Cys-230 and Cys-370. While this bond does not exist in the enzyme's native state, it appears when the enzyme is inactivated via oxidation. The presence of calcium protects against the formation of both disulfide bonds, thus making the enzyme more resistant to oxidation.
5057:
2088:
cultured human umbilical vein endothelial cells (HUVECs). ERp57 oxidized TG2 with a rate constant that was 400-2000-fold higher than those of the aforementioned small molecule oxidants. Moreover, its specificity for TG2 was also markedly higher than those of other secreted redox proteins, including protein disulfide isomerase (PDI), ERp72, TRX, and quiescin sulfhydryl oxidase 1 (QSOX1).
1696:
1668:
1461:, GTP-binding/hydrolyzing, and isopeptidase activities. Unlike other members of the transglutaminase family, tTG can be found both in the intracellular and the extracellular spaces of various types of tissues and is found in many different organs including the heart, the liver, and the small intestine. Intracellular tTG is abundant in the
1751:. Evidence shows that intracellular tTG crosslinks itself to myosin. It is also believed that tTG may stabilize the structure of the dying cells during apoptosis by polymerizing the components of the cytoskeleton, therefore preventing the leakage of the cellular contents into the extracellular space.
1838:
and tumor biology. tTG expression is elevated in multiple cancer cell types and is implicated in drug resistance and metastasis due to its ability to promote mesenchymal transition and stem cell like properties. In its GTP bound form, tTG contributes to cancer cell survival and appears to be a cancer
1758:
activity: In the presence of GTP, it suggested to function as a G protein participating in signaling processes. Besides its transglutaminase activity, tTG is proposed to also act as kinase, and protein disulfide isomerase, and deamidase. This latter activity is important in the deamidation of gliadin
1659:
residues, namely Cys 370 and Cys 371. When this disulfide bond forms, the enzyme remains in an open confirmation but becomes catalytically inactive. The, oxidation/reduction of the disulfide bond serves as a third allosteric regulatory mechanism (along with GTP/GDP and Ca) for the activation of tTG.
2087:
Endoplasmic reticulum (ER)-resident protein 57 (ERp57), a protein in the ER that promotes folding of nascent proteins and is also present in the extracellular environment, has the cellular and biochemical characteristics for inactivating TG2. We found that ERp57 colocalizes with extracellular TG2 in
1633:
Crosslinking activity by tTG requires the binding of Ca ions. Multiple Ca can bind to a single tTG molecule. Specifically, tTG binds up to 6 calcium ions at 5 different binding sites. Mutations to these binding sites causing lower calcium affinity, decrease the enzyme's transglutaminase activity. In
1892:
diseases by affecting transcription, differentiation and migration and adhesion . Such neurological diseases are characterized in part by the abnormal aggregation of proteins due to the increased activity of protein crosslinking in the affected brain. Additionally, specific proteins associated with
1676:
Recent studies have suggested that interferon-γ may serve as an activator of extracellular tTG in the small intestine; these studies have a direct implication to the pathogenesis of celiac disease. Activation of tTG has been shown to be accompanied by large conformational changes, switching from a
1782:
tTG is the most comprehensively studied transglutaminase and has been associated with many diseases. However, none of these diseases are related to an enzyme deficiency. Indeed, thus far no disease has been attributed to the lack of tTG activity and this has been attested through the study of tTG
1987:
of TG2 are known to affect TG2 activity, which enables it to subsequently execute diverse biological functions in the cell. However, the importance of non-enzymatic interactions in regulating TG2 activities is yet to be revealed. Recent studies indicate that non-enzymatic interactions play
1557:
have demonstrated that these forms of TG2 adopt a "closed" conformation, whereas TG2 with the active site occupied by an inhibitory gluten peptide mimic or other similar inhibitors adopts an "open" conformation. In the open conformation the four domains of TG2 are arranged in an extended
1642:
inhibits the crosslinking activity of the enzyme. Therefore, intracellular tTG is mostly inactive due to the relatively high concentration of GTP/GDP and the low levels of calcium inside the cell. Although extracellular tTG is expected to be active due to the low concentration of
1671:
Figure 2: Cystein residues relevant in tTG activity. The disulfide bond between Cys 370 and Cys 371 has formed, therefore the enzyme is in an active conformation. The distance between Cys 370 and Cys 230 is 11.3 Å. Cys 277 is the cystein located within the active site of the
73:
47:
1602:(i.e. deamidation). The deamidation of glutamine residues catalyzed by tTG is thought to be linked to the pathological immune response to gluten in celiac disease. A schematic for the crosslinking and the deamidation reactions is provided in Figure 1.
3634:
Rossin F, Villella VR, D'Eletto M, Farrace MG, Esposito S, Ferrari E, Monzani R, Occhigrossi L, Pagliarini V, Sette C, Cozza G, Barlev NA, Falasca L, Fimia GM, Kroemer G, Raia V, Maiuri L, Piacentini M (July 2018).
4088:
Sblattero D, Berti I, Trevisiol C, Marzari R, Tommasini A, Bradbury A, Fasano A, Ventura A, Not T (May 2000). "Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease".
3995:
Martin A, Giuliano A, Collaro D, De Vivo G, Sedia C, Serretiello E, Gentile V (January 2013). "Possible involvement of transglutaminase-catalyzed reactions in the physiopathology of neurodegenerative diseases".
1871:
have showed promise in using tTG inhibitors as anti-cancer therapeutic agents. However, other studies have noted that tTG transamidation activity could be linked to the inhibition of tumor cell invasiveness.
1650:
and the high levels of calcium in the extracellular space, evidence has shown that extracellular tTG is mostly inactive. Recent studies suggest that extracellular tTG is kept inactive by the formation of a
1692:, TG2 is "turned off", due primarily to the oxidizing activity of endoplasmic reticulum protein 57 (ERp57). Thus, tTG is allosterically regulated by two separate proteins, Erp57 and TRX-1. (See Figure 4).
1529:
which catalyze the crosslinking of proteins by epsilon-(gamma-glutamyl)lysine isopeptide bonds. Similarly to other transglutaminases, tTG consists of a GTP/ GDP binding site, a
5114:
1598:
bond between the two substrates (i.e. crosslinking). Alternatively, the thioester intermediate can be hydrolyzed, resulting in the net conversion of the glutamine residue to
1867:, lower survival rates and generally poor prognosis. Cancer cells can be killed by increasing calcium levels through the activation of tTG transamidation activity.
1774:(Heat Shock Factor 1) and thus the body's response to heat shock. In the absence of tTG, the response to heat shock is impaired since the necessary trimer is not formed.
3738:
Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, Schuppan D (July 1997). "Identification of tissue transglutaminase as the autoantigen of celiac disease".
357:
256:
1371:
2036:
These Mutant ES Cells can be studied directly or used to generate mice with this gene knocked out. Study of these mice can shed light on the function of Tgm2: see
2757:
1390:
2028:
4593:
4588:
2399:"Transglutaminase-2 interaction with heparin: identification of a heparin binding site that regulates cell adhesion to fibronectin-transglutaminase-2 matrix"
1481:. Extracellular tTG has been linked to cell adhesion, ECM stabilization, wound healing, receptor signaling, cellular proliferation, and cellular motility.
1500:
atrophy. It has also been implicated in the pathophysiology of many other diseases, including such as many different cancers and neurogenerative diseases.
4394:
4235:"Physiological, pathological, and structural implications of non-enzymatic protein-protein interactions of the multifunctional human transglutaminase 2"
2881:
Han BG, Cho JW, Cho YD, Jeong KC, Kim SY, Lee BI (August 2010). "Crystal structure of human transglutaminase 2 in complex with adenosine triphosphate".
917:
3440:"Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase"
898:
179:
1708:
tTG is expressed ubiquitously and is present in various cellular compartments, such as the cytosol, the nucleus, and the plasma membrane. It requires
4576:
1605:
2616:
Di
Sabatino A, Vanoli A, Giuffrida P, Luinetti O, Solcia E, Corazza GR (August 2012). "The function of tissue transglutaminase in celiac disease".
1795:. It was first associated with celiac disease in 1997 when the enzyme was found to be the antigen recognized by the antibodies specific to celiac.
1747:
tTG is thought to be involved in the regulation of the cytoskeleton by crosslinking various cytoskeletal proteins including myosin, actin, and
5089:
4583:
5337:
4333:
2164:
2146:
4571:
3889:"Presence of tissue transglutaminase in granular endoplasmic reticulum is characteristic of melanized neurons in Parkinson's disease brain"
1766:
tTG also presents PDI (Protein
Disulfide Isomerase) activity. Based on its PDI activity, tTG plays an important role in the regulation of
3246:"Cytosolic guanine nucledotide binding deficient form of transglutaminase 2 (R580a) potentiates cell death in oxygen glucose deprivation"
4743:
4353:
2660:"Functional significance of five noncanonical Ca2+-binding sites of human transglutaminase 2 characterized by site-directed mutagenesis"
2566:"Interferon-γ activates transglutaminase 2 via a phosphatidylinositol-3-kinase-dependent pathway: implications for celiac sprue therapy"
1622:. Once synthesized, most of the protein is found in the cytoplasm, plasma membrane and ECM, but a small fraction is translocated to the
2824:"Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity"
1699:
Figure 4: The proteins that allosterically regulate tTG. On the left Erp57 which oxidizes tTG and on the right TRX-1 which reduces tTG.
343:
4711:
1156:
1149:
808:
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway
4776:
4527:
4465:
3369:
2733:
1814:
responses that eventually result in the production of anti-transglutaminase antibodies IgA and IgG. tTG specifically deamidates the
1959:
diseases. This indicates that tTG inhibitors could also serve as a tool to mitigate the progression of tTG brain related diseases.
320:
5271:
4522:
4517:
1796:
1383:
4460:
2133:
2112:
4134:"Cardiac fibrosis can be attenuated by blocking the activity of transglutaminase 2 using a selective small-molecule inhibitor"
4041:"Tissue transglutaminase overexpression does not modify the disease phenotype of the R6/2 mouse model of Huntington's disease"
2069:
Transglutaminase 2 (TG2) is a ubiquitously expressed (intracellular as well as extracellular) protein, with multiple modes of
4475:
4387:
1334:
1310:
242:
217:
4932:
4295:
2658:
Király R, Csosz E, Kurtán T, Antus S, Szigeti K, Simon-Vecsei Z, Korponay-Szabó IR, Keresztessy Z, Fésüs L (December 2009).
2129:
1590:
intermediate. The thioester intermediate can then be attacked by the surface amine of a second substrate (typically from a
70:
5142:
4640:
4620:
4534:
2070:
2108:
2005:
4561:
4455:
4450:
2397:
Lortat-Jacob H, Burhan I, Scarpellini A, Thomas A, Imberty A, Vivès RR, Johnson T, Gutierrez A, Verderio EA (May 2012).
159:
3832:"Transglutaminase 2 takes center stage as a cancer cell survival factor and therapy target: Transglutaminase in cancer"
1477:. In the extracellular space, tTG binds to proteins of the extracellular matrix (ECM), binding particularly tightly to
4723:
4497:
2187:"Protein transamidation by transglutaminase 2 in cells: a disputed Ca2+-dependent action of a multifunctional protein"
1968:
1935:(>90%) for identifying celiac disease. Modern anti-tTG assays rely on a human recombinant protein as an antigen.
356:
255:
5119:
5082:
5047:
4733:
4566:
4440:
4917:
5033:
5020:
5007:
4994:
4981:
4968:
4955:
4681:
4657:
4608:
4492:
4445:
4420:
4380:
4326:
349:
248:
4927:
1328:
4881:
4824:
4686:
4411:
1908:. Specifically, in kidney fibrosis, tTG contributes to the stabilization and accumulation of the ECM affecting
1729:
1423:
1221:
962:
167:
31:
1457:(protein degradation). Aside from its crosslinking function, tTG catalyzes other types of reactions including
1315:
4829:
4728:
1943:
It's still experimental to use tTG as a form of surgical glue. It is also being studied as an attenuator of
1927:
has superseded older serological tests (anti-endomysium, anti-gliadin, and anti-reticulin) and has a strong
1889:
1864:
943:
5124:
4696:
4691:
4556:
2974:"Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease"
2353:
2012:
Mutant Mouse
Embryonic Stem Cell Clones. These are the known targeted mutations for this gene in a mouse.
1885:
1680:
1635:
1554:
1550:
1496:
causes a pathological immune response resulting in the inflammation of the small intestine and subsequent
1395:
5347:
5306:
5075:
4850:
4769:
4676:
4671:
3403:
Fesus L, Piacentini M (October 2002). "Transglutaminase 2: an enigmatic enzyme with diverse functions".
2074:
2063:
1984:
1639:
1546:
1303:
231:
4922:
2972:
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM (August 2015).
1736:. It has been noted that tTG may have very different activity in different cell types. For example, in
1238:
4738:
4625:
4612:
4512:
4319:
3257:
2835:
2059:
1725:
1689:
1627:
146:
3887:
Wilhelmus MM, Verhaar R, Andringa G, Bol JG, Cras P, Shan L, Hoozemans JJ, Drukarch B (March 2011).
3144:"Endoplasmic reticulum-resident protein 57 (ERp57) oxidatively inactivates human transglutaminase 2"
2358:
4886:
4470:
2051:
1932:
1928:
1619:
1331:
1233:
1103:
1099:
1095:
1048:
1044:
1040:
1578:
group from a Cys residue in the active site of tTG. The thiol group attacks the carboxamide of a
1255:
5245:
4819:
4272:
4114:
4039:
Kumar A, Kneynsberg A, Tucholski J, Perry G, van Groen T, Detloff PJ, Lesort M (September 2012).
4021:
3926:
3869:
3763:
3720:
2751:
2689:
2379:
2216:
1800:
1709:
1542:
191:
3479:
Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y (August 2003).
1132:
1111:
1107:
1069:
1036:
1032:
5342:
4701:
4430:
4264:
4215:
4163:
4106:
4070:
4013:
3977:
3918:
3861:
3812:
3755:
3712:
3666:
3613:
3559:
3510:
3461:
3420:
3385:
3365:
3334:
3285:
3226:
3175:
3115:
3055:
3005:
2954:
2898:
2863:
2804:
2739:
2729:
2681:
2633:
2595:
2535:
2479:
2430:
2371:
2344:
McConkey DJ, Orrenius S (October 1997). "The role of calcium in the regulation of apoptosis".
2326:
2280:
2208:
1956:
1955:. tTG inhibitors have also been shown to inhibit the formation of toxic inclusions related to
1868:
1852:
1804:
1526:
1497:
1322:
139:
63:
1609:
Figure 1: Transamidation (crosslinking) and deamidation mechanisms of tissue transglutaminase
5281:
5134:
4865:
4860:
4834:
4762:
4666:
4630:
4254:
4246:
4205:
4197:
4153:
4145:
4098:
4060:
4052:
4005:
3967:
3957:
3908:
3900:
3851:
3843:
3802:
3794:
3747:
3702:
3656:
3648:
3603:
3593:
3549:
3541:
3500:
3492:
3451:
3412:
3375:
3357:
3324:
3316:
3275:
3265:
3216:
3206:
3165:
3155:
3105:
3095:
3047:
3039:
2995:
2985:
2944:
2934:
2890:
2853:
2843:
2794:
2784:
2671:
2625:
2585:
2577:
2525:
2517:
2469:
2461:
2420:
2410:
2363:
2316:
2270:
2262:
2198:
1952:
1948:
1430:
601:
594:
436:
367:
311:
266:
3024:
1558:
configuration, allowing for catalytic activity, whereas in the closed conformation the two
1291:
5255:
5250:
5240:
5235:
5230:
5225:
5220:
5215:
4912:
4896:
4809:
4706:
4407:
4372:
4299:
3025:"Spotlight on the transglutaminase 2 gene: a focus on genomic and transcriptional aspects"
1971:. Enzymatic interactions are formed between TG2 and its substrate proteins containing the
1856:
1848:
1760:
608:
411:
187:
3582:"The Role of Tissue Transglutaminase in Cancer Cell Initiation, Survival and Progression"
2726:
Transglutaminases : multiple functional modifiers and targets for new drug discovery
1267:
4259:
4234:
3261:
2839:
1226:
46:
5205:
5200:
5195:
5190:
5185:
5180:
5175:
5170:
5165:
5061:
4950:
4891:
4210:
4185:
4158:
4133:
4065:
4040:
3972:
3945:
3913:
3888:
3856:
3831:
3807:
3782:
3661:
3636:
3608:
3581:
3554:
3529:
3505:
3480:
3380:
3361:
3329:
3304:
3280:
3245:
3221:
3194:
3170:
3143:
3110:
3083:
3000:
2973:
2949:
2922:
2799:
2772:
2590:
2565:
2530:
2505:
2474:
2449:
2425:
2398:
2275:
2250:
2037:
1839:
driver. tTG is upregulated in cancer cells and tissues in many cancer types, including
1792:
1626:, where it participates in the control of its own expression through the regulation of
1489:
1366:
3637:"TG2 regulates the heat-shock response by the post-translational modification of HSF1"
3416:
2858:
2823:
1346:
832:
827:
822:
817:
812:
807:
802:
797:
792:
787:
782:
777:
772:
767:
762:
757:
741:
736:
731:
726:
721:
716:
711:
706:
701:
696:
680:
675:
670:
665:
660:
655:
650:
5331:
5316:
5106:
4855:
4814:
4102:
3904:
3545:
2676:
2659:
2203:
2186:
2015:
1844:
1733:
1717:
1713:
1618:
The expression of tTG is regulated at the transcriptional level depending on complex
1599:
1538:
1514:
1341:
637:
4276:
4118:
4056:
4025:
3873:
3767:
3724:
3356:. International Review of Cell and Molecular Biology. Vol. 294. pp. 1–97.
2693:
2450:"Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin"
2383:
2220:
5301:
5098:
4804:
2894:
1988:
physiological roles and enable diverse TG2 functions in a context-specific manner.
1835:
1767:
1623:
1562:
are folded in on the catalytic core domain which includes the residue Cys-277. The
1485:
1470:
1466:
429:
208:
3930:
3305:"Transglutaminase 2: Friend or foe? The discordant role in neurons and astrocytes"
1999:
Marker Symbol for Mouse Gene. This symbol is assigned to the genomic locus by the
171:
17:
4292:
4201:
3798:
3270:
3211:
2789:
2629:
195:
150:, G-ALPHA-h, GNAH, HEL-S-45, TG2, TGC, TG(C), transglutaminase 2, G(h), hTG2, tTG
5286:
5157:
5028:
4963:
4799:
4645:
4403:
4304:
A collection of substrates and interaction partners of TG2 is accessible in the
3481:"A novel function of tissue-type transglutaminase: protein disulphide isomerase"
2169:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2151:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1947:
in certain tumors. tTG shows promise as a potential therapeutic target to treat
1661:
1647:
1534:
1530:
1478:
1458:
1454:
1446:
1350:
5056:
3580:
Tabolacci C, De
Martino A, Mischiati C, Feriotto G, Beninati S (January 2019).
3082:
Jin X, Stamnaes J, Klöck C, DiRaimondo TR, Sollid LM, Khosla C (October 2011).
2828:
Proceedings of the
National Academy of Sciences of the United States of America
1880:
tTG is believed to contribute to several neurodegenerative disorders including
1453:
residue, creating an inter- or intramolecular bond that is highly resistant to
512:
5291:
4305:
4250:
4149:
4009:
2266:
1944:
1909:
1860:
1595:
1563:
1559:
328:
225:
175:
4132:
Wang Z, Stuckey DJ, Murdoch CE, Camelliti P, Lip GY, Griffin M (April 2018).
3160:
2743:
1566:
only shows minor structural changes between the two different conformations.
5296:
5002:
4976:
4186:"New insight into transglutaminase 2 and link to neurodegenerative diseases"
3652:
3195:"Transglutaminase 2 undergoes a large conformational change upon activation"
3100:
2990:
2939:
2773:"Transglutaminase 2 undergoes a large conformational change upon activation"
2581:
2415:
2077:
1972:
1881:
1815:
1810:
that are crosslinked to tTG are able to stimulate transglutaminase specific
1741:
1721:
1652:
1587:
1579:
1474:
1450:
862:
572:
450:
395:
382:
294:
281:
183:
4268:
4219:
4167:
4110:
4074:
4017:
3981:
3962:
3922:
3865:
3816:
3716:
3670:
3617:
3563:
3514:
3465:
3456:
3439:
3424:
3389:
3338:
3289:
3230:
3179:
3119:
3059:
3009:
2958:
2902:
2867:
2848:
2808:
2685:
2637:
2599:
2539:
2483:
2434:
2367:
2330:
2284:
2212:
3759:
3707:
3690:
3598:
3530:"A role for tissue transglutaminase in alpha-gliadin peptide cytotoxicity"
2921:
Stamnaes J, Pinkas DM, Fleckenstein B, Khosla C, Sollid LM (August 2010).
2465:
2375:
1196:
1191:
5311:
4435:
3051:
2707:
1924:
1920:
1897:
1840:
1748:
1744:
knocking out the gene expression for tTG is beneficial to cell survival.
1712:
as a cofactor for transamidation activity. Transcription is increased by
1656:
1180:
1007:
988:
3043:
1677:
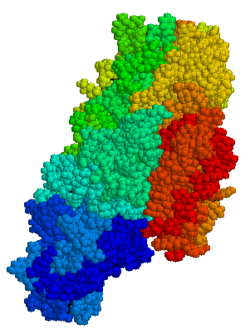
compact (inactive) to an extended (active) conformation. (see Figure 3)
1426:
1279:
3751:
3496:
2521:
2055:
1980:
1823:
1695:
1684:
Figure 3: Compact (inactive) and extended (active) conformations of tTG
1644:
1583:
1462:
1434:
1298:
974:
929:
763:
phospholipase C-activating G protein-coupled receptor signaling pathway
3320:
2022:
Example structure of targeted conditional mutant allele for this gene
773:
negative regulation of endoplasmic reticulum calcium ion concentration
116:
112:
108:
104:
100:
96:
92:
5015:
4785:
4716:
4544:
4539:
4480:
3847:
2305:"The role of transglutaminase-2 and its substrates in human diseases"
2081:
1976:
1905:
1819:
1818:
residues creating epitopes that increase the binding affinity of the
1811:
1755:
1737:
1591:
1493:
1442:
1419:
1378:
1274:
1262:
1250:
1164:
884:
1740:, tTG supports the survival of cells subjected to injury whereas in
1667:
1582:
residue on the surface of a protein or peptide substrate, releasing
1429:) of the protein-glutamine γ-glutamyltransferases family (or simply
3946:"Physio-pathological roles of transglutaminase-catalyzed reactions"
3244:
Colak G, Keillor JW, Johnson GV (January 2011). Polymenis M (ed.).
1574:
The catalytic mechanism for crosslinking in human tTG involves the
5276:
5210:
4989:
4598:
4485:
1716:. Among its many supposed functions, it appears to play a role in
1694:
1679:
1666:
1604:
1575:
1438:
2321:
2304:
1859:. Higher tTG expression also correlates with higher instances of
847:
843:
4502:
1901:
1771:
1286:
163:
5071:
5067:
4758:
4376:
4315:
3084:"Activation of extracellular transglutaminase 2 by thioredoxin"
2448:
Akimov SS, Krylov D, Fleischman LF, Belkin AM (February 2000).
617:
4311:
778:
positive regulation of mitochondrial calcium ion concentration
3691:"Transglutaminase diseases: from biochemistry to the bedside"
2000:
1473:. Intracellular tTG is thought to play an important role in
2251:"Role of transglutaminase 2 in celiac disease pathogenesis"
4754:
3193:
Pinkas DM, Strop P, Brunger AT, Khosla C (December 2007).
2771:
Pinkas DM, Strop P, Brunger AT, Khosla C (December 2007).
1759:
peptides, thus playing important role in the pathology of
803:
positive regulation of I-kappaB kinase/NF-kappaB signaling
3528:
Sakly W, Thomas V, Quash G, El Alaoui S (December 2006).
3142:
Yi MC, Melkonian AV, Ousey JA, Khosla C (February 2018).
2570:
The
Journal of Pharmacology and Experimental Therapeutics
1492:, a lifelong illness in which the consumption of dietary
813:
isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine
419:
828:
positive regulation of smooth muscle cell proliferation
3944:
Ricotta M, Iannuzzi M, Vivo GD, Gentile V (May 2010).
1594:
residue). The end product of the reaction is a stable
5045:
4308:, an interactive transglutaminase substrate database.
1967:
TG2 participates in both enzymatic and non-enzymatic
1834:
Recent studies suggest that tTG also plays a role in
1433:
family). Like other transglutaminases, it crosslinks
584:
5115:
Branched-chain alpha-keto acid dehydrogenase complex
2504:
Griffin M, Casadio R, Bergamini CM (December 2002).
661:
protein-glutamine gamma-glutamyltransferase activity
5264:
5156:
5133:
5105:
4941:
4905:
4874:
4843:
4792:
4656:
4607:
4419:
2346:
2303:Facchiano F, Facchiano A, Facchiano AM (May 2006).
1389:
1377:
1365:
1360:
1340:
1321:
1309:
1297:
1285:
1273:
1261:
1249:
1244:
1232:
1220:
1215:
1210:
1125:
1088:
1062:
1025:
3303:Quinn BR, Yunes-Medina L, Johnson GV (July 2018).
2883:International Journal of Biological Macromolecules
2125:
2123:
2121:
2104:
2102:
2100:
793:branching involved in salivary gland morphogenesis
4362:: HUMAN TISSUE TRANSGLUTAMINASE IN GDP BOUND FORM
3575:
3573:
2923:"Redox regulation of transglutaminase 2 activity"
1951:, through the activity of a highly selective tTG
366:
265:
3781:Murray JA, Frey MR, Oliva-Hemker M (June 2018).
3023:Bianchi N, Beninati S, Bergamini CM (May 2018).
2916:
2914:
2912:
1896:tTG has also been linked to the pathogenesis of
1525:TG2 is a multifunctional enzyme that belongs to
30:"TGM2" redirects here. For the Tetris game, see
4233:Kanchan K, Fuxreiter M, Fésüs L (August 2015).
2564:Diraimondo TR, Klöck C, Khosla C (April 2012).
2050:Endoplasmic reticulum protein 57 (Erp57), is a
27:Protein-coding gene in the species Homo sapiens
3684:
3682:
3680:
3629:
3627:
2719:
2717:
2653:
2651:
2649:
2647:
2506:"Transglutaminases: nature's biological glues"
2249:Klöck C, Diraimondo TR, Khosla C (July 2012).
2130:GRCm38: Ensembl release 89: ENSMUSG00000037820
2080:bond between Cys-370-Cys-371 that renders the
5083:
4770:
4388:
4327:
3354:Cellular functions of tissue transglutaminase
2559:
2557:
2555:
2553:
2551:
2549:
2185:Király R, Demény M, Fésüs L (December 2011).
1465:but smaller amounts can also be found in the
8:
4594:2-acylglycerol-3-phosphate O-acyltransferase
4589:1-acylglycerol-3-phosphate O-acyltransferase
798:positive regulation of inflammatory response
4179:
4177:
2611:
2609:
2298:
2296:
2294:
2109:GRCh38: Ensembl release 89: ENSG00000198959
1211:Protein-glutamine gamma-glutamyltransferase
5090:
5076:
5068:
4777:
4763:
4755:
4395:
4381:
4373:
4334:
4320:
4312:
3137:
3135:
3133:
3131:
3129:
2822:Liu S, Cerione RA, Clardy J (March 2002).
2756:: CS1 maint: location missing publisher (
2499:
2497:
2495:
2493:
1826:, initiating an adaptive immune response.
1357:
858:
633:
407:
306:
203:
81:
4258:
4209:
4157:
4064:
3971:
3961:
3912:
3855:
3806:
3706:
3660:
3607:
3597:
3553:
3504:
3455:
3379:
3328:
3279:
3269:
3220:
3210:
3169:
3159:
3109:
3099:
2999:
2989:
2948:
2938:
2857:
2847:
2798:
2788:
2675:
2589:
2529:
2473:
2424:
2414:
2357:
2320:
2274:
2202:
1634:contrast, the binding of one molecule of
4091:The American Journal of Gastroenterology
2180:
2178:
1990:
818:positive regulation of apoptotic process
742:collagen-containing extracellular matrix
5052:
4349:
2244:
2242:
2240:
2238:
2236:
2234:
2232:
2230:
2096:
4584:Glycerol-3-phosphate O-acyltransferase
3077:
3075:
3073:
3071:
3069:
2749:
2708:"Entrez Gene: TGM2 transglutaminase 2"
2084:inactive in the extracellular matrix.
1207:
702:intrinsic component of plasma membrane
36:
4577:Lecithin—cholesterol acyltransferase
3950:World Journal of Biological Chemistry
1770:, by catalyzing the trimerization of
1513:The human tTG gene is located on the
371:
332:
327:
270:
229:
224:
7:
4572:Glyceronephosphate O-acyltransferase
4239:Cellular and Molecular Life Sciences
3689:Lorand L, Iismaa SE (January 2019).
3534:Clinical and Experimental Immunology
2724:Hitomi K, Kojima S, Fesus L (2015).
1791:tTG is best known for its link with
788:positive regulation of cell adhesion
4744:Sulfoacetaldehyde acetyltransferase
4436:Acetyl-Coenzyme A acetyltransferase
3444:The Journal of Biological Chemistry
3148:The Journal of Biological Chemistry
3088:The Journal of Biological Chemistry
2978:The Journal of Biological Chemistry
2927:The Journal of Biological Chemistry
2403:The Journal of Biological Chemistry
4660:: converted into alkyl on transfer
3362:10.1016/B978-0-12-394305-7.00001-X
3352:Nurminskaya MV, Belkin AM (2012).
1822:peptide to the antigen presenting
1122:
1085:
1059:
1022:
998:
979:
953:
934:
908:
889:
589:
507:
445:
424:
25:
4466:Chloramphenicol acetyltransferase
3830:Eckert, Richard L. (2019-01-29).
3438:Mishra S, Murphy LJ (June 2004).
1418:) is a 78-kDa, calcium-dependent
5055:
4518:Carnitine O-palmitoyltransferase
4352:
4184:Min B, Chung KC (January 2018).
4103:10.1111/j.1572-0241.2000.02018.x
3905:10.1111/j.1750-3639.2010.00429.x
3546:10.1111/j.1365-2249.2006.03236.x
3309:Journal of Neuroscience Research
2677:10.1111/j.1742-4658.2009.07420.x
2204:10.1111/j.1742-4658.2011.08345.x
2027:
1979:donor groups in the presence of
1900:in various organs including the
1803:in which a cellular response to
1797:Anti-transglutaminase antibodies
607:
600:
593:
355:
348:
342:
319:
254:
247:
241:
216:
45:
4461:Beta-galactoside transacetylase
4057:10.1016/j.expneurol.2012.05.015
681:protein domain specific binding
4476:Serotonin N-acetyl transferase
4423:: other than amino-acyl groups
3405:Trends in Biochemical Sciences
2895:10.1016/j.ijbiomac.2010.04.023
1994:Mouse Mutant Alleles for Tgm2
618:More reference expression data
573:More reference expression data
1:
4641:Keratinocyte transglutaminase
4621:Gamma-glutamyl transpeptidase
4535:Serine C-palmitoyltransferase
3417:10.1016/S0968-0004(02)02182-5
2071:Post-translational regulation
2054:molecule involved in loading
531:tunica media of zone of aorta
340:
239:
5338:Genes on human chromosome 20
4562:Aminolevulinic acid synthase
4456:Acetyl-CoA C-acyltransferase
4451:Dihydrolipoyl transacetylase
4202:10.5483/BMBRep.2018.51.1.227
3799:10.1053/j.gastro.2017.12.026
3271:10.1371/journal.pone.0016665
3212:10.1371/journal.pbio.0050327
2790:10.1371/journal.pbio.0050327
2630:10.1016/j.autrev.2012.01.007
4724:2-hydroxyglutarate synthase
2454:The Journal of Cell Biology
2255:Seminars in Immunopathology
823:protein homooligomerization
5364:
5143:Epidermal transglutaminase
5120:Oxoglutarate dehydrogenase
4734:2-isopropylmalate synthase
4567:Beta-ketoacyl-ACP synthase
4441:N-Acetylglutamate synthase
29:
4933:Michaelis–Menten kinetics
4682:Decylhomocitrate synthase
4493:Histone acetyltransferase
4446:Choline acetyltransferase
4347:
4251:10.1007/s00018-015-1909-z
4150:10.1038/s41419-018-0573-2
4010:10.1007/s00726-011-1081-1
2267:10.1007/s00281-012-0305-0
2165:"Mouse PubMed Reference:"
2147:"Human PubMed Reference:"
2035:
2026:
2021:
1993:
1655:bond between two vicinal
1356:
1195:
1190:
1186:
1179:
1163:
1157:Chr 2: 157.96 – 157.99 Mb
1144:
1129:
1092:
1081:
1066:
1029:
1018:
1005:
1001:
986:
982:
973:
960:
956:
941:
937:
928:
915:
911:
896:
892:
883:
868:
861:
857:
841:
768:salivary gland cavitation
636:
632:
615:
592:
583:
570:
519:
510:
457:
448:
418:
410:
406:
389:
376:
339:
318:
309:
305:
288:
275:
238:
215:
206:
202:
157:
154:
144:
137:
132:
89:
84:
67:
62:
57:
53:
44:
39:
5297:Filaggrin (Citrullinate)
4825:Diffusion-limited enzyme
4687:2-methylcitrate synthase
4138:Cell Death & Disease
3836:Molecular Carcinogenesis
3161:10.1074/jbc.RA117.001382
1150:Chr 20: 38.13 – 38.17 Mb
758:apoptotic cell clearance
656:acyltransferase activity
32:Tetris: The Grand Master
5148:Tissue transglutaminase
4729:3-propylmalate synthase
4636:Tissue transglutaminase
3653:10.15252/embr.201745067
3485:The Biochemical Journal
3101:10.1074/jbc.M111.287490
3032:The Biochemical Journal
2991:10.1074/jbc.M115.669895
2940:10.1074/jbc.M109.097162
2582:10.1124/jpet.111.187385
2510:The Biochemical Journal
2416:10.1074/jbc.M111.337089
2309:Frontiers in Bioscience
1865:chemotherapy resistance
1728:development as well as
1408:Tissue transglutaminase
833:blood vessel remodeling
559:external carotid artery
493:upper lobe of left lung
5272:Acetylcholine receptor
5125:Pyruvate dehydrogenase
4697:3-ethylmalate synthase
4692:2-ethylmalate synthase
4557:Acyltransferase like 2
4045:Experimental Neurology
3963:10.4331/wjbc.v1.i5.181
3457:10.1074/jbc.M311919200
2849:10.1073/pnas.042454899
2368:10.1006/bbrc.1997.7409
1700:
1685:
1673:
1610:
547:mesenteric lymph nodes
5307:Sp100 nuclear antigen
4918:Eadie–Hofstee diagram
4851:Allosteric regulation
4677:Citrate (Re)-synthase
4672:Decylcitrate synthase
4613:Aminoacyltransferases
4513:palmitoyltransferases
4293:Endomysial antibodies
3708:10.1096/fj.201801544R
3599:10.3390/medsci7020019
2466:10.1083/jcb.148.4.825
2064:endoplasmic reticulum
2060:MHC class I molecules
1778:Clinical significance
1698:
1683:
1670:
1628:transcription factors
1608:
783:peptide cross-linking
727:endoplasmic reticulum
712:extracellular exosome
465:right coronary artery
232:Chromosome 20 (human)
4928:Lineweaver–Burk plot
4739:Homocitrate synthase
4626:Peptidyl transferase
2618:Autoimmunity Reviews
1799:result in a form of
1726:extracellular matrix
1690:extracellular matrix
737:extracellular matrix
651:transferase activity
469:left coronary artery
373:2 H1|2 78.72 cM
334:Chromosome 2 (mouse)
85:List of PDB id codes
58:Available structures
4471:N-acetyltransferase
3262:2011PLoSO...616665C
3044:10.1042/BCJ20170601
2840:2002PNAS...99.2743L
523:conjunctival fornix
473:canal of the cervix
4887:Enzyme superfamily
4820:Enzyme promiscuity
4431:acetyltransferases
4298:2021-05-12 at the
3752:10.1038/nm0797-797
3497:10.1042/BJ20021084
2522:10.1042/BJ20021234
1869:Preclinical trials
1801:gluten sensitivity
1701:
1686:
1674:
1611:
1586:, and producing a
1560:C-terminal domains
1545:of TG2 with bound
1543:Crystal structures
963:ENSMUSG00000037820
751:Biological process
690:Cellular component
644:Molecular function
489:right uterine tube
18:Transglutaminase 2
5325:
5324:
5043:
5042:
4752:
4751:
4702:ATP citrate lyase
4370:
4369:
3491:(Pt 3): 793–803.
3321:10.1002/jnr.24239
2043:
2042:
1957:neurodegenerative
1853:pancreatic cancer
1564:N-terminal domain
1527:transglutaminases
1405:
1404:
1401:
1400:
1304:metabolic pathway
1206:
1205:
1202:
1201:
1175:
1174:
1140:
1139:
1119:
1118:
1077:
1076:
1056:
1055:
1014:
1013:
995:
994:
969:
968:
950:
949:
924:
923:
905:
904:
853:
852:
671:metal ion binding
628:
627:
624:
623:
579:
578:
566:
565:
504:
503:
402:
401:
301:
300:
128:
127:
124:
123:
68:Ortholog search:
16:(Redirected from
5355:
5282:Apolipoprotein H
5135:Transglutaminase
5092:
5085:
5078:
5069:
5060:
5059:
5051:
4923:Hanes–Woolf plot
4866:Enzyme activator
4861:Enzyme inhibitor
4835:Enzyme catalysis
4779:
4772:
4765:
4756:
4712:HMG-CoA synthase
4667:Citrate synthase
4631:Transglutaminase
4408:acyltransferases
4397:
4390:
4383:
4374:
4356:
4336:
4329:
4322:
4313:
4281:
4280:
4262:
4230:
4224:
4223:
4213:
4181:
4172:
4171:
4161:
4129:
4123:
4122:
4085:
4079:
4078:
4068:
4036:
4030:
4029:
3992:
3986:
3985:
3975:
3965:
3941:
3935:
3934:
3916:
3884:
3878:
3877:
3859:
3848:10.1002/mc.22986
3827:
3821:
3820:
3810:
3793:(8): 2005–2008.
3787:Gastroenterology
3783:"Celiac Disease"
3778:
3772:
3771:
3735:
3729:
3728:
3710:
3686:
3675:
3674:
3664:
3631:
3622:
3621:
3611:
3601:
3586:Medical Sciences
3577:
3568:
3567:
3557:
3525:
3519:
3518:
3508:
3476:
3470:
3469:
3459:
3435:
3429:
3428:
3400:
3394:
3393:
3383:
3349:
3343:
3342:
3332:
3315:(7): 1150–1158.
3300:
3294:
3293:
3283:
3273:
3241:
3235:
3234:
3224:
3214:
3190:
3184:
3183:
3173:
3163:
3154:(8): 2640–2649.
3139:
3124:
3123:
3113:
3103:
3094:(43): 37866–73.
3079:
3064:
3063:
3038:(9): 1643–1667.
3029:
3020:
3014:
3013:
3003:
2993:
2984:(35): 21365–75.
2969:
2963:
2962:
2952:
2942:
2918:
2907:
2906:
2878:
2872:
2871:
2861:
2851:
2819:
2813:
2812:
2802:
2792:
2768:
2762:
2761:
2755:
2747:
2721:
2712:
2711:
2704:
2698:
2697:
2679:
2664:The FEBS Journal
2655:
2642:
2641:
2613:
2604:
2603:
2593:
2561:
2544:
2543:
2533:
2516:(Pt 2): 377–96.
2501:
2488:
2487:
2477:
2445:
2439:
2438:
2428:
2418:
2409:(22): 18005–17.
2394:
2388:
2387:
2361:
2341:
2335:
2334:
2324:
2300:
2289:
2288:
2278:
2246:
2225:
2224:
2206:
2191:The FEBS Journal
2182:
2173:
2172:
2161:
2155:
2154:
2143:
2137:
2127:
2116:
2106:
2031:
1991:
1949:cardiac fibrosis
1531:catalytic domain
1445:residue and a γ-
1431:transglutaminase
1410:(abbreviated as
1358:
1208:
1188:
1187:
1159:
1152:
1135:
1123:
1114:
1086:
1082:RefSeq (protein)
1072:
1060:
1051:
1023:
999:
980:
954:
935:
909:
890:
859:
634:
620:
611:
604:
597:
590:
575:
551:efferent ductule
515:
513:Top expressed in
508:
477:popliteal artery
453:
451:Top expressed in
446:
425:
408:
398:
385:
374:
359:
352:
346:
335:
323:
307:
297:
284:
273:
258:
251:
245:
234:
220:
204:
198:
196:TGM2 - orthologs
149:
142:
119:
82:
76:
55:
54:
49:
37:
21:
5363:
5362:
5358:
5357:
5356:
5354:
5353:
5352:
5328:
5327:
5326:
5321:
5260:
5152:
5129:
5101:
5096:
5066:
5054:
5046:
5044:
5039:
4951:Oxidoreductases
4937:
4913:Enzyme kinetics
4901:
4897:List of enzymes
4870:
4839:
4810:Catalytic triad
4788:
4783:
4753:
4748:
4707:Malate synthase
4652:
4603:
4415:
4401:
4371:
4366:
4363:
4357:
4343:
4340:
4300:Wayback Machine
4289:
4284:
4245:(16): 3009–35.
4232:
4231:
4227:
4183:
4182:
4175:
4131:
4130:
4126:
4087:
4086:
4082:
4038:
4037:
4033:
3994:
3993:
3989:
3943:
3942:
3938:
3893:Brain Pathology
3886:
3885:
3881:
3829:
3828:
3824:
3780:
3779:
3775:
3740:Nature Medicine
3737:
3736:
3732:
3688:
3687:
3678:
3633:
3632:
3625:
3579:
3578:
3571:
3527:
3526:
3522:
3478:
3477:
3473:
3450:(23): 23863–8.
3437:
3436:
3432:
3402:
3401:
3397:
3372:
3351:
3350:
3346:
3302:
3301:
3297:
3243:
3242:
3238:
3192:
3191:
3187:
3141:
3140:
3127:
3081:
3080:
3067:
3027:
3022:
3021:
3017:
2971:
2970:
2966:
2933:(33): 25402–9.
2920:
2919:
2910:
2880:
2879:
2875:
2821:
2820:
2816:
2770:
2769:
2765:
2748:
2736:
2723:
2722:
2715:
2706:
2705:
2701:
2670:(23): 7083–96.
2657:
2656:
2645:
2615:
2614:
2607:
2563:
2562:
2547:
2503:
2502:
2491:
2447:
2446:
2442:
2396:
2395:
2391:
2359:10.1.1.483.2738
2343:
2342:
2338:
2302:
2301:
2292:
2248:
2247:
2228:
2197:(24): 4717–39.
2184:
2183:
2176:
2163:
2162:
2158:
2145:
2144:
2140:
2128:
2119:
2107:
2098:
2094:
2073:, including an
2048:
1965:
1941:
1918:
1878:
1857:cervical cancer
1849:prostate cancer
1832:
1789:
1783:knockout mice.
1780:
1761:coeliac disease
1730:differentiation
1706:
1620:signal cascades
1616:
1572:
1523:
1517:(20q11.2-q12).
1515:20th chromosome
1511:
1506:
1197:View/Edit Mouse
1192:View/Edit Human
1155:
1148:
1145:Location (UCSC)
1131:
1110:
1106:
1102:
1098:
1094:
1068:
1047:
1043:
1039:
1035:
1031:
944:ENSG00000198959
837:
746:
732:plasma membrane
685:
666:protein binding
616:
606:
605:
599:
598:
571:
562:
557:
553:
549:
545:
541:
539:right lung lobe
537:
535:ascending aorta
533:
529:
525:
511:
500:
495:
491:
487:
483:
481:tibial arteries
479:
475:
471:
467:
463:
449:
393:
380:
372:
362:
361:
360:
353:
333:
310:Gene location (
292:
279:
271:
261:
260:
259:
252:
230:
207:Gene location (
158:
145:
138:
91:
69:
35:
28:
23:
22:
15:
12:
11:
5:
5361:
5359:
5351:
5350:
5345:
5340:
5330:
5329:
5323:
5322:
5320:
5319:
5314:
5309:
5304:
5299:
5294:
5289:
5284:
5279:
5274:
5268:
5266:
5262:
5261:
5259:
5258:
5253:
5248:
5243:
5238:
5233:
5228:
5223:
5218:
5213:
5208:
5203:
5198:
5193:
5188:
5183:
5178:
5173:
5168:
5162:
5160:
5154:
5153:
5151:
5150:
5145:
5139:
5137:
5131:
5130:
5128:
5127:
5122:
5117:
5111:
5109:
5103:
5102:
5097:
5095:
5094:
5087:
5080:
5072:
5065:
5064:
5041:
5040:
5038:
5037:
5024:
5011:
4998:
4985:
4972:
4959:
4945:
4943:
4939:
4938:
4936:
4935:
4930:
4925:
4920:
4915:
4909:
4907:
4903:
4902:
4900:
4899:
4894:
4889:
4884:
4878:
4876:
4875:Classification
4872:
4871:
4869:
4868:
4863:
4858:
4853:
4847:
4845:
4841:
4840:
4838:
4837:
4832:
4827:
4822:
4817:
4812:
4807:
4802:
4796:
4794:
4790:
4789:
4784:
4782:
4781:
4774:
4767:
4759:
4750:
4749:
4747:
4746:
4741:
4736:
4731:
4726:
4721:
4720:
4719:
4709:
4704:
4699:
4694:
4689:
4684:
4679:
4674:
4669:
4663:
4661:
4654:
4653:
4651:
4650:
4649:
4648:
4643:
4638:
4628:
4623:
4617:
4615:
4605:
4604:
4602:
4601:
4596:
4591:
4586:
4580:
4579:
4574:
4569:
4564:
4559:
4550:
4549:
4548:
4547:
4542:
4532:
4531:
4530:
4525:
4508:
4507:
4506:
4505:
4500:
4490:
4489:
4488:
4483:
4478:
4468:
4463:
4458:
4453:
4448:
4443:
4438:
4426:
4424:
4417:
4416:
4402:
4400:
4399:
4392:
4385:
4377:
4368:
4367:
4365:
4364:
4358:
4351:
4348:
4345:
4344:
4341:
4339:
4338:
4331:
4324:
4316:
4310:
4309:
4302:
4288:
4287:External links
4285:
4283:
4282:
4225:
4173:
4124:
4080:
4031:
3987:
3936:
3879:
3842:(6): 837–853.
3822:
3773:
3746:(7): 797–801.
3730:
3676:
3623:
3569:
3520:
3471:
3430:
3395:
3370:
3344:
3295:
3236:
3185:
3125:
3065:
3015:
2964:
2908:
2873:
2814:
2763:
2734:
2713:
2699:
2643:
2624:(10): 746–53.
2605:
2545:
2489:
2440:
2389:
2336:
2290:
2226:
2174:
2156:
2138:
2117:
2095:
2093:
2090:
2047:
2044:
2041:
2040:
2038:Knockout mouse
2033:
2032:
2024:
2023:
2019:
2018:
2013:
2009:
2008:
2003:
1996:
1995:
1964:
1961:
1940:
1937:
1917:
1914:
1877:
1876:Other Diseases
1874:
1831:
1828:
1793:celiac disease
1788:
1787:Celiac Disease
1785:
1779:
1776:
1705:
1702:
1615:
1612:
1571:
1568:
1522:
1519:
1510:
1507:
1505:
1502:
1490:celiac disease
1403:
1402:
1399:
1398:
1393:
1387:
1386:
1381:
1375:
1374:
1369:
1363:
1362:
1354:
1353:
1344:
1338:
1337:
1326:
1319:
1318:
1313:
1307:
1306:
1301:
1295:
1294:
1289:
1283:
1282:
1277:
1271:
1270:
1265:
1259:
1258:
1253:
1247:
1246:
1242:
1241:
1236:
1230:
1229:
1224:
1218:
1217:
1213:
1212:
1204:
1203:
1200:
1199:
1194:
1184:
1183:
1177:
1176:
1173:
1172:
1170:
1168:
1161:
1160:
1153:
1146:
1142:
1141:
1138:
1137:
1127:
1126:
1120:
1117:
1116:
1090:
1089:
1083:
1079:
1078:
1075:
1074:
1064:
1063:
1057:
1054:
1053:
1027:
1026:
1020:
1016:
1015:
1012:
1011:
1003:
1002:
996:
993:
992:
984:
983:
977:
971:
970:
967:
966:
958:
957:
951:
948:
947:
939:
938:
932:
926:
925:
922:
921:
913:
912:
906:
903:
902:
894:
893:
887:
881:
880:
875:
870:
866:
865:
855:
854:
851:
850:
839:
838:
836:
835:
830:
825:
820:
815:
810:
805:
800:
795:
790:
785:
780:
775:
770:
765:
760:
754:
752:
748:
747:
745:
744:
739:
734:
729:
724:
719:
717:focal adhesion
714:
709:
704:
699:
693:
691:
687:
686:
684:
683:
678:
673:
668:
663:
658:
653:
647:
645:
641:
640:
630:
629:
626:
625:
622:
621:
613:
612:
587:
581:
580:
577:
576:
568:
567:
564:
563:
561:
560:
556:
552:
548:
544:
540:
536:
532:
528:
524:
520:
517:
516:
505:
502:
501:
499:
498:
494:
490:
486:
482:
478:
474:
470:
466:
462:
458:
455:
454:
442:
441:
433:
422:
416:
415:
412:RNA expression
404:
403:
400:
399:
391:
387:
386:
378:
375:
370:
364:
363:
354:
347:
341:
337:
336:
331:
325:
324:
316:
315:
303:
302:
299:
298:
290:
286:
285:
277:
274:
269:
263:
262:
253:
246:
240:
236:
235:
228:
222:
221:
213:
212:
200:
199:
156:
152:
151:
143:
135:
134:
130:
129:
126:
125:
122:
121:
87:
86:
78:
77:
66:
60:
59:
51:
50:
42:
41:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
5360:
5349:
5346:
5344:
5341:
5339:
5336:
5335:
5333:
5318:
5317:Topoisomerase
5315:
5313:
5310:
5308:
5305:
5303:
5300:
5298:
5295:
5293:
5290:
5288:
5285:
5283:
5280:
5278:
5275:
5273:
5270:
5269:
5267:
5263:
5257:
5254:
5252:
5249:
5247:
5244:
5242:
5239:
5237:
5234:
5232:
5229:
5227:
5224:
5222:
5219:
5217:
5214:
5212:
5209:
5207:
5204:
5202:
5199:
5197:
5194:
5192:
5189:
5187:
5184:
5182:
5179:
5177:
5174:
5172:
5169:
5167:
5164:
5163:
5161:
5159:
5155:
5149:
5146:
5144:
5141:
5140:
5138:
5136:
5132:
5126:
5123:
5121:
5118:
5116:
5113:
5112:
5110:
5108:
5107:Dehydrogenase
5104:
5100:
5093:
5088:
5086:
5081:
5079:
5074:
5073:
5070:
5063:
5058:
5053:
5049:
5035:
5031:
5030:
5025:
5022:
5018:
5017:
5012:
5009:
5005:
5004:
4999:
4996:
4992:
4991:
4986:
4983:
4979:
4978:
4973:
4970:
4966:
4965:
4960:
4957:
4953:
4952:
4947:
4946:
4944:
4940:
4934:
4931:
4929:
4926:
4924:
4921:
4919:
4916:
4914:
4911:
4910:
4908:
4904:
4898:
4895:
4893:
4892:Enzyme family
4890:
4888:
4885:
4883:
4880:
4879:
4877:
4873:
4867:
4864:
4862:
4859:
4857:
4856:Cooperativity
4854:
4852:
4849:
4848:
4846:
4842:
4836:
4833:
4831:
4828:
4826:
4823:
4821:
4818:
4816:
4815:Oxyanion hole
4813:
4811:
4808:
4806:
4803:
4801:
4798:
4797:
4795:
4791:
4787:
4780:
4775:
4773:
4768:
4766:
4761:
4760:
4757:
4745:
4742:
4740:
4737:
4735:
4732:
4730:
4727:
4725:
4722:
4718:
4715:
4714:
4713:
4710:
4708:
4705:
4703:
4700:
4698:
4695:
4693:
4690:
4688:
4685:
4683:
4680:
4678:
4675:
4673:
4670:
4668:
4665:
4664:
4662:
4659:
4655:
4647:
4644:
4642:
4639:
4637:
4634:
4633:
4632:
4629:
4627:
4624:
4622:
4619:
4618:
4616:
4614:
4610:
4606:
4600:
4597:
4595:
4592:
4590:
4587:
4585:
4582:
4581:
4578:
4575:
4573:
4570:
4568:
4565:
4563:
4560:
4558:
4555:
4552:
4551:
4546:
4543:
4541:
4538:
4537:
4536:
4533:
4529:
4526:
4524:
4521:
4520:
4519:
4516:
4514:
4510:
4509:
4504:
4501:
4499:
4496:
4495:
4494:
4491:
4487:
4484:
4482:
4479:
4477:
4474:
4473:
4472:
4469:
4467:
4464:
4462:
4459:
4457:
4454:
4452:
4449:
4447:
4444:
4442:
4439:
4437:
4434:
4432:
4428:
4427:
4425:
4422:
4418:
4413:
4409:
4405:
4398:
4393:
4391:
4386:
4384:
4379:
4378:
4375:
4361:
4355:
4350:
4346:
4337:
4332:
4330:
4325:
4323:
4318:
4317:
4314:
4307:
4303:
4301:
4297:
4294:
4291:
4290:
4286:
4278:
4274:
4270:
4266:
4261:
4256:
4252:
4248:
4244:
4240:
4236:
4229:
4226:
4221:
4217:
4212:
4207:
4203:
4199:
4195:
4191:
4187:
4180:
4178:
4174:
4169:
4165:
4160:
4155:
4151:
4147:
4143:
4139:
4135:
4128:
4125:
4120:
4116:
4112:
4108:
4104:
4100:
4097:(5): 1253–7.
4096:
4092:
4084:
4081:
4076:
4072:
4067:
4062:
4058:
4054:
4050:
4046:
4042:
4035:
4032:
4027:
4023:
4019:
4015:
4011:
4007:
4003:
3999:
3991:
3988:
3983:
3979:
3974:
3969:
3964:
3959:
3955:
3951:
3947:
3940:
3937:
3932:
3928:
3924:
3920:
3915:
3910:
3906:
3902:
3898:
3894:
3890:
3883:
3880:
3875:
3871:
3867:
3863:
3858:
3853:
3849:
3845:
3841:
3837:
3833:
3826:
3823:
3818:
3814:
3809:
3804:
3800:
3796:
3792:
3788:
3784:
3777:
3774:
3769:
3765:
3761:
3757:
3753:
3749:
3745:
3741:
3734:
3731:
3726:
3722:
3718:
3714:
3709:
3704:
3700:
3696:
3695:FASEB Journal
3692:
3685:
3683:
3681:
3677:
3672:
3668:
3663:
3658:
3654:
3650:
3647:(7): e45067.
3646:
3642:
3638:
3630:
3628:
3624:
3619:
3615:
3610:
3605:
3600:
3595:
3591:
3587:
3583:
3576:
3574:
3570:
3565:
3561:
3556:
3551:
3547:
3543:
3539:
3535:
3531:
3524:
3521:
3516:
3512:
3507:
3502:
3498:
3494:
3490:
3486:
3482:
3475:
3472:
3467:
3463:
3458:
3453:
3449:
3445:
3441:
3434:
3431:
3426:
3422:
3418:
3414:
3411:(10): 534–9.
3410:
3406:
3399:
3396:
3391:
3387:
3382:
3377:
3373:
3371:9780123943057
3367:
3363:
3359:
3355:
3348:
3345:
3340:
3336:
3331:
3326:
3322:
3318:
3314:
3310:
3306:
3299:
3296:
3291:
3287:
3282:
3277:
3272:
3267:
3263:
3259:
3256:(1): e16665.
3255:
3251:
3247:
3240:
3237:
3232:
3228:
3223:
3218:
3213:
3208:
3204:
3200:
3196:
3189:
3186:
3181:
3177:
3172:
3167:
3162:
3157:
3153:
3149:
3145:
3138:
3136:
3134:
3132:
3130:
3126:
3121:
3117:
3112:
3107:
3102:
3097:
3093:
3089:
3085:
3078:
3076:
3074:
3072:
3070:
3066:
3061:
3057:
3053:
3052:11392/2388638
3049:
3045:
3041:
3037:
3033:
3026:
3019:
3016:
3011:
3007:
3002:
2997:
2992:
2987:
2983:
2979:
2975:
2968:
2965:
2960:
2956:
2951:
2946:
2941:
2936:
2932:
2928:
2924:
2917:
2915:
2913:
2909:
2904:
2900:
2896:
2892:
2888:
2884:
2877:
2874:
2869:
2865:
2860:
2855:
2850:
2845:
2841:
2837:
2834:(5): 2743–7.
2833:
2829:
2825:
2818:
2815:
2810:
2806:
2801:
2796:
2791:
2786:
2782:
2778:
2774:
2767:
2764:
2759:
2753:
2745:
2741:
2737:
2735:9784431558255
2731:
2727:
2720:
2718:
2714:
2709:
2703:
2700:
2695:
2691:
2687:
2683:
2678:
2673:
2669:
2665:
2661:
2654:
2652:
2650:
2648:
2644:
2639:
2635:
2631:
2627:
2623:
2619:
2612:
2610:
2606:
2601:
2597:
2592:
2587:
2583:
2579:
2576:(1): 104–14.
2575:
2571:
2567:
2560:
2558:
2556:
2554:
2552:
2550:
2546:
2541:
2537:
2532:
2527:
2523:
2519:
2515:
2511:
2507:
2500:
2498:
2496:
2494:
2490:
2485:
2481:
2476:
2471:
2467:
2463:
2460:(4): 825–38.
2459:
2455:
2451:
2444:
2441:
2436:
2432:
2427:
2422:
2417:
2412:
2408:
2404:
2400:
2393:
2390:
2385:
2381:
2377:
2373:
2369:
2365:
2360:
2355:
2352:(2): 357–66.
2351:
2347:
2340:
2337:
2332:
2328:
2323:
2318:
2314:
2310:
2306:
2299:
2297:
2295:
2291:
2286:
2282:
2277:
2272:
2268:
2264:
2261:(4): 513–22.
2260:
2256:
2252:
2245:
2243:
2241:
2239:
2237:
2235:
2233:
2231:
2227:
2222:
2218:
2214:
2210:
2205:
2200:
2196:
2192:
2188:
2181:
2179:
2175:
2170:
2166:
2160:
2157:
2152:
2148:
2142:
2139:
2135:
2131:
2126:
2124:
2122:
2118:
2114:
2110:
2105:
2103:
2101:
2097:
2091:
2089:
2085:
2083:
2079:
2076:
2072:
2067:
2065:
2061:
2057:
2053:
2045:
2039:
2034:
2030:
2025:
2020:
2017:
2014:
2011:
2010:
2007:
2004:
2002:
1998:
1997:
1992:
1989:
1986:
1982:
1978:
1974:
1970:
1962:
1960:
1958:
1954:
1950:
1946:
1938:
1936:
1934:
1930:
1926:
1923:for anti-tTG
1922:
1915:
1913:
1911:
1907:
1903:
1899:
1894:
1891:
1887:
1883:
1875:
1873:
1870:
1866:
1862:
1858:
1854:
1850:
1846:
1845:breast cancer
1842:
1837:
1829:
1827:
1825:
1821:
1817:
1813:
1809:
1807:
1802:
1798:
1794:
1786:
1784:
1777:
1775:
1773:
1769:
1764:
1762:
1757:
1754:tTG also has
1752:
1750:
1745:
1743:
1739:
1735:
1734:cell adhesion
1731:
1727:
1723:
1719:
1718:wound healing
1715:
1714:retinoic acid
1711:
1703:
1697:
1693:
1691:
1682:
1678:
1669:
1665:
1663:
1658:
1654:
1649:
1646:
1641:
1637:
1631:
1629:
1625:
1621:
1613:
1607:
1603:
1601:
1600:glutamic acid
1597:
1593:
1589:
1585:
1581:
1577:
1569:
1567:
1565:
1561:
1556:
1552:
1548:
1544:
1540:
1539:beta-sandwich
1536:
1532:
1528:
1520:
1518:
1516:
1508:
1503:
1501:
1499:
1495:
1491:
1487:
1482:
1480:
1476:
1472:
1468:
1464:
1460:
1456:
1452:
1448:
1444:
1440:
1437:between an ε-
1436:
1432:
1428:
1425:
1421:
1417:
1413:
1409:
1397:
1394:
1392:
1388:
1385:
1382:
1380:
1376:
1373:
1370:
1368:
1364:
1359:
1355:
1352:
1348:
1345:
1343:
1342:Gene Ontology
1339:
1336:
1333:
1330:
1327:
1324:
1320:
1317:
1314:
1312:
1308:
1305:
1302:
1300:
1296:
1293:
1290:
1288:
1284:
1281:
1280:NiceZyme view
1278:
1276:
1272:
1269:
1266:
1264:
1260:
1257:
1254:
1252:
1248:
1243:
1240:
1237:
1235:
1231:
1228:
1225:
1223:
1219:
1214:
1209:
1198:
1193:
1189:
1185:
1182:
1178:
1171:
1169:
1166:
1162:
1158:
1154:
1151:
1147:
1143:
1136:
1134:
1128:
1124:
1121:
1115:
1113:
1109:
1105:
1101:
1097:
1091:
1087:
1084:
1080:
1073:
1071:
1065:
1061:
1058:
1052:
1050:
1046:
1042:
1038:
1034:
1028:
1024:
1021:
1019:RefSeq (mRNA)
1017:
1010:
1009:
1004:
1000:
997:
991:
990:
985:
981:
978:
976:
972:
965:
964:
959:
955:
952:
946:
945:
940:
936:
933:
931:
927:
920:
919:
914:
910:
907:
901:
900:
895:
891:
888:
886:
882:
879:
876:
874:
871:
867:
864:
860:
856:
849:
845:
840:
834:
831:
829:
826:
824:
821:
819:
816:
814:
811:
809:
806:
804:
801:
799:
796:
794:
791:
789:
786:
784:
781:
779:
776:
774:
771:
769:
766:
764:
761:
759:
756:
755:
753:
750:
749:
743:
740:
738:
735:
733:
730:
728:
725:
723:
722:mitochondrion
720:
718:
715:
713:
710:
708:
705:
703:
700:
698:
695:
694:
692:
689:
688:
682:
679:
677:
674:
672:
669:
667:
664:
662:
659:
657:
654:
652:
649:
648:
646:
643:
642:
639:
638:Gene ontology
635:
631:
619:
614:
610:
603:
596:
591:
588:
586:
582:
574:
569:
558:
554:
550:
546:
542:
538:
534:
530:
526:
522:
521:
518:
514:
509:
506:
496:
492:
488:
485:right auricle
484:
480:
476:
472:
468:
464:
460:
459:
456:
452:
447:
444:
443:
440:
438:
434:
432:
431:
427:
426:
423:
421:
417:
413:
409:
405:
397:
392:
388:
384:
379:
369:
365:
358:
351:
345:
338:
330:
326:
322:
317:
313:
308:
304:
296:
291:
287:
283:
278:
268:
264:
257:
250:
244:
237:
233:
227:
223:
219:
214:
210:
205:
201:
197:
193:
189:
185:
181:
177:
173:
169:
165:
161:
153:
148:
141:
136:
131:
120:
118:
114:
110:
106:
102:
98:
94:
88:
83:
80:
79:
75:
72:
65:
61:
56:
52:
48:
43:
38:
33:
19:
5348:Autoantigens
5302:Gangliosides
5158:Nucleoporins
5147:
5099:Autoantigens
5029:Translocases
5026:
5013:
5000:
4987:
4974:
4964:Transferases
4961:
4948:
4805:Binding site
4635:
4553:
4511:
4429:
4404:Transferases
4359:
4242:
4238:
4228:
4193:
4189:
4141:
4137:
4127:
4094:
4090:
4083:
4051:(1): 78–89.
4048:
4044:
4034:
4004:(1): 111–8.
4001:
3997:
3990:
3956:(5): 181–7.
3953:
3949:
3939:
3899:(2): 130–9.
3896:
3892:
3882:
3839:
3835:
3825:
3790:
3786:
3776:
3743:
3739:
3733:
3698:
3694:
3644:
3641:EMBO Reports
3640:
3589:
3585:
3540:(3): 550–8.
3537:
3533:
3523:
3488:
3484:
3474:
3447:
3443:
3433:
3408:
3404:
3398:
3353:
3347:
3312:
3308:
3298:
3253:
3249:
3239:
3205:(12): e327.
3202:
3199:PLOS Biology
3198:
3188:
3151:
3147:
3091:
3087:
3035:
3031:
3018:
2981:
2977:
2967:
2930:
2926:
2889:(2): 190–5.
2886:
2882:
2876:
2831:
2827:
2817:
2783:(12): e327.
2780:
2777:PLOS Biology
2776:
2766:
2725:
2702:
2667:
2663:
2621:
2617:
2573:
2569:
2513:
2509:
2457:
2453:
2443:
2406:
2402:
2392:
2349:
2345:
2339:
2322:10.2741/1921
2312:
2308:
2258:
2254:
2194:
2190:
2168:
2159:
2150:
2141:
2086:
2068:
2049:
1969:interactions
1966:
1963:Interactions
1942:
1919:
1895:
1879:
1836:inflammation
1833:
1805:
1790:
1781:
1768:proteostasis
1765:
1753:
1746:
1707:
1687:
1675:
1632:
1617:
1573:
1524:
1512:
1483:
1471:mitochondria
1415:
1411:
1407:
1406:
1268:BRENDA entry
1130:
1104:NP_001310247
1100:NP_001310246
1096:NP_001310245
1093:
1067:
1049:NM_001323318
1045:NM_001323317
1041:NM_001323316
1030:
1006:
987:
961:
942:
916:
897:
877:
872:
555:carotid body
543:ciliary body
435:
428:
394:157,988,356
381:157,958,322
155:External IDs
90:
5287:Cardiolipin
4800:Active site
4646:Factor XIII
4342:PDB gallery
4196:(1): 5–13.
4190:BMB Reports
3998:Amino Acids
3701:(1): 3–12.
2315:: 1758–73.
1939:Therapeutic
1933:specificity
1929:sensitivity
1662:Thioredoxin
1648:nucleotides
1535:beta barrel
1486:autoantigen
1484:tTG is the
1479:fibronectin
1459:deamidation
1455:proteolysis
1447:carboxamide
1441:group of a
1256:IntEnz view
1239:80146-85-6
1216:Identifiers
676:GTP binding
527:Paneth cell
293:38,166,578
280:38,127,385
133:Identifiers
5332:Categories
5292:Centromere
5003:Isomerases
4977:Hydrolases
4844:Regulation
4144:(6): 613.
2136:, May 2017
2115:, May 2017
2092:References
2075:allosteric
1985:Substrates
1975:donor and
1945:metastasis
1931:(99%) and
1925:antibodies
1916:Diagnostic
1912:activity.
1890:Huntington
1861:metastasis
1742:astrocytes
1614:Regulation
1596:isopeptide
1325:structures
1292:KEGG entry
439:(ortholog)
176:HomoloGene
4882:EC number
3592:(2): 19.
2752:cite book
2744:937392418
2728:. Tokyo.
2354:CiteSeerX
2078:disulfide
2052:chaperone
1973:glutamine
1953:inhibitor
1886:Parkinson
1882:Alzheimer
1816:glutamine
1806:Triticeae
1722:apoptosis
1653:disulfide
1588:thioester
1580:glutamine
1570:Mechanism
1504:Structure
1475:apoptosis
1451:glutamine
1449:group of
1245:Databases
1133:NP_033399
1112:NP_945189
1108:NP_004604
1070:NM_009373
1037:NM_198951
1033:NM_004613
863:Orthologs
697:cytoplasm
461:beta cell
184:GeneCards
5343:EC 2.3.2
5312:Thrombin
4906:Kinetics
4830:Cofactor
4793:Activity
4498:P300/CBP
4306:TRANSDAB
4296:Archived
4277:14849506
4269:25943306
4260:11113818
4220:29187283
4168:29795262
4119:11018740
4111:10811336
4075:22698685
4026:16143202
4018:21938398
3982:21541002
3923:20731657
3874:59341070
3866:30693974
3817:29550590
3768:20033968
3725:58551851
3717:30593123
3671:29752334
3618:30691081
3564:17100777
3515:12737632
3466:15069073
3425:12368090
3390:22364871
3339:29570839
3290:21304968
3250:PLOS ONE
3231:18092889
3180:29305423
3120:21908620
3060:29764956
3010:26160175
2959:20547769
2903:20450932
2868:11867708
2809:18092889
2694:21883387
2686:19878304
2638:22326684
2600:22228808
2540:12366374
2484:10684262
2435:22442151
2384:11242870
2331:16368554
2285:22437759
2221:19217277
2213:21902809
2132:–
2111:–
1921:Serology
1910:TGF beta
1904:and the
1898:fibrosis
1841:leukemia
1749:spectrin
1704:Function
1657:cysteine
1469:and the
1435:proteins
1427:2.3.2.13
1396:proteins
1384:articles
1372:articles
1329:RCSB PDB
1227:2.3.2.13
1181:Wikidata
842:Sources:
272:20q11.23
5062:Biology
5016:Ligases
4786:Enzymes
4211:5796628
4159:5966415
4066:3418489
3973:3083958
3914:8094245
3857:7754084
3808:6203336
3760:9212111
3662:6030705
3609:6409630
3555:1810403
3506:1223550
3381:3746560
3330:5980740
3281:3031627
3258:Bibcode
3222:2140088
3171:5827427
3111:3199528
3001:4571865
2950:2919103
2836:Bibcode
2800:2140088
2591:3310700
2531:1223021
2475:2169362
2426:3365763
2376:9344835
2276:3712867
2134:Ensembl
2113:Ensembl
2062:in the
2056:peptide
1981:calcium
1824:T cells
1808:glutens
1738:neurons
1710:calcium
1688:In the
1672:enzyme.
1645:guanine
1624:nucleus
1584:ammonia
1521:Protein
1498:villous
1467:nucleus
1463:cytosol
1351:QuickGO
1316:profile
1299:MetaCyc
1234:CAS no.
975:UniProt
930:Ensembl
869:Species
848:QuickGO
707:cytosol
497:decidua
414:pattern
140:Aliases
5256:NUP214
5251:NUP205
5246:NUP210
5241:NUP188
5236:NUP160
5231:NUP155
5226:NUP153
5221:NUP133
5216:NUP107
5048:Portal
4990:Lyases
4717:HMGCS2
4554:other:
4545:SPTLC2
4540:SPTLC1
4481:HGSNAT
4275:
4267:
4257:
4218:
4208:
4166:
4156:
4117:
4109:
4073:
4063:
4024:
4016:
3980:
3970:
3931:586174
3929:
3921:
3911:
3872:
3864:
3854:
3815:
3805:
3766:
3758:
3723:
3715:
3669:
3659:
3616:
3606:
3562:
3552:
3513:
3503:
3464:
3423:
3388:
3378:
3368:
3337:
3327:
3288:
3278:
3229:
3219:
3178:
3168:
3118:
3108:
3058:
3008:
2998:
2957:
2947:
2901:
2866:
2859:122418
2856:
2807:
2797:
2742:
2732:
2692:
2684:
2636:
2598:
2588:
2538:
2528:
2482:
2472:
2433:
2423:
2382:
2374:
2356:
2329:
2283:
2273:
2219:
2211:
2082:enzyme
1977:lysine
1906:kidney
1830:Cancer
1820:gluten
1812:B-cell
1756:GTPase
1724:, and
1592:lysine
1537:and a
1533:, two
1494:gluten
1443:lysine
1420:enzyme
1379:PubMed
1361:Search
1347:AmiGO
1335:PDBsum
1275:ExPASy
1263:BRENDA
1251:IntEnz
1222:EC no.
1167:search
1165:PubMed
1008:P21981
989:P21980
885:Entrez
585:BioGPS
164:190196
5277:Actin
5265:Other
5211:NUP98
5206:NUP93
5201:NUP88
5196:NUP85
5191:NUP62
5186:NUP54
5181:NUP50
5176:NUP43
5171:NUP37
5166:NUP35
4942:Types
4658:2.3.3
4609:2.3.2
4599:ABHD5
4486:ARD1A
4421:2.3.1
4273:S2CID
4115:S2CID
4022:S2CID
3927:S2CID
3870:S2CID
3764:S2CID
3721:S2CID
3028:(PDF)
2690:S2CID
2380:S2CID
2217:S2CID
2058:onto
2046:Erp57
1576:thiol
1553:, or
1439:amino
1311:PRIAM
918:21817
878:Mouse
873:Human
844:Amigo
437:Mouse
430:Human
377:Start
312:Mouse
276:Start
209:Human
172:98731
5034:list
5027:EC7
5021:list
5014:EC6
5008:list
5001:EC5
4995:list
4988:EC4
4982:list
4975:EC3
4969:list
4962:EC2
4956:list
4949:EC1
4528:CPT2
4523:CPT1
4503:NAT2
4414:2.3)
4360:1kv3
4265:PMID
4216:PMID
4164:PMID
4107:PMID
4071:PMID
4014:PMID
3978:PMID
3919:PMID
3862:PMID
3813:PMID
3756:PMID
3713:PMID
3667:PMID
3614:PMID
3560:PMID
3511:PMID
3462:PMID
3421:PMID
3386:PMID
3366:ISBN
3335:PMID
3286:PMID
3227:PMID
3176:PMID
3116:PMID
3056:PMID
3006:PMID
2955:PMID
2899:PMID
2864:PMID
2805:PMID
2758:link
2740:OCLC
2730:ISBN
2682:PMID
2634:PMID
2596:PMID
2536:PMID
2480:PMID
2431:PMID
2372:PMID
2327:PMID
2281:PMID
2209:PMID
2016:Tgm2
2006:Tgm2
1902:lung
1888:and
1855:and
1772:HSF1
1732:and
1509:Gene
1391:NCBI
1332:PDBe
1287:KEGG
899:7052
420:Bgee
368:Band
329:Chr.
267:Band
226:Chr.
188:TGM2
180:3391
160:OMIM
147:TGM2
117:3S3S
113:3S3P
109:3S3J
105:3LY6
101:2Q3Z
97:1KV3
93:4PYG
74:RCSB
71:PDBe
40:TGM2
4255:PMC
4247:doi
4206:PMC
4198:doi
4154:PMC
4146:doi
4099:doi
4061:PMC
4053:doi
4049:237
4006:doi
3968:PMC
3958:doi
3909:PMC
3901:doi
3852:PMC
3844:doi
3803:PMC
3795:doi
3791:154
3748:doi
3703:doi
3657:PMC
3649:doi
3604:PMC
3594:doi
3550:PMC
3542:doi
3538:146
3501:PMC
3493:doi
3489:373
3452:doi
3448:279
3413:doi
3376:PMC
3358:doi
3325:PMC
3317:doi
3276:PMC
3266:doi
3217:PMC
3207:doi
3166:PMC
3156:doi
3152:293
3106:PMC
3096:doi
3092:286
3048:hdl
3040:doi
3036:475
2996:PMC
2986:doi
2982:290
2945:PMC
2935:doi
2931:285
2891:doi
2854:PMC
2844:doi
2795:PMC
2785:doi
2672:doi
2668:276
2626:doi
2586:PMC
2578:doi
2574:341
2526:PMC
2518:doi
2514:368
2470:PMC
2462:doi
2458:148
2421:PMC
2411:doi
2407:287
2364:doi
2350:239
2317:doi
2271:PMC
2263:doi
2199:doi
2195:278
2001:MGI
1640:GDP
1638:or
1636:GTP
1555:ATP
1551:GTP
1547:GDP
1488:in
1416:TG2
1414:or
1412:tTG
1367:PMC
1323:PDB
390:End
289:End
192:OMA
168:MGI
64:PDB
5334::
4611::
4412:EC
4406::
4271:.
4263:.
4253:.
4243:72
4241:.
4237:.
4214:.
4204:.
4194:51
4192:.
4188:.
4176:^
4162:.
4152:.
4140:.
4136:.
4113:.
4105:.
4095:95
4093:.
4069:.
4059:.
4047:.
4043:.
4020:.
4012:.
4002:44
4000:.
3976:.
3966:.
3952:.
3948:.
3925:.
3917:.
3907:.
3897:21
3895:.
3891:.
3868:.
3860:.
3850:.
3840:58
3838:.
3834:.
3811:.
3801:.
3789:.
3785:.
3762:.
3754:.
3742:.
3719:.
3711:.
3699:33
3697:.
3693:.
3679:^
3665:.
3655:.
3645:19
3643:.
3639:.
3626:^
3612:.
3602:.
3588:.
3584:.
3572:^
3558:.
3548:.
3536:.
3532:.
3509:.
3499:.
3487:.
3483:.
3460:.
3446:.
3442:.
3419:.
3409:27
3407:.
3384:.
3374:.
3364:.
3333:.
3323:.
3313:96
3311:.
3307:.
3284:.
3274:.
3264:.
3252:.
3248:.
3225:.
3215:.
3201:.
3197:.
3174:.
3164:.
3150:.
3146:.
3128:^
3114:.
3104:.
3090:.
3086:.
3068:^
3054:.
3046:.
3034:.
3030:.
3004:.
2994:.
2980:.
2976:.
2953:.
2943:.
2929:.
2925:.
2911:^
2897:.
2887:47
2885:.
2862:.
2852:.
2842:.
2832:99
2830:.
2826:.
2803:.
2793:.
2779:.
2775:.
2754:}}
2750:{{
2738:.
2716:^
2688:.
2680:.
2666:.
2662:.
2646:^
2632:.
2622:11
2620:.
2608:^
2594:.
2584:.
2572:.
2568:.
2548:^
2534:.
2524:.
2512:.
2508:.
2492:^
2478:.
2468:.
2456:.
2452:.
2429:.
2419:.
2405:.
2401:.
2378:.
2370:.
2362:.
2348:.
2325:.
2313:11
2311:.
2307:.
2293:^
2279:.
2269:.
2259:34
2257:.
2253:.
2229:^
2215:.
2207:.
2193:.
2189:.
2177:^
2167:.
2149:.
2120:^
2099:^
2066:.
1983:.
1884:,
1863:,
1851:,
1847:,
1843:,
1763:.
1720:,
1630:.
1549:,
1541:.
1424:EC
1349:/
846:/
396:bp
383:bp
295:bp
282:bp
190:;
186::
182:;
178::
174:;
170::
166:;
162::
115:,
111:,
107:,
103:,
99:,
95:,
5091:e
5084:t
5077:v
5050::
5036:)
5032:(
5023:)
5019:(
5010:)
5006:(
4997:)
4993:(
4984:)
4980:(
4971:)
4967:(
4958:)
4954:(
4778:e
4771:t
4764:v
4515::
4433::
4410:(
4396:e
4389:t
4382:v
4335:e
4328:t
4321:v
4279:.
4249::
4222:.
4200::
4170:.
4148::
4142:9
4121:.
4101::
4077:.
4055::
4028:.
4008::
3984:.
3960::
3954:1
3933:.
3903::
3876:.
3846::
3819:.
3797::
3770:.
3750::
3744:3
3727:.
3705::
3673:.
3651::
3620:.
3596::
3590:7
3566:.
3544::
3517:.
3495::
3468:.
3454::
3427:.
3415::
3392:.
3360::
3341:.
3319::
3292:.
3268::
3260::
3254:6
3233:.
3209::
3203:5
3182:.
3158::
3122:.
3098::
3062:.
3050::
3042::
3012:.
2988::
2961:.
2937::
2905:.
2893::
2870:.
2846::
2838::
2811:.
2787::
2781:5
2760:)
2746:.
2710:.
2696:.
2674::
2640:.
2628::
2602:.
2580::
2542:.
2520::
2486:.
2464::
2437:.
2413::
2386:.
2366::
2333:.
2319::
2287:.
2265::
2223:.
2201::
2171:.
2153:.
1422:(
314:)
211:)
194::
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.