1639:
1580:
1746:
1806:
1429:
1351:
1687:
1527:
1465:
1387:
336:
40:
473:
1446:
1368:
1614:
1550:
1508:
1404:
1710:
1482:
1781:
1670:
2765:
1326:
523:
1829:
1638:
1579:
736:
The structure of triose phosphate isomerase contributes to its function. Besides the precisely placed glutamate and histidine residues to form the enediol, a ten- or eleven-amino acid chain of TPI acts as a loop to stabilize the intermediate. The loop, formed by residues 166 to 176, closes and
760:
Studies suggest that a lysine close to the active site (at position 12) is also crucial for enzyme function. The lysine, protonated at physiological pH, may help neutralize the negative charge of the phosphate group. When this lysine residue is replaced with a neutral amino acid, TPI loses all
490:
histidine 95 residue donates a proton to form the enediol intermediate. When deprotonated, the enediolate then collapses and, abstracting a proton from protonated glutamate 165, forms the GAP product. Catalysis of the reverse reaction proceeds analogously, forming the same enediol but with
1745:
494:
TPI is diffusion-limited. In terms of thermodynamics, DHAP formation is favored 20:1 over GAP production. However, in glycolysis, the use of GAP in the subsequent steps of metabolism drives the reaction toward its production. TPI is inhibited by
752:
and inorganic phosphate. The hydrogen bond between the enzyme and the phosphate group of the substrate makes such decomposition stereoelectronically unfavorable. Methylglyoxal is a toxin and, if formed, is removed through the
1805:
1428:
1350:
972:
Nickbarg EB, Davenport RC, Petsko GA, Knowles JR (August 1988). "Triosephosphate isomerase: removal of a putatively electrophilic histidine residue results in a subtle change in catalytic mechanism".
448:
Triose phosphate isomerase is a highly efficient enzyme, performing the reaction billions of times faster than it would occur naturally in solution. The reaction is so efficient that it is said to be
2212:
1158:
Lolis E, Petsko GA (July 1990). "Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis".
709:
on the inside. In the illustration, the ribbon backbone of each subunit is colored in blue to red from N-terminus to C-terminus. This structural motif is called an αβ-barrel, or a
2374:
1686:
1526:
1464:
1386:
657:
205:
1007:
Komives EA, Chang LC, Lolis E, Tilton RF, Petsko GA, Knowles JR (March 1991). "Electrophilic catalysis in triosephosphate isomerase: the role of histidine-95".
224:
2205:
1863:
1193:
Creighton DJ, Hamilton DS (March 2001). "Brief history of glyoxalase I and what we have learned about metal ion-dependent, enzyme-catalyzed isomerizations".
321:
1228:
Lodi PJ, Chang LC, Knowles JR, Komives EA (March 1994). "Triosephosphate isomerase requires a positively charged active site: the role of lysine-12".
1997:
1558:
2329:
2198:
406:
and is essential for efficient energy production. TPI has been found in nearly every organism searched for the enzyme, including animals such as
311:
748:
In addition to making the reaction kinetically feasible, the TPI loop sequesters the reactive enediol intermediate to prevent decomposition to
775:
605:
434:
1293:
468:. The relative free energy of each ground state and transition state has been determined experimentally, and is displayed in the figure.
1119:"Kinetic properties of triose-phosphate isomerase from Trypanosoma brucei brucei. A comparison with the rabbit muscle and yeast enzymes"
757:. The loss of a high-energy phosphate bond and the substrate for the rest of glycolysis makes formation of methylglyoxal inefficient.
478:
The structure of TPI facilitates the conversion between dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). The
2484:
2062:
2369:
1856:
472:
217:
2379:
2354:
2334:
2389:
1966:
1454:
144:
2640:
677:
168:
1082:
Harris TK, Cole RN, Comer FI, Mildvan AS (November 1998). "Proton transfer in the mechanism of triosephosphate isomerase".
2339:
1931:
1841:
1376:
335:
2755:
2305:
2259:
2790:
2408:
2344:
2264:
1952:
1849:
861:
Albery WJ, Knowles JR (December 1976). "Free-energy profile of the reaction catalyzed by triosephosphate isomerase".
39:
2295:
2179:
1539:
1500:
1474:
357:
302:
293:
288:
279:
275:
2625:
2741:
2728:
2715:
2702:
2689:
2676:
2663:
2432:
2423:
2399:
2359:
2315:
2279:
2238:
1718:
1438:
2635:
2589:
2532:
2441:
2436:
2348:
2300:
2229:
1286:
257:
162:
55:
1268:
provides an overview of all the structure information available in the PDB for Human
Triosephosphate isomerase
665:
445:
that cause this disease, most include the replacement of glutamic acid at position 104 with an aspartic acid.
2537:
2324:
2007:
2002:
1876:
1678:
1622:
1603:
512:
149:
896:
Rose IA, Fung WJ, Warms JV (May 1990). "Proton diffusion in the active site of triosephosphate isomerase".
2451:
2384:
2135:
1699:
1659:
2558:
2477:
2446:
2155:
2086:
1936:
1925:
1412:
741:
to the phosphate group of the substrate. This action stabilizes the enediol intermediate and the other
229:
2630:
733:. The sequence around the active site residues is conserved in all known triose phosphate isomerases.
661:
137:
2190:
433:
In humans, deficiencies in TPI are associated with a progressive, severe neurological disorder called
320:
2110:
2057:
1396:
618:
72:
1445:
1367:
2795:
2594:
2364:
1770:
1360:
1279:
730:
67:
165:
2527:
785:
449:
89:
1549:
1507:
2785:
1403:
1305:
1245:
1210:
1175:
1140:
1099:
1064:
1024:
989:
954:
913:
878:
840:
754:
652:
156:
2573:
2568:
2542:
2470:
2269:
2048:
1709:
1613:
1237:
1202:
1167:
1130:
1091:
1056:
1016:
981:
944:
905:
870:
830:
761:
function, but variants with a different positively charged amino acid retain some function.
742:
516:
483:
438:
644:
2620:
2604:
2517:
2130:
2017:
1884:
1789:
1780:
1481:
933:"On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase"
714:
694:
391:
369:
310:
125:
931:
Alber T, Banner DW, Bloomer AC, Petsko GA, Phillips D, Rivers PS, Wilson IA (June 1981).
380:
937:
Philosophical
Transactions of the Royal Society of London. Series B, Biological Sciences
101:
17:
2769:
2658:
2599:
2225:
1135:
1118:
260:
200:
60:
1669:
180:
2779:
2563:
2522:
2101:
1265:
749:
738:
722:
690:
610:
175:
561:
2512:
1818:
640:
487:
574:
586:
2736:
2671:
2507:
2073:
1921:
789:
718:
702:
508:
479:
184:
2764:
1271:
2077:
1978:
1974:
1970:
1905:
1880:
1872:
1334:
1302:
835:
818:
770:
710:
706:
698:
427:
403:
2710:
2684:
2221:
1325:
726:
522:
500:
1214:
1206:
949:
932:
844:
1249:
1179:
1144:
1103:
1068:
1028:
993:
958:
917:
794:
614:
2121:
1047:
Knowles JR (March 1991). "Enzyme catalysis: not different, just better".
882:
581:
504:
442:
423:
264:
1241:
1171:
1020:
985:
909:
874:
2097:
2012:
1724:
1318:
798:
598:
593:
496:
453:
407:
132:
113:
1095:
701:
residues. The three-dimensional structure of a subunit contains eight
2723:
2493:
2283:
2246:
2242:
2172:
2168:
2164:
2160:
1887:
1060:
672:
437:. Triose phosphate isomerase deficiency is characterized by chronic
411:
272:
268:
253:
212:
108:
96:
84:
1828:
2697:
2412:
521:
419:
415:
44:
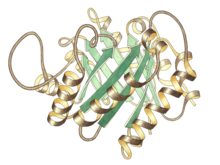
Side view of triose P isomerase monomer, active site at top center
2287:
2025:
2021:
1948:
1944:
1940:
780:
634:
568:
556:
465:
395:
384:
373:
120:
2466:
2194:
1845:
1275:
1917:
1913:
1909:
426:. However, some bacteria that do not perform glycolysis, like
2462:
819:"Triosephosphate isomerase deficiency: facts and doubts"
464:
The mechanism involves the intermediate formation of an
1117:
Lambeir AM, Opperdoes FR, Wierenga RK (October 1987).
2753:
452:: It is limited only by the rate the substrate can
2649:
2613:
2582:
2551:
2500:
2422:
2398:
2314:
2278:
2237:
2148:
2119:
2095:
2071:
2046:
2039:
1894:
671:
651:
633:
628:
604:
592:
580:
567:
555:
547:
542:
537:
223:
211:
199:
194:
174:
155:
143:
131:
119:
107:
95:
83:
78:
66:
54:
49:
32:
1042:
1040:
1038:
511:. Other inhibitors include 2-phosphoglycolate, a
2156:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
721:of this enzyme is in the center of the barrel. A
526:Side view of triose phosphate isomerase dimer.
482:glutamate 165 residue of TPI deprotonates the
2478:
2375:5-carboxymethyl-2-hydroxymuconate D-isomerase
2206:
1857:
1287:
8:
713:, and is by far the most commonly observed
491:enediolate collapse from the oxygen at C2.
2485:
2471:
2463:
2213:
2199:
2191:
2043:
1898:
1864:
1850:
1842:
1294:
1280:
1272:
817:Orosz F, Oláh J, Ovádi J (December 2006).
625:
456:into and out of the enzyme's active site.
191:
38:
1134:
948:
834:
1998:Glyceraldehyde 3-phosphate dehydrogenase
856:
854:
697:, each of which is made up of about 250
2760:
2330:Isopentenyl-diphosphate delta isomerase
1195:Archives of Biochemistry and Biophysics
809:
795:Triosephosphate isomerase (TIM) family
534:
333:
292:
267:the reversible interconversion of the
29:
776:Triose Phosphate Isomerase deficiency
435:triose phosphate isomerase deficiency
7:
356:
348:
315:
1136:10.1111/j.1432-1033.1987.tb13388.x
705:on the outside and eight parallel
25:
2063:Phosphoenolpyruvate carboxykinase
2763:
2370:Prostaglandin-A1 Delta-isomerase
1827:
1804:
1779:
1744:
1708:
1685:
1668:
1637:
1612:
1578:
1548:
1525:
1506:
1480:
1463:
1444:
1427:
1402:
1385:
1366:
1349:
1324:
1123:European Journal of Biochemistry
689:Triose phosphate isomerase is a
515:, and D-glycerol-1-phosphate, a
471:
334:
319:
309:
402:TPI plays an important role in
2390:Polyenoic fatty acid isomerase
1967:Fructose-bisphosphate aldolase
1:
629:Available protein structures:
27:Enzyme involved in glycolysis
2306:4-Oxalocrotonate tautomerase
2260:Ribose-5-phosphate isomerase
2409:Protein disulfide-isomerase
2380:Isopiperitenone D-isomerase
2355:Methylitaconate D-isomerase
2345:Cholestenol Delta-isomerase
2335:Vinylacetyl-CoA D-isomerase
2265:Mannose phosphate isomerase
1953:Fructose 1,6-bisphosphatase
441:. While there are various
2812:
2296:Phenylpyruvate tautomerase
2180:Bisphosphoglycerate mutase
1559:Glyceraldehyde-3-phosphate
1540:Glyceraldehyde 3-phosphate
1501:Glyceraldehyde 3-phosphate
1475:Dihydroxyacetone phosphate
358:triose phosphate isomerase
344:
342:
329:
327:
303:glyceraldehyde 3-phosphate
294:triose phosphate isomerase
289:Dihydroxyacetone phosphate
280:glyceraldehyde 3-phosphate
276:dihydroxyacetone phosphate
242:Triose-phosphate isomerase
2641:Michaelis–Menten kinetics
2433:Prostaglandin D2 synthase
2360:Aconitate Delta-isomerase
2340:Muconolactone D-isomerase
2255:Triosephosphate isomerase
1993:
1985:Triosephosphate isomerase
1962:
1901:
1439:Fructose 1,6-bisphosphate
1312:
836:10.1080/15216540601115960
786:Triosephosphate isomerase
745:on the reaction pathway.
624:
538:Triosephosphate isomerase
318:
308:
190:
37:
33:triosephosphate isomerase
2533:Diffusion-limited enzyme
2442:Prostaglandin E synthase
2437:Prostaglandin-D synthase
2301:Oxaloacetate tautomerase
507:ions, which bind to the
18:Triosephosphateisomerase
2008:Phosphoglycerate mutase
2003:Phosphoglycerate kinase
1877:carbohydrate metabolism
1679:Phosphoglycerate mutase
1623:Phosphoglycerate kinase
1604:1,3-Bisphosphoglycerate
513:transition state analog
2452:Thromboxane-A synthase
2385:L-dopachrome isomerase
2136:Glycerol dehydrogenase
1207:10.1006/abbi.2000.2253
950:10.1098/rstb.1981.0069
527:
2626:Eadie–Hofstee diagram
2559:Allosteric regulation
2447:Prostacyclin synthase
2087:Lactate dehydrogenase
1937:Phosphofructokinase 1
1926:Glucose 6-phosphatase
1455:Fructose-bisphosphate
1413:Phosphofructokinase-1
788:in interactive 3D at
525:
450:catalytically perfect
2636:Lineweaver–Burk plot
2111:Alanine transaminase
2058:Pyruvate carboxylase
2040:Gluconeogenesis only
1397:Fructose 6-phosphate
729:are involved in the
2365:Enoyl CoA isomerase
2325:Steroid D-isomerase
1771:Phosphoenolpyruvate
1377:Glucose-6-phosphate
1361:Glucose 6-phosphate
1242:10.1021/bi00176a009
1172:10.1021/bi00480a010
1021:10.1021/bi00226a005
986:10.1021/bi00416a019
910:10.1021/bi00470a008
875:10.1021/bi00670a031
731:catalytic mechanism
2791:Glycolysis enzymes
2595:Enzyme superfamily
2528:Enzyme promiscuity
1826:
1817:
1778:
1769:
1707:
1700:2-Phosphoglycerate
1698:
1667:
1660:3-Phosphoglycerate
1658:
1611:
1602:
1547:
1538:
528:
2751:
2750:
2460:
2459:
2270:Glucose isomerase
2224:: intramolecular
2188:
2187:
2144:
2143:
2035:
2034:
1932:Glucose isomerase
1839:
1838:
1824:
1815:
1776:
1767:
1705:
1696:
1665:
1656:
1609:
1600:
1545:
1536:
1306:metabolic pathway
1096:10.1021/bi982089f
755:glyoxalase system
743:transition states
687:
686:
683:
682:
678:structure summary
398:Pathway Database.
387:Pathway Database.
376:Pathway Database.
365:
364:
300:
239:
238:
235:
234:
138:metabolic pathway
16:(Redirected from
2803:
2768:
2767:
2759:
2631:Hanes–Woolf plot
2574:Enzyme activator
2569:Enzyme inhibitor
2543:Enzyme catalysis
2487:
2480:
2473:
2464:
2215:
2208:
2201:
2192:
2044:
1899:
1866:
1859:
1852:
1843:
1831:
1808:
1783:
1748:
1712:
1689:
1672:
1641:
1616:
1582:
1552:
1529:
1510:
1484:
1467:
1448:
1431:
1406:
1389:
1370:
1353:
1328:
1296:
1289:
1282:
1273:
1254:
1253:
1225:
1219:
1218:
1190:
1184:
1183:
1155:
1149:
1148:
1138:
1114:
1108:
1107:
1090:(47): 16828–38.
1079:
1073:
1072:
1061:10.1038/350121a0
1044:
1033:
1032:
1004:
998:
997:
969:
963:
962:
952:
943:(1063): 159–71.
928:
922:
921:
893:
887:
886:
858:
849:
848:
838:
814:
626:
535:
517:substrate analog
475:
439:hemolytic anemia
399:
388:
377:
338:
323:
313:
298:
285:
284:
192:
42:
30:
21:
2811:
2810:
2806:
2805:
2804:
2802:
2801:
2800:
2776:
2775:
2774:
2762:
2754:
2752:
2747:
2659:Oxidoreductases
2645:
2621:Enzyme kinetics
2609:
2605:List of enzymes
2578:
2547:
2518:Catalytic triad
2496:
2491:
2461:
2456:
2418:
2394:
2310:
2274:
2233:
2226:oxidoreductases
2219:
2189:
2184:
2140:
2131:Glycerol kinase
2115:
2091:
2067:
2031:
2018:Pyruvate kinase
1989:
1958:
1890:
1885:gluconeogenesis
1870:
1840:
1835:
1834:
1833:
1832:
1822:
1811:
1810:
1809:
1801:
1800:
1797:
1793:
1790:Pyruvate kinase
1785:
1784:
1774:
1763:
1762:
1761:
1759:
1754:
1750:
1749:
1741:
1740:
1738:
1733:
1729:
1720:
1719:Phosphopyruvate
1714:
1713:
1703:
1692:
1691:
1690:
1682:
1674:
1673:
1663:
1652:
1651:
1650:
1647:
1643:
1642:
1634:
1633:
1630:
1626:
1618:
1617:
1607:
1596:
1595:
1594:
1591:
1590:
1584:
1583:
1575:
1574:
1571:
1570:
1564:
1560:
1554:
1553:
1543:
1532:
1531:
1530:
1522:
1518:
1517:Triosephosphate
1512:
1511:
1504:
1496:
1495:
1492:
1486:
1485:
1478:
1470:
1469:
1468:
1460:
1456:
1450:
1449:
1442:
1434:
1433:
1432:
1424:
1423:
1420:
1416:
1408:
1407:
1400:
1392:
1391:
1390:
1382:
1378:
1372:
1371:
1364:
1356:
1355:
1354:
1346:
1345:
1342:
1338:
1330:
1329:
1322:
1308:
1300:
1262:
1257:
1236:(10): 2809–14.
1227:
1226:
1222:
1192:
1191:
1187:
1166:(28): 6619–25.
1157:
1156:
1152:
1116:
1115:
1111:
1081:
1080:
1076:
1055:(6314): 121–4.
1046:
1045:
1036:
1006:
1005:
1001:
980:(16): 5948–60.
971:
970:
966:
930:
929:
925:
895:
894:
890:
869:(25): 5627–31.
860:
859:
852:
816:
815:
811:
807:
767:
533:
462:
389:
378:
367:
45:
28:
23:
22:
15:
12:
11:
5:
2809:
2807:
2799:
2798:
2793:
2788:
2778:
2777:
2773:
2772:
2749:
2748:
2746:
2745:
2732:
2719:
2706:
2693:
2680:
2667:
2653:
2651:
2647:
2646:
2644:
2643:
2638:
2633:
2628:
2623:
2617:
2615:
2611:
2610:
2608:
2607:
2602:
2597:
2592:
2586:
2584:
2583:Classification
2580:
2579:
2577:
2576:
2571:
2566:
2561:
2555:
2553:
2549:
2548:
2546:
2545:
2540:
2535:
2530:
2525:
2520:
2515:
2510:
2504:
2502:
2498:
2497:
2492:
2490:
2489:
2482:
2475:
2467:
2458:
2457:
2455:
2454:
2449:
2444:
2439:
2429:
2427:
2420:
2419:
2417:
2416:
2405:
2403:
2396:
2395:
2393:
2392:
2387:
2382:
2377:
2372:
2367:
2362:
2357:
2352:
2342:
2337:
2332:
2327:
2321:
2319:
2312:
2311:
2309:
2308:
2303:
2298:
2292:
2290:
2276:
2275:
2273:
2272:
2267:
2262:
2257:
2251:
2249:
2235:
2234:
2220:
2218:
2217:
2210:
2203:
2195:
2186:
2185:
2183:
2182:
2177:
2176:
2175:
2152:
2150:
2146:
2145:
2142:
2141:
2139:
2138:
2133:
2127:
2125:
2117:
2116:
2114:
2113:
2107:
2105:
2093:
2092:
2090:
2089:
2083:
2081:
2069:
2068:
2066:
2065:
2060:
2054:
2052:
2041:
2037:
2036:
2033:
2032:
2030:
2029:
2015:
2010:
2005:
2000:
1994:
1991:
1990:
1988:
1987:
1982:
1963:
1960:
1959:
1957:
1956:
1934:
1929:
1902:
1896:
1892:
1891:
1871:
1869:
1868:
1861:
1854:
1846:
1837:
1836:
1825:2 ×
1823:
1813:
1812:
1803:
1802:
1798:
1795:
1794:
1787:
1786:
1777:2 ×
1775:
1765:
1764:
1757:
1755:
1752:
1751:
1743:
1742:
1736:
1734:
1731:
1730:
1716:
1715:
1706:2 ×
1704:
1694:
1693:
1684:
1683:
1676:
1675:
1666:2 ×
1664:
1654:
1653:
1648:
1645:
1644:
1636:
1635:
1631:
1628:
1627:
1620:
1619:
1610:2 ×
1608:
1598:
1597:
1592:
1588:
1586:
1585:
1577:
1576:
1572:
1568:
1566:
1565:
1556:
1555:
1546:2 ×
1544:
1534:
1533:
1524:
1523:
1514:
1513:
1505:
1498:
1497:
1493:
1488:
1487:
1479:
1472:
1471:
1462:
1461:
1452:
1451:
1443:
1436:
1435:
1426:
1425:
1421:
1418:
1417:
1410:
1409:
1401:
1394:
1393:
1384:
1383:
1374:
1373:
1365:
1358:
1357:
1348:
1347:
1343:
1340:
1339:
1332:
1331:
1323:
1316:
1315:
1314:
1313:
1310:
1309:
1301:
1299:
1298:
1291:
1284:
1276:
1270:
1269:
1261:
1260:External links
1258:
1256:
1255:
1220:
1185:
1150:
1109:
1074:
1034:
1015:(12): 3011–9.
999:
964:
923:
904:(18): 4312–7.
888:
850:
829:(12): 703–15.
808:
806:
803:
802:
801:
792:
783:
778:
773:
766:
763:
725:residue and a
685:
684:
681:
680:
675:
669:
668:
655:
649:
648:
638:
631:
630:
622:
621:
608:
602:
601:
596:
590:
589:
584:
578:
577:
572:
565:
564:
559:
553:
552:
549:
545:
544:
540:
539:
532:
529:
461:
458:
414:as well as in
363:
362:
360:
355:
351:
350:
346:
345:
343:
340:
339:
331:
330:
328:
325:
324:
317:
314:
306:
305:
296:
291:
237:
236:
233:
232:
227:
221:
220:
215:
209:
208:
203:
197:
196:
188:
187:
178:
172:
171:
160:
153:
152:
147:
141:
140:
135:
129:
128:
123:
117:
116:
111:
105:
104:
99:
93:
92:
87:
81:
80:
76:
75:
70:
64:
63:
58:
52:
51:
47:
46:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2808:
2797:
2794:
2792:
2789:
2787:
2784:
2783:
2781:
2771:
2766:
2761:
2757:
2743:
2739:
2738:
2733:
2730:
2726:
2725:
2720:
2717:
2713:
2712:
2707:
2704:
2700:
2699:
2694:
2691:
2687:
2686:
2681:
2678:
2674:
2673:
2668:
2665:
2661:
2660:
2655:
2654:
2652:
2648:
2642:
2639:
2637:
2634:
2632:
2629:
2627:
2624:
2622:
2619:
2618:
2616:
2612:
2606:
2603:
2601:
2600:Enzyme family
2598:
2596:
2593:
2591:
2588:
2587:
2585:
2581:
2575:
2572:
2570:
2567:
2565:
2564:Cooperativity
2562:
2560:
2557:
2556:
2554:
2550:
2544:
2541:
2539:
2536:
2534:
2531:
2529:
2526:
2524:
2523:Oxyanion hole
2521:
2519:
2516:
2514:
2511:
2509:
2506:
2505:
2503:
2499:
2495:
2488:
2483:
2481:
2476:
2474:
2469:
2468:
2465:
2453:
2450:
2448:
2445:
2443:
2440:
2438:
2434:
2431:
2430:
2428:
2425:
2421:
2414:
2410:
2407:
2406:
2404:
2401:
2397:
2391:
2388:
2386:
2383:
2381:
2378:
2376:
2373:
2371:
2368:
2366:
2363:
2361:
2358:
2356:
2353:
2350:
2346:
2343:
2341:
2338:
2336:
2333:
2331:
2328:
2326:
2323:
2322:
2320:
2317:
2313:
2307:
2304:
2302:
2299:
2297:
2294:
2293:
2291:
2289:
2285:
2281:
2277:
2271:
2268:
2266:
2263:
2261:
2258:
2256:
2253:
2252:
2250:
2248:
2244:
2240:
2236:
2231:
2227:
2223:
2216:
2211:
2209:
2204:
2202:
2197:
2196:
2193:
2181:
2178:
2174:
2170:
2166:
2162:
2159:
2158:
2157:
2154:
2153:
2151:
2147:
2137:
2134:
2132:
2129:
2128:
2126:
2123:
2118:
2112:
2109:
2108:
2106:
2103:
2102:Alanine cycle
2099:
2094:
2088:
2085:
2084:
2082:
2079:
2075:
2070:
2064:
2061:
2059:
2056:
2055:
2053:
2050:
2045:
2042:
2038:
2027:
2023:
2019:
2016:
2014:
2011:
2009:
2006:
2004:
2001:
1999:
1996:
1995:
1992:
1986:
1983:
1980:
1976:
1972:
1968:
1965:
1964:
1961:
1954:
1950:
1946:
1942:
1938:
1935:
1933:
1930:
1927:
1923:
1919:
1915:
1911:
1907:
1904:
1903:
1900:
1897:
1893:
1889:
1886:
1882:
1878:
1874:
1867:
1862:
1860:
1855:
1853:
1848:
1847:
1844:
1830:
1821:
1820:
1807:
1792:
1791:
1782:
1773:
1772:
1747:
1728:
1726:
1722:
1711:
1702:
1701:
1688:
1681:
1680:
1671:
1662:
1661:
1640:
1625:
1624:
1615:
1606:
1605:
1581:
1563:
1562:
1561:dehydrogenase
1551:
1542:
1541:
1528:
1521:
1520:
1509:
1503:
1502:
1491:
1483:
1477:
1476:
1466:
1459:
1458:
1447:
1441:
1440:
1430:
1415:
1414:
1405:
1399:
1398:
1388:
1381:
1380:
1369:
1363:
1362:
1352:
1337:
1336:
1327:
1321:
1320:
1311:
1307:
1304:
1297:
1292:
1290:
1285:
1283:
1278:
1277:
1274:
1267:
1264:
1263:
1259:
1251:
1247:
1243:
1239:
1235:
1231:
1224:
1221:
1216:
1212:
1208:
1204:
1200:
1196:
1189:
1186:
1181:
1177:
1173:
1169:
1165:
1161:
1154:
1151:
1146:
1142:
1137:
1132:
1128:
1124:
1120:
1113:
1110:
1105:
1101:
1097:
1093:
1089:
1085:
1078:
1075:
1070:
1066:
1062:
1058:
1054:
1050:
1043:
1041:
1039:
1035:
1030:
1026:
1022:
1018:
1014:
1010:
1003:
1000:
995:
991:
987:
983:
979:
975:
968:
965:
960:
956:
951:
946:
942:
938:
934:
927:
924:
919:
915:
911:
907:
903:
899:
892:
889:
884:
880:
876:
872:
868:
864:
857:
855:
851:
846:
842:
837:
832:
828:
824:
820:
813:
810:
804:
800:
796:
793:
791:
787:
784:
782:
779:
777:
774:
772:
769:
768:
764:
762:
758:
756:
751:
750:methylglyoxal
746:
744:
740:
739:hydrogen bond
734:
732:
728:
724:
723:glutamic acid
720:
716:
712:
708:
704:
700:
696:
693:of identical
692:
679:
676:
674:
670:
667:
663:
659:
656:
654:
650:
646:
642:
639:
636:
632:
627:
623:
620:
616:
612:
609:
607:
603:
600:
597:
595:
591:
588:
585:
583:
579:
576:
573:
570:
566:
563:
560:
558:
554:
550:
546:
541:
536:
530:
524:
520:
518:
514:
510:
506:
502:
498:
492:
489:
488:electrophilic
485:
481:
476:
474:
469:
467:
459:
457:
455:
451:
446:
444:
440:
436:
431:
429:
425:
421:
417:
413:
409:
405:
400:
397:
393:
386:
382:
375:
371:
361:
359:
353:
352:
347:
341:
337:
332:
326:
322:
312:
307:
304:
297:
295:
290:
287:
286:
283:
281:
277:
274:
270:
266:
262:
259:
255:
251:
247:
243:
231:
228:
226:
222:
219:
216:
214:
210:
207:
204:
202:
198:
193:
189:
186:
182:
179:
177:
176:Gene Ontology
173:
170:
167:
164:
161:
158:
154:
151:
148:
146:
142:
139:
136:
134:
130:
127:
124:
122:
118:
115:
114:NiceZyme view
112:
110:
106:
103:
100:
98:
94:
91:
88:
86:
82:
77:
74:
71:
69:
65:
62:
59:
57:
53:
48:
41:
36:
31:
19:
2737:Translocases
2734:
2721:
2708:
2695:
2682:
2672:Transferases
2669:
2656:
2513:Binding site
2254:
2049:oxaloacetate
1984:
1814:
1788:
1766:
1717:
1695:
1677:
1655:
1621:
1599:
1557:
1535:
1516:
1515:
1499:
1489:
1473:
1453:
1437:
1411:
1395:
1375:
1359:
1333:
1317:
1233:
1230:Biochemistry
1229:
1223:
1198:
1194:
1188:
1163:
1160:Biochemistry
1159:
1153:
1129:(1): 69–74.
1126:
1122:
1112:
1087:
1084:Biochemistry
1083:
1077:
1052:
1048:
1012:
1009:Biochemistry
1008:
1002:
977:
974:Biochemistry
973:
967:
940:
936:
926:
901:
898:Biochemistry
897:
891:
866:
863:Biochemistry
862:
826:
822:
812:
759:
747:
735:
715:protein fold
688:
493:
480:nucleophilic
477:
470:
463:
447:
432:
430:, lack TPI.
401:
366:
249:
245:
241:
240:
102:BRENDA entry
2508:Active site
1922:Glucokinase
1201:(1): 1–10.
790:Proteopedia
719:active site
543:Identifiers
509:active site
428:ureaplasmas
90:IntEnz view
50:Identifiers
2796:Glycolysis
2780:Categories
2711:Isomerases
2685:Hydrolases
2552:Regulation
2222:Isomerases
2149:Regulatory
2078:Cori cycle
1969:(Aldolase
1906:Hexokinase
1895:Glycolysis
1881:glycolysis
1873:Metabolism
1335:Hexokinase
1303:Glycolysis
823:IUBMB Life
805:References
771:TIM barrel
711:TIM-barrel
699:amino acid
641:structures
486:, and the
404:glycolysis
271:phosphate
159:structures
126:KEGG entry
73:9023-78-3
2590:EC number
1721:hydratase
1519:isomerase
1379:isomerase
727:histidine
707:β-strands
703:α-helices
599:PDOC00155
587:IPR000652
531:Structure
501:phosphate
484:substrate
460:Mechanism
443:mutations
390:Compound
368:Compound
265:catalyzes
79:Databases
2786:EC 5.3.1
2614:Kinetics
2538:Cofactor
2501:Activity
2122:glycerol
1949:Platelet
1819:Pyruvate
1593:NADH + H
1573:NADH + H
1457:aldolase
1215:11368170
845:17424909
765:See also
737:forms a
695:subunits
658:RCSB PDB
582:InterPro
505:arsenate
424:bacteria
252:) is an
230:proteins
218:articles
206:articles
163:RCSB PDB
2770:Biology
2724:Ligases
2494:Enzymes
2426:: other
2318:: C = C
2247:Ketoses
2243:Aldoses
2098:alanine
2074:lactate
2013:Enolase
1888:enzymes
1725:enolase
1319:Glucose
1266:PDBe-KB
1250:8130193
1180:2204418
1145:3311744
1104:9843453
1069:2005961
1029:2007138
994:2847777
959:6115415
918:2161683
799:PROSITE
594:PROSITE
562:PF00121
497:sulfate
466:enediol
454:diffuse
412:insects
408:mammals
381:5.3.1.1
379:Enzyme
354:
349:
316:
273:isomers
263:) that
261:5.3.1.1
185:QuickGO
150:profile
133:MetaCyc
68:CAS no.
61:5.3.1.1
2756:Portal
2698:Lyases
2424:5.3.99
2173:PFKFB4
2169:PFKFB3
2165:PFKFB2
2161:PFKFB1
1945:Muscle
1753:
1732:
1587:NAD+ P
1567:NAD+ P
1248:
1213:
1178:
1143:
1102:
1067:
1049:Nature
1027:
992:
957:
916:
883:999838
881:
843:
717:. The
673:PDBsum
647:
637:
619:SUPFAM
575:CL0036
548:Symbol
503:, and
422:, and
420:plants
392:C00118
370:C00111
278:and D-
269:triose
254:enzyme
213:PubMed
195:Search
181:AmiGO
169:PDBsum
109:ExPASy
97:BRENDA
85:IntEnz
56:EC no.
2650:Types
2413:PDIA3
2402:: S-S
2400:5.3.4
2316:5.3.3
2280:5.3.2
2239:5.3.1
2120:from
2096:from
2072:from
1941:Liver
691:dimer
615:SCOPe
606:SCOP2
416:fungi
145:PRIAM
2742:list
2735:EC7
2729:list
2722:EC6
2716:list
2709:EC5
2703:list
2696:EC4
2690:list
2683:EC3
2677:list
2670:EC2
2664:list
2657:EC1
2288:Enol
2284:Keto
2232:5.3)
2026:PKM2
2022:PKLR
1816:2 ×
1768:2 ×
1697:2 ×
1657:2 ×
1601:2 ×
1537:2 ×
1246:PMID
1211:PMID
1176:PMID
1141:PMID
1100:PMID
1065:PMID
1025:PMID
990:PMID
955:PMID
914:PMID
879:PMID
841:PMID
781:TPI1
666:PDBj
662:PDBe
645:ECOD
635:Pfam
611:1tph
571:clan
569:Pfam
557:Pfam
410:and
396:KEGG
385:KEGG
374:KEGG
225:NCBI
166:PDBe
121:KEGG
2349:EBP
2047:to
1951:)→/
1924:)→/
1918:HK3
1914:HK2
1910:HK1
1799:ATP
1796:ADP
1649:ATP
1646:ADP
1632:ATP
1629:ADP
1422:ADP
1419:ATP
1344:ADP
1341:ATP
1238:doi
1203:doi
1199:387
1168:doi
1131:doi
1127:168
1092:doi
1057:doi
1053:350
1017:doi
982:doi
945:doi
941:293
906:doi
871:doi
831:doi
797:in
653:PDB
551:TIM
394:at
383:at
372:at
250:TIM
248:or
246:TPI
201:PMC
157:PDB
2782::
2282::
2241::
2230:EC
2171:,
2167:,
2163:,
2104:):
2080:):
2024:,
1977:,
1973:,
1947:,
1943:,
1920:,
1916:,
1912:,
1879::
1875::
1244:.
1234:33
1232:.
1209:.
1197:.
1174:.
1164:29
1162:.
1139:.
1125:.
1121:.
1098:.
1088:37
1086:.
1063:.
1051:.
1037:^
1023:.
1013:30
1011:.
988:.
978:27
976:.
953:.
939:.
935:.
912:.
902:29
900:.
877:.
867:15
865:.
853:^
839:.
827:58
825:.
821:.
664:;
660:;
643:/
617:/
613:/
519:.
499:,
418:,
282:.
258:EC
183:/
2758::
2744:)
2740:(
2731:)
2727:(
2718:)
2714:(
2705:)
2701:(
2692:)
2688:(
2679:)
2675:(
2666:)
2662:(
2486:e
2479:t
2472:v
2435:/
2415:)
2411:(
2351:)
2347:(
2286:/
2245:/
2228:(
2214:e
2207:t
2200:v
2124::
2100:(
2076:(
2051::
2028:)
2020:(
1981:)
1979:C
1975:B
1971:A
1955:←
1939:(
1928:←
1908:(
1883:/
1865:e
1858:t
1851:v
1760:O
1758:2
1756:H
1739:O
1737:2
1735:H
1727:)
1723:(
1589:i
1569:i
1494:+
1490:+
1295:e
1288:t
1281:v
1252:.
1240::
1217:.
1205::
1182:.
1170::
1147:.
1133::
1106:.
1094::
1071:.
1059::
1031:.
1019::
996:.
984::
961:.
947::
920:.
908::
885:.
873::
847:.
833::
301:-
299:D
256:(
244:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.