384:, in which the repeat sequence is generally 1 to 6 nucleotides. The two types of repeat sequences are both tandem but are specified by the length of the repeat sequence. VNTRs, therefore, because they have repeat sequences of ten to one hundred nucleotides in which every repeat is exactly the same, are considered minisatellites. However, while all VNTRs are minisatellites, not all minisatellites are VNTRs. VNTRs can vary in number of repeats from individual to individual, as where some non-VNTR minisatellites have repeat sequences that repeat the same number of times in all individuals containing the tandem repeats in their genomes.
36:
364:
380:, meaning that the sequences repeat one after another without other sequences or nucleotides in between them. Minisatellites are characterized by a repeat sequence of about ten to one hundred nucleotides, and the number of times the sequence repeats varies from about five to fifty times. The sequences of minisatellites are larger than those of
319:– the VNTR alleles must follow the rules of inheritance. In matching an individual with his parents or children, a person must have an allele that matches one from each parent. If the relationship is more distant, such as a grandparent or sibling, then matches must be consistent with the degree of relatedness.
367:
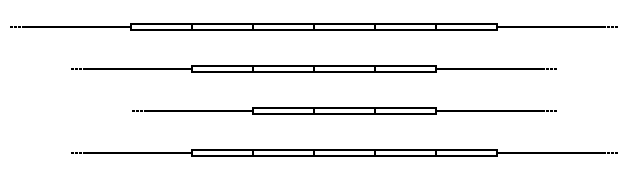
This shows a theoretical example of a VNTR in two different individuals. A single strand of DNA from each individual is displayed in which there is tandem repeat sequence that the individuals share. The sequence presence is a VNTR because one individual has five repeats, while the other has seven
211:
In the schematic above, the rectangular blocks represent each of the repeated DNA sequences at a particular VNTR location. The repeats are in tandem – i.e. they are clustered together and oriented in the same direction. Individual repeats can be removed from (or added to) the VNTR via
272:, they produce a pattern of bands unique to each individual. When tested with a group of independent VNTR markers, the likelihood of two unrelated individuals' having the same allelic pattern is extremely low. VNTR analysis is also being used to study
292:
202:
220:
errors, leading to alleles with different numbers of repeats. Flanking regions are segments of repetitive sequence (shown here as thin lines), allowing the VNTR blocks to be extracted with
368:
repeats (number of repeats varies in different individuals). Each repeat is ten nucleotides, making it a minisatellite, rather than a microsatellite in which each repeat is 1-6 nucleotides.
280:
of wild or domesticated animals. As such, VNTRs can be used to distinguish strains of bacterial pathogens. In this microbial forensics context, such assays are usually called
465:
53:
610:
561:
225:
331:, representing over 40% of the human genome, is arranged in a bewildering array of patterns. Repeats were first identified by the extraction of
393:
355:(which may interfere with DNA replication). VNTRs are the class of clustered tandem repeats that exhibit allelic variation in their lengths.
313:– both VNTR alleles from a specific location must match. If two samples are from the same individual, they must show the same allele pattern.
100:
440:
72:
796:
783:
572:
Phobos – a tandem repeat search tool for perfect and imperfect repeats – the maximum pattern size depends only on computational power
79:
1099:
806:
119:
472:
603:
86:
268:
database. When removed from surrounding DNA by the PCR or RFLP methods, and their size determined by gel electrophoresis or
57:
133:
837:
683:
68:
566:
376:
in which the size of the repeat sequence is generally ten to one hundred base pairs. Minisatellites are a type of DNA
281:
556:
954:
596:
46:
842:
265:
229:
582:
578:
648:
623:
588:
328:
93:
1078:
1006:
899:
167:
924:
874:
213:
934:
1056:
974:
1051:
964:
879:
765:
627:
253:
829:
700:
403:
340:
277:
233:
155:
1001:
909:
884:
728:
335:, which does not reveal how they are organized. The use of restriction enzymes showed that some
221:
190:
177:, allowing them to be used for personal or parental identification. Their analysis is useful in
17:
433:
979:
273:
944:
894:
889:
531:
1063:
993:
919:
869:
631:
914:
819:
939:
774:
746:
715:
710:
352:
261:
249:
217:
1046:
904:
861:
689:
679:
381:
344:
245:
532:
TAPO: A combined method for the identification of tandem repeats in protein structures
1093:
1011:
751:
674:
665:
657:
408:
377:
373:
348:
332:
269:
159:
1023:
336:
551:
1018:
929:
791:
495:
413:
35:
1030:
847:
163:
571:
949:
640:
186:
516:
291:
466:"Minisatellites and microsatellites – similar names but different biology"
347:
later showed that other repeats are clustered at specific locations, with
170:
in length (number of repeats) among individuals. Each variant acts as an
1073:
959:
723:
619:
257:
178:
171:
363:
969:
182:
201:
1068:
174:
151:
536:
132:
506:
814:
306:
In analyzing VNTR data, two basic genetic principles can be used:
290:
200:
131:
398:
285:
592:
541:
252:(mapping) of diploid genomes. Now that many genomes have been
137:
Schematic of a
Variable Number of Tandem Repeats in 4 alleles.
29:
296:
Chromosomal locations of the 13 VNTR loci in the CODIS panel.
546:
206:
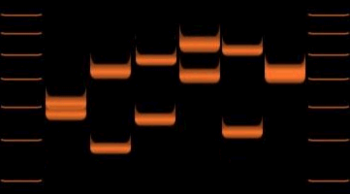
Variations of VNTR (D1S80) allele lengths in 6 individuals.
511:
521:
1039:
992:
860:
828:
805:
782:
773:
764:
739:
699:
656:
647:
567:
JSTRING – Java Search for Tandem
Repeats in genomes
60:. Unsourced material may be challenged and removed.
604:
324:Relationship to other types of repetitive DNA
232:(PCR) technique and their size determined by
8:
779:
770:
653:
611:
597:
589:
507:The Microorganisms Tandem Repeats Database
581:at the U.S. National Library of Medicine
120:Learn how and when to remove this message
362:
425:
244:VNTRs were an important source of RFLP
7:
58:adding citations to reliable sources
680:Short tandem repeat/Microsatellite
25:
579:Variable+Number+of+Tandem+Repeats
256:, VNTRs have become essential to
18:Variable number of tandem repeats
34:
446:from the original on 2017-05-01
197:Structure and allelic variation
69:"Variable number tandem repeat"
45:needs additional citations for
684:Trinucleotide repeat disorders
522:Tandem Repeats Database (TRDB)
162:. These can be found on many
1:
671:Variable number tandem repeat
562:Microsatellite repeats finder
517:Short Tandem Repeats Database
144:variable number tandem repeat
498:– info and animated example
282:Multiple Loci VNTR Analysis
1116:
260:crime investigations, via
638:
276:and breeding patterns in
230:polymerase chain reaction
1100:Repetitive DNA sequences
583:Medical Subject Headings
343:throughout the genome.
351:being more common than
240:Use in genetic analysis
1079:Protein tandem repeats
1007:Tandemly arrayed genes
378:tandem repeat sequence
369:
298:
228:, or amplified by the
208:
139:
537:Tandem Repeats Finder
366:
294:
204:
150:) is a location in a
135:
1052:Pathogenicity island
528:Search tools :
372:VNTRs are a type of
317:Inheritance Matching
54:improve this article
27:Location in a genome
404:Short tandem repeat
234:gel electrophoresis
222:restriction enzymes
156:nucleotide sequence
1002:Gene amplification
370:
299:
262:DNA fingerprinting
209:
191:DNA fingerprinting
158:is organized as a
140:
1087:
1086:
988:
987:
856:
855:
760:
759:
649:Repeated sequence
624:repeated sequence
503:Databases :
464:Dubrova, Yuri E.
311:Identity Matching
274:genetic diversity
270:Southern blotting
166:, and often show
130:
129:
122:
104:
16:(Redirected from
1107:
1064:Low copy repeats
1057:Symbiosis island
994:Gene duplication
780:
771:
654:
632:gene duplication
613:
606:
599:
590:
492:Examples :
480:
479:
477:
471:. Archived from
470:
461:
455:
454:
452:
451:
445:
438:
430:
353:inverted repeats
250:linkage analysis
224:and analyzed by
125:
118:
114:
111:
105:
103:
62:
38:
30:
21:
1115:
1114:
1110:
1109:
1108:
1106:
1105:
1104:
1090:
1089:
1088:
1083:
1035:
984:
852:
824:
801:
775:Retrotransposon
756:
747:Inverted repeat
735:
720:DNA transposon
716:Retrotransposon
711:Gene conversion
702:
695:
692:
643:
634:
617:
489:
484:
483:
475:
468:
463:
462:
458:
449:
447:
443:
436:
432:
431:
427:
422:
390:
382:microsatellites
361:
326:
304:
297:
246:genetic markers
242:
207:
199:
138:
126:
115:
109:
106:
63:
61:
51:
39:
28:
23:
22:
15:
12:
11:
5:
1113:
1111:
1103:
1102:
1092:
1091:
1085:
1084:
1082:
1081:
1076:
1071:
1066:
1061:
1060:
1059:
1054:
1047:Genomic island
1043:
1041:
1037:
1036:
1034:
1033:
1028:
1027:
1026:
1016:
1015:
1014:
1004:
998:
996:
990:
989:
986:
985:
983:
982:
977:
972:
967:
962:
957:
952:
947:
942:
937:
932:
927:
922:
917:
912:
907:
902:
897:
892:
887:
882:
877:
872:
866:
864:
862:DNA transposon
858:
857:
854:
853:
851:
850:
845:
840:
834:
832:
826:
825:
823:
822:
817:
811:
809:
803:
802:
800:
799:
794:
788:
786:
777:
768:
762:
761:
758:
757:
755:
754:
749:
743:
741:
737:
736:
734:
733:
732:
731:
726:
718:
713:
707:
705:
697:
696:
694:
693:
690:Macrosatellite
687:
677:
668:
662:
660:
658:Tandem repeats
651:
645:
644:
639:
636:
635:
618:
616:
615:
608:
601:
593:
587:
586:
576:
575:
574:
569:
564:
559:
554:
549:
544:
539:
534:
526:
525:
524:
519:
514:
509:
501:
500:
499:
488:
487:External links
485:
482:
481:
478:on 2017-12-15.
456:
424:
423:
421:
418:
417:
416:
411:
406:
401:
396:
389:
386:
360:
357:
349:tandem repeats
345:DNA sequencing
329:Repetitive DNA
325:
322:
321:
320:
314:
303:
300:
295:
241:
238:
205:
198:
195:
154:where a short
136:
128:
127:
42:
40:
33:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1112:
1101:
1098:
1097:
1095:
1080:
1077:
1075:
1072:
1070:
1067:
1065:
1062:
1058:
1055:
1053:
1050:
1049:
1048:
1045:
1044:
1042:
1038:
1032:
1029:
1025:
1022:
1021:
1020:
1017:
1013:
1012:Ribosomal DNA
1010:
1009:
1008:
1005:
1003:
1000:
999:
997:
995:
991:
981:
978:
976:
973:
971:
968:
966:
963:
961:
958:
956:
953:
951:
948:
946:
943:
941:
938:
936:
933:
931:
928:
926:
923:
921:
918:
916:
913:
911:
908:
906:
903:
901:
898:
896:
893:
891:
888:
886:
883:
881:
878:
876:
873:
871:
868:
867:
865:
863:
859:
849:
846:
844:
841:
839:
836:
835:
833:
831:
827:
821:
818:
816:
813:
812:
810:
808:
804:
798:
795:
793:
790:
789:
787:
785:
781:
778:
776:
772:
769:
767:
763:
753:
752:Direct repeat
750:
748:
745:
744:
742:
738:
730:
727:
725:
722:
721:
719:
717:
714:
712:
709:
708:
706:
704:
698:
691:
688:
685:
681:
678:
676:
675:Minisatellite
672:
669:
667:
666:Satellite DNA
664:
663:
661:
659:
655:
652:
650:
646:
642:
637:
633:
629:
625:
621:
614:
609:
607:
602:
600:
595:
594:
591:
584:
580:
577:
573:
570:
568:
565:
563:
560:
558:
555:
553:
550:
548:
545:
543:
540:
538:
535:
533:
530:
529:
527:
523:
520:
518:
515:
513:
510:
508:
505:
504:
502:
497:
494:
493:
491:
490:
486:
474:
467:
460:
457:
442:
435:
429:
426:
419:
415:
412:
410:
409:Tandem repeat
407:
405:
402:
400:
397:
395:
392:
391:
387:
385:
383:
379:
375:
374:minisatellite
365:
358:
356:
354:
350:
346:
342:
338:
337:repeat blocks
334:
333:Satellite DNA
330:
323:
318:
315:
312:
309:
308:
307:
301:
293:
289:
287:
283:
279:
275:
271:
267:
263:
259:
255:
251:
247:
239:
237:
235:
231:
227:
223:
219:
215:
214:recombination
203:
196:
194:
192:
188:
184:
180:
176:
173:
169:
165:
161:
160:tandem repeat
157:
153:
149:
145:
134:
124:
121:
113:
102:
99:
95:
92:
88:
85:
81:
78:
74:
71: –
70:
66:
65:Find sources:
59:
55:
49:
48:
43:This article
41:
37:
32:
31:
19:
1024:Gene cluster
792:Alu sequence
701:Interspersed
670:
512:The MLVAbank
473:the original
459:
448:. Retrieved
428:
371:
341:interspersed
327:
316:
310:
305:
243:
210:
147:
143:
141:
116:
107:
97:
90:
83:
76:
64:
52:Please help
47:verification
44:
1019:Gene family
930:Tc1/mariner
885:EnSpm/CACTA
414:BioNumerics
302:Inheritance
278:populations
218:replication
164:chromosomes
1031:Pseudogene
848:retroposon
766:Transposon
628:transposon
557:TandemSWAN
450:2024-01-13
420:References
185:research,
168:variations
80:newspapers
950:P element
900:Harbinger
641:Repeatome
254:sequenced
187:forensics
172:inherited
1094:Category
1074:Telomere
1040:See also
980:Zisupton
960:Polinton
955:PiggyBac
910:Helitron
729:Helitron
724:Polinton
620:Genetics
441:Archived
388:See also
264:and the
258:forensic
248:used in
179:genetics
110:May 2017
970:Transib
945:Novosib
925:Kolobok
895:Ginger2
890:Ginger1
875:Crypton
359:Classes
183:biology
94:scholar
1069:CRISPR
935:Merlin
920:ISL2EU
870:Academ
703:repeat
585:(MeSH)
434:"VNTR"
189:, and
175:allele
152:genome
96:
89:
82:
75:
67:
975:Zator
915:IS3EU
820:LINE2
815:LINE1
807:LINEs
784:SINEs
740:Other
542:Mreps
496:VNTRs
476:(PDF)
469:(PDF)
444:(PDF)
437:(PDF)
339:were
266:CODIS
101:JSTOR
87:books
965:Sola
940:MuDR
880:Dada
843:MER4
838:HERV
830:LTRs
552:TRED
547:STAR
399:MLVA
394:AFLP
286:MLVA
226:RFLP
181:and
148:VNTR
146:(or
73:news
905:hAT
797:MIR
284:or
216:or
56:by
1096::
630:,
626:,
622::
439:.
288:.
236:.
193:.
142:A
686:)
682:(
673:/
612:e
605:t
598:v
453:.
123:)
117:(
112:)
108:(
98:·
91:·
84:·
77:·
50:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.