318:
33:
115:
214:(paired) or non-homologous chromosomes. Owing to the acrocentric nature of the chromosomes involved, the long arms of these chromosomes contain the majority of genetic material contained on the original chromosomes. The short arms also join to form a smaller reciprocal product, which typically contains only nonessential genes also present elsewhere in the genome, and is usually lost within a few
264:
A Robertsonian translocation results when the long arms of two acrocentric chromosomes fuse at the centromere and the two short arms are lost. If, for example, the long arms of chromosomes 13 and 14 fuse, no significant genetic material is lost—and the person is completely normal in spite of the
252:
A Robertsonian translocation in balanced form results in no excess or deficit of genetic material and causes no health difficulties. In unbalanced forms, Robertsonian translocations cause chromosomal deletions or addition and result in syndromes of multiple malformations, including trisomy 13
272:
Most people with
Robertsonian translocations have only 45 chromosomes in each of their cells, yet all essential genetic material is present, and they appear normal. Their children, however, may either be normal, carry the fusion chromosome (depending which chromosome is represented in the
105:
is present in our DNA in all four great apes this is split into two separate chromosomes typically numbered 2a and 2b. Similarly, the fact that horses have 64 chromosomes and donkeys 62, and that they can still have common, albeit usually infertile, offspring, may be due to a
Robertsonian
175:. Robertsonian translocations can only occur between chromosomes which have the centromere very close to one end. This means these chromosomes have a long arm which is particularly long, and a short arm which is particularly short. These are known as
288:
Rarely, the same translocation may be present homozygously if heterozygous parents with the same
Robertsonian translocation have children. The result may be viable offspring with 44 chromosomes. Outside of humans,
743:
Guarracino A, Buonaiuto S, Potapova T, Rhie A, Koren S, Rubinstein B, Fischer C, Gerton J, Phillippy A, Colonna V, Garrison E (2022). "Recombination between heterologous human acrocentric chromosomes".
75:. Robertsonian translocations result in a reduction in the number of chromosomes. A Robertsonian evolutionary fusion, which may have occurred in the common ancestor of humans and other
222:
visible, and can reduce chromosome number (in humans, from 23 to 22). However, the smaller chromosome carries so few essential genes that its loss is usually clinically insignificant.
359:
842:
Martinez-Castro P, Ramos MC, Rey JA, Benitez J, Sanchez Cascos A (1984). "Homozygosity for a
Robertsonian translocation (13q14q) in three offspring of heterozygous parents".
265:
translocation. Common
Robertsonian translocations are confined to the acrocentric chromosomes 13, 14, 15, 21 and 22, because the short arms of these chromosomes encode for
441:
238:
normal because there are two copies of all major chromosome arms and hence two copies of all essential genes. However, the progeny of this carrier may inherit an
369:, which includes band names, symbols and abbreviated terms used in the description of human chromosome and chromosome abnormalities. Abbreviations include
787:"Practice Guidelines for Communicating a Prenatal or Postnatal Diagnosis of Down Syndrome: Recommendations of the National Society of Genetic Counselors"
785:
Sheets KB, Crissman BG, Feist CD, Sell SL, Johnson LR, Donahue KC, Masser-Frye D, Brookshire GS, Carre AM, LaGrave D, Brasington CK (October 2011).
669:
613:
901:
Rajasekhar M, Rekharao RM, Shetty H, Gopinath PM, Satyamoorthy K (2010). "Cytogenetic
Analysis of 1400 Referral Cases: Manipal Experience".
1072:
325:
with annotated bands and sub-bands as used for the nomenclature of chromosome abnormalities. It shows dark and white regions as seen on
1067:
424:
E. Therman, B. Susman and C. Denniston. The nonrandom participation of human acrocentric chromosomes in
Robertsonian translocations.
59:
in humans, affecting 1 out of every 1,000 babies born. It does not usually cause medical problems, though some people may produce
474:"Centromere Destiny in Dicentric Chromosomes: New Insights from the Evolution of Human Chromosome 2 Ancestral Centromeric Region"
603:
230:
In humans, when a
Robertsonian translocation joins the long arm of chromosome 21 with the long arm of chromosomes 14 or 15, the
478:
448:
393:
345:
261:). The most frequent forms of Robertsonian translocations are between chromosomes 13 and 14, 14 and 21, and 14 and 15.
239:
64:
56:
317:
877:
48:
1062:
334:
211:
196:
192:
188:
184:
180:
290:
277:), or they may inherit a missing or extra long arm of an acrocentric chromosome (phenotype affected).
538:
79:, is the reason humans have 46 chromosomes while all other primates have 48. Detailed DNA studies of
1038:
926:
824:
767:
529:
278:
106:
evolutionary fusion at some point in the descent of today's donkeys from their common ancestor.
63:
with an incorrect number of chromosomes, resulting in a risk of miscarriage. In rare cases this
36:
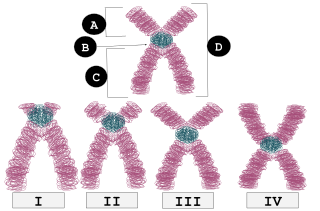
A Robertsonian translocation. The short arms of the chromosomes (shown on right) are often lost
1030:
1022:
983:
965:
946:"Uniparental disomy in Robertsonian translocations: strategies for uniparental disomy testing"
918:
859:
816:
808:
725:
707:
665:
609:
564:
525:"An alignment-free method to find and visualise rearrangements between pairs of DNA sequences"
505:
157:
1014:
973:
957:
910:
851:
798:
757:
749:
715:
554:
546:
495:
487:
363:
32:
282:
961:
542:
203:, the two resulting long arms may fuse. The result is a single, large chromosome with a
978:
720:
695:
559:
524:
500:
473:
390:
341:
254:
118:
Chromosome arms can have different length ratios. Robertsonian translocation occurs in
72:
1056:
771:
374:
258:
246:
215:
68:
1042:
930:
828:
114:
914:
366:
309:
have 62; it is thought that the difference is due to a
Robertsonian translocation.
231:
102:
803:
786:
753:
330:
302:
204:
200:
176:
172:
141:
119:
80:
52:
1026:
969:
945:
922:
812:
711:
472:
Chiatante G, Giannuzzi G, Calabrese FM, Eichler EE, Ventura M (1 July 2017).
491:
397:
386:
351:
337:
326:
322:
235:
84:
76:
17:
987:
820:
729:
633:
Chromosome studies. I. Taxonomic relationships shown in the chromosomes of
568:
509:
285:
is offered to families that may be carriers of chromosomal translocations.
1034:
1018:
863:
128:
in the image), where the short arms are fairly short but not very short.
373:
for
Robertsonian translocations. For example, rob(21;21)(q10;q10) causes
340:
chromosome pairs, both the female (XX) and male (XY) versions of the two
219:
1002:
207:
centromere. This form of rearrangement is a Robertsonian translocation.
762:
242:
88:
855:
550:
306:
298:
274:
92:
60:
586:
294:
149:
133:
99:
31:
582:
396:(1881–1941) who first described a Robertsonian translocation in
266:
179:
chromosomes. Humans have five of these acrocentric chromosomes:
95:
660:
Hartwell L, Hood L, Goldberg M, Reynolds A, Silver L (2011).
55:
become fused to each other. It is the most common form of
523:
Pratas D, Silva RM, Pinho AJ, Ferreira PJ (18 May 2015).
385:
Robertsonian translocations are named after the American
360:
International System for Human Cytogenomic Nomenclature
163:
All chromosomes in animals have a long arm (known as
1003:"Telomeres and mechanisms of Robertsonian fusion"
641:. V-shaped chromosomes and their significance in
442:"Unique: Rare Chromosome Disorder Support Group"
293:has 66 chromosomes, while both of domesticated
696:"Down Syndrome - Genetics and Cardiogenetics"
8:
664:. New York: McGraw-Hill. pp. 443, 454.
51:where the entire long arms of two different
977:
802:
761:
719:
558:
499:
684:Peter J. Russel; Essential Genetics 2003
362:(ISCN) is an international standard for
316:
199:. When these chromosomes break at their
113:
903:International Journal of Human Genetics
597:
595:
417:
210:This type of translocation may involve
882:Coriell Institute for Medical Research
909:(1–3). Kamla Raj Enterprises: 49–55.
269:which is present in multiple copies.
7:
878:"ISCN Symbols and Abbreviated Terms"
647:Gryllidae: chromosome and variation.
627:
625:
436:
434:
329:. Each row is vertically aligned at
171:), separated by a region called the
962:10.3978/j.issn.2224-4336.2014.03.03
662:Genetics From Genes to Genomes, 4e
25:
218:. This type of translocation is
479:Molecular Biology and Evolution
915:10.1080/09723757.2010.11886084
400:in 1916. They are also called
394:William Rees Brebner Robertson
1:
791:Journal of Genetic Counseling
406:centric-fusion translocations
27:Human chromosomal abnormality
1001:Slijepcevic P (1998-05-01).
694:Plaiasu V (September 2017).
167:) and a short arm (known as
602:Chowdhary BP (2013-01-22).
1089:
1073:Chromosomal translocations
349:
98:has determined that where
41:Robertsonian translocation
1068:Chromosomal abnormalities
804:10.1007/s10897-011-9375-8
754:10.1101/2022.08.15.504037
608:. John Wiley & Sons.
124:chromosome pairs (number
57:chromosomal translocation
950:Translational Pediatrics
649:J Morph 1916;27:179-331.
426:Annals of Human Genetics
402:whole-arm translocations
49:chromosomal abnormality
643:Acrididae, Locustidae
355:
160:
37:
1019:10.1007/s004120050289
944:Yip MY (April 2014).
492:10.1093/molbev/msx108
350:Further information:
320:
117:
35:
844:Cytogenet Cell Genet
346:mitochondrial genome
581:More details under
543:2015NatSR...510203P
333:level. It shows 22
530:Scientific Reports
356:
348:(at bottom left).
291:Przewalski's horse
279:Genetic counseling
257:) and trisomy 21 (
161:
38:
856:10.1159/000132080
671:978-0-07-352526-6
615:978-1-118-52212-7
551:10.1038/srep10203
344:, as well as the
158:Sister chromatids
134:Short arm (p arm)
16:(Redirected from
1080:
1047:
1046:
998:
992:
991:
981:
956:(2): 9807–9107.
941:
935:
934:
898:
892:
891:
889:
888:
874:
868:
867:
839:
833:
832:
806:
782:
776:
775:
765:
740:
734:
733:
723:
691:
685:
682:
676:
675:
657:
651:
629:
620:
619:
599:
590:
579:
573:
572:
562:
520:
514:
513:
503:
486:(7): 1669–1681.
469:
463:
462:
460:
459:
453:
447:. Archived from
446:
438:
429:
422:
364:human chromosome
150:Long arm (q arm)
127:
21:
1088:
1087:
1083:
1082:
1081:
1079:
1078:
1077:
1053:
1052:
1051:
1050:
1000:
999:
995:
943:
942:
938:
900:
899:
895:
886:
884:
876:
875:
871:
841:
840:
836:
784:
783:
779:
742:
741:
737:
693:
692:
688:
683:
679:
672:
659:
658:
654:
631:Robertson WRB.
630:
623:
616:
605:Equine Genomics
601:
600:
593:
580:
576:
522:
521:
517:
471:
470:
466:
457:
455:
451:
444:
440:
439:
432:
423:
419:
414:
383:
354:
342:sex chromosomes
315:
283:genetic testing
228:
152:
144:
136:
125:
112:
28:
23:
22:
15:
12:
11:
5:
1086:
1084:
1076:
1075:
1070:
1065:
1055:
1054:
1049:
1048:
1013:(2): 136–140.
993:
936:
893:
869:
834:
797:(5): 432–441.
777:
735:
706:(3): 208–213.
686:
677:
670:
652:
621:
614:
591:
574:
515:
464:
430:
428:1989;53:49-65.
416:
415:
413:
410:
391:cytogeneticist
382:
379:
314:
311:
255:Patau syndrome
236:phenotypically
227:
224:
216:cell divisions
111:
108:
73:Patau syndrome
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1085:
1074:
1071:
1069:
1066:
1064:
1061:
1060:
1058:
1044:
1040:
1036:
1032:
1028:
1024:
1020:
1016:
1012:
1008:
1004:
997:
994:
989:
985:
980:
975:
971:
967:
963:
959:
955:
951:
947:
940:
937:
932:
928:
924:
920:
916:
912:
908:
904:
897:
894:
883:
879:
873:
870:
865:
861:
857:
853:
849:
845:
838:
835:
830:
826:
822:
818:
814:
810:
805:
800:
796:
792:
788:
781:
778:
773:
769:
764:
759:
755:
751:
747:
739:
736:
731:
727:
722:
717:
713:
709:
705:
701:
697:
690:
687:
681:
678:
673:
667:
663:
656:
653:
650:
646:
642:
638:
634:
628:
626:
622:
617:
611:
607:
606:
598:
596:
592:
588:
584:
578:
575:
570:
566:
561:
556:
552:
548:
544:
540:
536:
532:
531:
526:
519:
516:
511:
507:
502:
497:
493:
489:
485:
481:
480:
475:
468:
465:
454:on 2019-02-18
450:
443:
437:
435:
431:
427:
421:
418:
411:
409:
407:
403:
399:
395:
392:
388:
380:
378:
376:
375:Down syndrome
372:
368:
365:
361:
353:
347:
343:
339:
336:
332:
328:
324:
319:
312:
310:
308:
304:
300:
296:
292:
286:
284:
280:
276:
270:
268:
262:
260:
259:Down syndrome
256:
250:
248:
247:Down syndrome
244:
241:
237:
233:
225:
223:
221:
220:cytologically
217:
213:
208:
206:
202:
198:
194:
190:
186:
182:
178:
174:
170:
166:
159:
155:
151:
147:
143:
139:
135:
131:
123:
122:
116:
109:
107:
104:
101:
97:
94:
90:
86:
82:
78:
74:
70:
69:Down syndrome
66:
65:translocation
62:
58:
54:
50:
46:
42:
34:
30:
19:
1063:Cytogenetics
1010:
1006:
996:
953:
949:
939:
906:
902:
896:
885:. Retrieved
881:
872:
850:(4): 310–2.
847:
843:
837:
794:
790:
780:
745:
738:
703:
699:
689:
680:
661:
655:
648:
644:
640:
636:
632:
604:
577:
534:
528:
518:
483:
477:
467:
456:. Retrieved
449:the original
425:
420:
405:
401:
398:grasshoppers
384:
370:
367:nomenclature
357:
313:Nomenclature
287:
271:
263:
251:
245:21, causing
232:heterozygous
229:
226:Consequences
209:
168:
164:
162:
153:
145:
137:
129:
120:
103:chromosome 2
44:
40:
39:
29:
18:Robertsonian
763:2117/393184
537:(1): 1203.
303:chromosomes
234:carrier is
205:metacentric
201:centromeres
177:acrocentric
121:acrocentric
67:results in
53:chromosomes
1057:Categories
1007:Chromosoma
887:2022-10-27
635:Tettigidae
458:2019-02-17
412:References
335:homologous
331:centromere
240:unbalanced
212:homologous
173:centromere
142:Centromere
81:chimpanzee
77:great apes
1027:1432-0886
970:2224-4344
923:0972-3757
813:1059-7700
772:251647679
712:1841-9038
639:Acrididae
387:zoologist
352:Karyotype
338:autosomal
327:G banding
323:karyotype
110:Mechanism
85:orangutan
1043:11712171
988:26835328
931:55971437
829:19308113
821:21618060
730:29218069
569:25984837
510:28333343
301:have 64
297:and the
1035:9601982
979:4729106
864:6510025
746:bioRxiv
721:5706761
700:Maedica
560:4434998
539:Bibcode
501:5722054
307:donkeys
243:trisomy
89:gorilla
61:gametes
47:) is a
1041:
1033:
1025:
986:
976:
968:
929:
921:
862:
827:
819:
811:
770:
728:
718:
710:
668:
612:
567:
557:
508:
498:
321:Human
299:tarpan
295:horses
275:gamete
93:bonobo
1039:S2CID
927:S2CID
825:S2CID
768:S2CID
587:Hinny
452:(PDF)
445:(PDF)
100:human
1031:PMID
1023:ISSN
984:PMID
966:ISSN
919:ISSN
860:PMID
817:PMID
809:ISSN
726:PMID
708:ISSN
666:ISBN
637:and
610:ISBN
585:and
583:Mule
565:PMID
506:PMID
389:and
381:Name
358:The
305:and
281:and
267:rRNA
195:and
96:apes
91:and
71:and
1015:doi
1011:107
974:PMC
958:doi
911:doi
852:doi
799:doi
758:hdl
750:doi
716:PMC
645:and
555:PMC
547:doi
496:PMC
488:doi
404:or
371:rob
249:.
45:ROB
1059::
1037:.
1029:.
1021:.
1009:.
1005:.
982:.
972:.
964:.
952:.
948:.
925:.
917:.
907:10
905:.
880:.
858:.
848:38
846:.
823:.
815:.
807:.
795:20
793:.
789:.
766:.
756:.
748:.
724:.
714:.
704:12
702:.
698:.
624:^
594:^
563:.
553:.
545:.
533:.
527:.
504:.
494:.
484:34
482:.
476:.
433:^
408:.
377:.
197:22
193:21
191:,
189:15
187:,
185:14
183:,
181:13
156::
148::
140::
132::
126:II
87:,
83:,
1045:.
1017::
990:.
960::
954:3
933:.
913::
890:.
866:.
854::
831:.
801::
774:.
760::
752::
732:.
674:.
618:.
589:.
571:.
549::
541::
535:5
512:.
490::
461:.
253:(
169:p
165:q
154:D
146:C
138:B
130:A
43:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.