469:
332:
36:
133:
539:
region is a region of DNA that is not copied into the mature mRNA, but which is present adjacent to 3′-end of the gene. It was originally thought that the 3′-flanking DNA was not transcribed at all, but it was discovered to be transcribed into RNA and quickly removed during processing of the primary
452:, but is usually involved in the regulation of translation. The 5′-untranslated region is the portion of the DNA starting from the cap site and extending to the base just before the AUG translation initiation codon of the main coding sequence. This region may have sequences, such as the
563:
is considered to be 3′-untranslated. The 3′-untranslated region may affect the translation efficiency of the mRNA or the stability of the mRNA. It also has sequences which are required for the addition of the poly(A) tail to the message, including the hexanucleotide AAUAAA.
288:(5′-ATG-3′) is a DNA sequence within the sense strand. Transcription begins at an upstream site (relative to the sense strand), and as it proceeds through the region it copies the 3′-TAC-5′ from the template strand to produce 5′-AUG-3′ within a
540:
transcript to form the mature mRNA. The 3′-flanking region often contains sequences that affect the formation of the 3′-end of the message. It may also contain enhancers or other sites to which proteins may bind.
260:. Transcription of single-stranded RNA from a double-stranded DNA template requires the selection of one strand of the DNA template as the template strand that directly interacts with the nascent RNA due to
268:
generally occur on both strands of an organism's DNA, and specify the location, direction, and circumstances under which transcription will occur. If the transcript encodes one or (rarely) more
53:
532:
residues to produce mature messenger RNA. This chain helps in determining how long the messenger RNA lasts in the cell, influencing how much protein is produced from it.
670:
100:
599:
72:
394:
occurs, a process which is vital to producing mature messenger RNA. Capping increases the stability of the messenger RNA while it undergoes
555:
transcribed into mRNA and becomes the 3′-end of the message, but which does not contain protein coding sequence. Everything between the
261:
79:
119:
86:
544:
437:
68:
57:
444:(5′-UTR) is a region of a gene which is transcribed into mRNA, and is located at the 5′-end of the mRNA. This region of an
250:
690:
339:
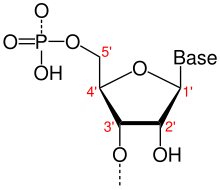
The 5′-end (pronounced "five prime end") designates the end of the DNA or RNA strand that has the fifth carbon in the
367:. Removal of the 5′-phosphate prevents ligation. To prevent unwanted nucleic acid ligation (e.g. self-ligation of a
140:(sugar-ring) molecule with carbon atoms labeled using standard notation. The 5′ is upstream; the 3′ is downstream.
292:(mRNA). The mRNA is scanned by the ribosome from the 5′ end, where the start codon directs the incorporation of a
46:
620:"Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro"
573:
257:
495:(joined) to the 5′-phosphate of a separate nucleotide, allowing the formation of strands of linked nucleotides.
93:
426:
265:
264:. The other strand is not copied directly, but necessarily its sequence will be similar to that of the RNA.
222:
207:
198:(usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a
414:
395:
323:
sequences are written in a 5′-to-3′ direction except as needed to illustrate the pattern of base pairing.
460:, which determine the translation efficiency of the mRNA, or which may affect the stability of the mRNA.
363:, i.e., the covalent binding of a 5′-phosphate to the 3′-hydroxyl group of another nucleotide, to form a
453:
449:
472:
364:
335:
In the DNA segment shown, the 5′ to 3′ directions are down the left strand and up the right strand.
230:
309:
649:
595:
498:
468:
376:
156:
221:
that assemble various types of new strands generally rely on the energy produced by breaking
639:
631:
199:
525:
407:
644:
619:
514:
491:. The 3′-hydroxyl is necessary in the synthesis of new nucleic acid molecules as it is
457:
331:
635:
679:
588:
521:
510:
430:
387:
289:
695:
685:
301:
238:
203:
164:
160:
479:
The 3′-end (three prime end) of a strand is so named due to it terminating at the
505:
that lack a 3′-hydroxyl (dideoxyribonucleotides) to interrupt the replication of
502:
399:
380:
372:
344:
285:
233:. The relative positions of structures along strands of nucleic acid, including
35:
391:
560:
556:
492:
403:
360:
356:
293:
281:
277:
218:
176:
529:
352:
315:
instead) at the N terminus of the protein. By convention, single strands of
187:
653:
276:
will proceed in a 5′-to-3′ direction, and will extend the protein from its
132:
17:
410:) attached to the messenger RNA in a rare 5′- to 5′-triphosphate linkage.
509:. This technique is known as the dideoxy chain-termination method or the
484:
480:
340:
305:
297:
273:
226:
137:
368:
269:
214:
179:
586:
Harvey Lodish; Arnold Berk; Paul
Matsudaira; Chris A. Kaiser (2004).
348:
191:
186:(usually pronounced "five-prime end"), which frequently contains a
467:
330:
131:
27:
End-to-end chemical orientation of a single strand of nucleic acid
433:, and may also contain enhancers or other protein binding sites.
445:
422:
234:
163:, is the end-to-end chemical orientation of a single strand of
506:
320:
316:
172:
168:
145:
141:
29:
594:(5th ed.). New York City: W.H. Freeman and Company.
225:
bonds to attach new nucleoside monophosphates to the 3′-
175:, the chemical convention of naming carbon atoms in the
398:, providing resistance to the degradative effects of
206:
between them, which is essential for replication or
202:, the strands run in opposite directions to permit
60:. Unsourced material may be challenged and removed.
587:
256:Directionality is related to, but different from,
618:Gao M, Fritz DT, Ford LP, Wilusz J (March 2000).
429:into RNA. The 5′-flanking region contains the
8:
425:often denotes a region of DNA which is not
148:are synthesized in the 5′-to-3′ direction.
643:
515:determine the order of nucleotides in DNA
120:Learn how and when to remove this message
379:commonly remove the 5′-phosphate with a
610:
190:group attached to the 5′ carbon of the
69:"Directionality" molecular biology
551:(3′-UTR) is a region of the DNA which
528:, which attaches a chain of 50 to 250
213:Nucleic acids can only be synthesized
355:group attached to the 5′-end permits
272:, translation of each protein by the
7:
526:post-transcriptional polyadenylation
241:, are usually noted as being either
58:adding citations to reliable sources
284:. For example, in a typical gene a
217:in the 5′-to-3′ direction, as the
25:
483:group of the third carbon in the
249:(towards the 3′-end). (See also
34:
45:needs additional citations for
266:Transcription initiation sites
1:
636:10.1016/S1097-2765(00)80442-6
475:(circled) between nucleotides
671:A Molecular Biology Glossary
392:post-transcriptional capping
210:of the encoded information.
182:means that there will be a
712:
574:Sense (molecular biology)
245:(towards the 5′-end) or
167:. In a single strand of
251:upstream and downstream
223:nucleoside triphosphate
590:Molecular Cell Biology
520:The 3′-end of nascent
487:, and is known as the
476:
386:The 5′-end of nascent
336:
262:complementary sequence
149:
471:
454:ribosome binding site
390:is the site at which
334:
135:
499:Molecular biologists
473:Phosphodiester bonds
377:molecular biologists
237:and various protein
54:improve this article
402:. It consists of a
365:phosphodiester bond
351:at its terminus. A
231:phosphodiester bond
229:(−OH) group, via a
691:Molecular genetics
477:
448:may or may not be
337:
180:pentose-sugar-ring
150:
601:978-0-7167-4366-8
513:, and is used to
313:-formylmethionine
157:molecular biology
130:
129:
122:
104:
16:(Redirected from
703:
658:
657:
647:
615:
605:
593:
200:DNA double helix
125:
118:
114:
111:
105:
103:
62:
38:
30:
21:
711:
710:
706:
705:
704:
702:
701:
700:
676:
675:
667:
662:
661:
617:
616:
612:
602:
585:
582:
580:Further reading
570:
524:is the site of
466:
408:methylguanosine
329:
126:
115:
109:
106:
63:
61:
51:
39:
28:
23:
22:
15:
12:
11:
5:
709:
707:
699:
698:
693:
688:
678:
677:
674:
673:
666:
665:External links
663:
660:
659:
630:(3): 479–488.
624:Molecular Cell
609:
608:
607:
606:
600:
581:
578:
577:
576:
569:
566:
465:
462:
458:Kozak sequence
369:plasmid vector
328:
325:
153:Directionality
128:
127:
42:
40:
33:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
708:
697:
694:
692:
689:
687:
684:
683:
681:
672:
669:
668:
664:
655:
651:
646:
641:
637:
633:
629:
625:
621:
614:
611:
603:
597:
592:
591:
584:
583:
579:
575:
572:
571:
567:
565:
562:
558:
554:
550:
548:
541:
538:
533:
531:
527:
523:
522:messenger RNA
518:
516:
512:
511:Sanger method
508:
504:
500:
496:
494:
490:
486:
482:
474:
470:
463:
461:
459:
455:
451:
447:
443:
441:
434:
432:
431:gene promoter
428:
424:
420:
418:
411:
409:
405:
401:
397:
393:
389:
388:messenger RNA
384:
382:
378:
374:
370:
366:
362:
358:
354:
350:
346:
342:
333:
326:
324:
322:
318:
314:
312:
307:
303:
299:
295:
291:
290:messenger RNA
287:
283:
279:
275:
271:
267:
263:
259:
254:
252:
248:
244:
240:
239:binding sites
236:
232:
228:
224:
220:
216:
211:
209:
208:transcription
205:
201:
197:
193:
189:
185:
181:
178:
174:
170:
166:
162:
158:
154:
147:
143:
139:
134:
124:
121:
113:
102:
99:
95:
92:
88:
85:
81:
78:
74:
71: –
70:
66:
65:Find sources:
59:
55:
49:
48:
43:This article
41:
37:
32:
31:
19:
627:
623:
613:
589:
552:
547:untranslated
546:
542:
536:
534:
519:
497:
488:
478:
440:untranslated
439:
435:
416:
412:
406:nucleotide (
400:exonucleases
385:
338:
310:
302:mitochondria
255:
246:
242:
212:
204:base pairing
195:
194:ring, and a
183:
165:nucleic acid
161:biochemistry
152:
151:
116:
107:
97:
90:
83:
76:
64:
52:Please help
47:verification
44:
503:nucleotides
427:transcribed
396:translation
381:phosphatase
373:DNA cloning
361:nucleotides
345:deoxyribose
286:start codon
280:toward its
219:polymerases
196:3′ end
184:5′ end
18:3' end
680:Categories
561:polyA tail
557:stop codon
485:sugar-ring
450:translated
404:methylated
341:sugar-ring
294:methionine
282:C-terminus
278:N-terminus
247:downstream
177:nucleotide
80:newspapers
530:adenosine
353:phosphate
188:phosphate
654:10882133
568:See also
559:and the
537:flanking
501:can use
489:tail end
481:hydroxyl
417:flanking
357:ligation
306:plastids
298:bacteria
274:ribosome
270:proteins
243:upstream
227:hydroxyl
138:furanose
110:May 2023
645:2811581
535:The 3′-
493:ligated
359:of two
343:of the
215:in vivo
94:scholar
652:
642:
598:
549:region
464:3′-end
442:region
419:region
349:ribose
327:5′-end
304:, and
192:ribose
96:
89:
82:
75:
67:
421:of a
258:sense
235:genes
155:, in
101:JSTOR
87:books
650:PMID
596:ISBN
543:The
456:and
446:mRNA
436:The
423:gene
413:The
319:and
308:use
159:and
144:and
73:news
696:RNA
686:DNA
640:PMC
632:doi
545:3′-
507:DNA
438:5′-
415:5′-
375:),
371:in
347:or
321:RNA
317:DNA
253:.)
173:RNA
171:or
169:DNA
146:RNA
142:DNA
56:by
682::
648:.
638:.
626:.
622:.
553:is
517:.
383:.
300:,
136:A
656:.
634::
628:5
604:.
311:N
296:(
123:)
117:(
112:)
108:(
98:·
91:·
84:·
77:·
50:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.