17:
155:
and thus negatively affect the host cell. Additionally, with traditional gene insertion methods, scientists have had less ability to control where the newly inserted genes are located on the host cell chromosomes, which makes it difficult to predict inheritance of multiple genes from generation to generation. Minichromosome technology allows for the stacking of genes side-by-side on the same chromosome thus reducing likelihood of segregation of novel traits.
178:
The use of minichromosomes as a means for generating more desirable crop traits is actively being explored. Major advantages include the ability to introduce genetic information which is highly compatible with the host genome. This eliminates the risk of disrupting various important processes such as
154:
Unlike traditional methods of genetic engineering, minichromosomes can be used to transfer and express multiple sets of genes onto one engineered chromosome package. Traditional methods which involve the insertion of novel genes into existing sequences may result in the disruption of endogenous genes
145:
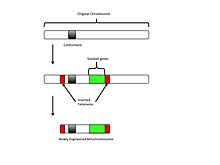
by selective transformation of telomeric sequences into a host genome. This insertion causes the generation of more telomeric sequences and eventual truncation. The newly synthesized truncated chromosome can then be altered through the insertion of new genes for desired traits. The top-down approach
163:
In 2006, scientists demonstrated the successful use of telomere truncation in maize plants to produce minichromosomes that could be utilized as a platform for inserting genes into the plant genome. In plants, the telomere sequence is conserved, which implies that this strategy can be utilized to
146:
is generally considered as the more plausible means of generating extra-numerary chromosomes for the use of genetic engineering of plants. In particular it is useful because their stability during cell division has been demonstrated. The limitation of this approach is that it is labor-intensive.
116:
into a host which is capable of assembling the components (typically yeast or mammalian cells) into a functional chromosome. This approach has been attempted for the introduction of minichromosomes into
179:
cell division and gene expression. With continued development, the future for use of minichromosomes may make a huge impact on the productivity of major crops.
72:(centromere, telomeres, and replication sequences), molecular biologists aim to construct a chromosomal platform which can be utilized to insert or present new
392:
Carlson, Shawn R.; Rudgers, Gary W.; Zieler, Helge; Mach, Jennifer M.; Luo, Song; Grunden, Eric; Krol, Cheryl; Copenhaver, Gregory P.; Preuss, Daphne (2007).
559:
262:
20:
Through the insertion of multiple genes and telomeres, a shortened minichromosome is produced, which can then be inserted into a host cell
187:
Minichromosomes have also been successfully inserted into yeast and animal cells. These minichromosomes were constructed using the
68:. By minimizing the amount of unnecessary genetic information on the chromosome and including the basic components necessary for
104:
The minimum constituent parts of a chromosome (centromere, telomeres, and DNA replication sequences) are assembled by using
200:
16:
113:
121:
for the possibility of genetic engineering, but success has been limited and questionable. In general, the
563:
598:
456:
53:
125:
approach is more difficult than the top-down method due to species incompatibility issues and the
523:
266:
45:
691:
667:
626:
541:
484:
425:
371:
105:
88:
Producing minichromosomes by genetic engineering techniques involves two primary methods, the
49:
350:"Plant artificial chromosome technology and its potential application in genetic engineering"
657:
616:
606:
531:
515:
474:
464:
415:
405:
361:
244:
205:
126:
69:
646:"Gene stacking in transgenic plants - the challenge for 21st century plant biotechnology"
602:
460:
621:
586:
536:
503:
479:
444:
420:
393:
298:
77:
52:. Minichromosomes may be created by natural processes as chromosomal aberrations or by
48:
but little additional genetic material. They replicate autonomously in the cell during
685:
662:
645:
210:
410:
394:"Meiotic Transmission of an in Vitro–Assembled Autonomous Maize Minichromosome"
248:
142:
37:
33:
324:
611:
469:
29:
671:
630:
545:
519:
488:
429:
375:
141:-mediated chromosomal truncation (TMCT). This process is the generation of
239:
Xu, Chunhui; Yu, Weichang (2009). "Engineered minichromosomes in plants".
164:
successfully construct additional minichromosomes in other plant species.
90:
138:
41:
560:"Researchers to study minichromosomes in maize with $ 1.9 million grant"
527:
502:
Houben, Andreas; Dawe, R. Kelly; Jiang, Jiming; Schubert, Ingo (2008).
325:"Minichromosomes: The Next Generation Technology for Plant Engineering"
366:
349:
167:
In 2007, scientists reported success in assembling minichromosomes
445:"Construction and behavior of engineered minichromosomes in maize"
118:
15:
299:"Minichromosomes: The second generation genetic engineering tool"
297:
Goyal, Aakash; Bhowmik, Pankaj Kumar; Basu, Saikat Kumar (2009).
73:
443:
Yu, W.; Han, F.; Gao, Z.; Vega, J. M.; Birchler, J. A. (2007).
65:
112:. Next, the desired contents of the minichromosome must be
64:
Minichromosomes can be either linear or circular pieces of
108:
techniques to construct the desired chromosomal contents
504:"Engineered Plant Minichromosomes: A Bottom-Up Success?"
348:
Yu, Weichang; Yau, Yuan-Yeu; Birchler, James A. (2016).
585:
Yu, W.; Lamb, J. C.; Han, F.; Birchler, J. A. (2006).
387:
385:
587:"Telomere-mediated chromosomal truncation in maize"
292:
290:
288:
286:
284:
591:Proceedings of the National Academy of Sciences
449:Proceedings of the National Academy of Sciences
343:
341:
323:Yu, Weichang; Birchler, James (August 2007).
8:
234:
232:
230:
228:
226:
661:
620:
610:
535:
478:
468:
419:
409:
365:
222:
94:(bottom-up) and the top-down approach.
137:This method utilizes the mechanism of
7:
201:Minichromosome maintenance proteins
14:
263:"Attach Genes To Minichromosomes"
663:10.1111/j.1467-7652.2004.00113.x
129:nature of centromeric regions.
1:
32:-like structure resembling a
411:10.1371/journal.pgen.0030179
650:Plant Biotechnology Journal
354:Plant Biotechnology Journal
211:Y chromosome, §Degeneration
150:Role in genetic engineering
708:
249:10.1036/1097-8542.YB090068
243:. McGraw-Hill Education.
644:Halpin, Claire (2005).
612:10.1073/pnas.0605750103
470:10.1073/pnas.0700932104
520:10.1105/tpc.107.056622
21:
508:The Plant Cell Online
19:
603:2006PNAS..10317331Y
461:2007PNAS..104.8924Y
306:Plant Omics Journal
54:genetic engineering
46:replication origins
36:and consisting of
22:
367:10.1111/pbi.12466
106:molecular cloning
50:cellular division
699:
676:
675:
665:
641:
635:
634:
624:
614:
582:
576:
575:
573:
571:
562:. Archived from
556:
550:
549:
539:
499:
493:
492:
482:
472:
440:
434:
433:
423:
413:
389:
380:
379:
369:
345:
336:
335:
333:
331:
320:
314:
313:
303:
294:
279:
278:
276:
274:
269:on June 10, 2010
265:. Archived from
259:
253:
252:
236:
707:
706:
702:
701:
700:
698:
697:
696:
682:
681:
680:
679:
643:
642:
638:
597:(46): 17331–6.
584:
583:
579:
569:
567:
566:on June 5, 2010
558:
557:
553:
501:
500:
496:
442:
441:
437:
404:(10): 1965–74.
391:
390:
383:
347:
346:
339:
329:
327:
322:
321:
317:
301:
296:
295:
282:
272:
270:
261:
260:
256:
238:
237:
224:
219:
206:Microchromosome
197:
185:
183:Other organisms
161:
152:
135:
127:heterochromatic
102:
86:
70:DNA replication
62:
12:
11:
5:
705:
703:
695:
694:
684:
683:
678:
677:
636:
577:
551:
494:
455:(21): 8924–9.
435:
381:
360:(5): 1175–82.
337:
315:
280:
254:
221:
220:
218:
215:
214:
213:
208:
203:
196:
193:
184:
181:
160:
157:
151:
148:
134:
131:
101:
96:
85:
82:
61:
58:
26:minichromosome
13:
10:
9:
6:
4:
3:
2:
704:
693:
690:
689:
687:
673:
669:
664:
659:
656:(2): 141–55.
655:
651:
647:
640:
637:
632:
628:
623:
618:
613:
608:
604:
600:
596:
592:
588:
581:
578:
565:
561:
555:
552:
547:
543:
538:
533:
529:
525:
521:
517:
513:
509:
505:
498:
495:
490:
486:
481:
476:
471:
466:
462:
458:
454:
450:
446:
439:
436:
431:
427:
422:
417:
412:
407:
403:
399:
398:PLOS Genetics
395:
388:
386:
382:
377:
373:
368:
363:
359:
355:
351:
344:
342:
338:
326:
319:
316:
311:
307:
300:
293:
291:
289:
287:
285:
281:
268:
264:
258:
255:
250:
246:
242:
241:AccessScience
235:
233:
231:
229:
227:
223:
216:
212:
209:
207:
204:
202:
199:
198:
194:
192:
190:
182:
180:
176:
174:
170:
165:
158:
156:
149:
147:
144:
140:
132:
130:
128:
124:
120:
115:
111:
107:
100:
97:
95:
93:
92:
83:
81:
79:
75:
71:
67:
59:
57:
55:
51:
47:
43:
39:
35:
31:
27:
18:
653:
649:
639:
594:
590:
580:
568:. Retrieved
564:the original
554:
511:
507:
497:
452:
448:
438:
401:
397:
357:
353:
328:. Retrieved
318:
309:
305:
271:. Retrieved
267:the original
257:
240:
188:
186:
177:
172:
168:
166:
162:
153:
136:
122:
109:
103:
98:
89:
87:
63:
25:
23:
514:(1): 8–10.
114:transformed
38:centromeres
28:is a small
217:References
191:approach.
171:using the
143:truncation
84:Production
34:chromosome
312:(1): 1–8.
78:host cell
60:Structure
42:telomeres
30:chromatin
692:Genetics
686:Category
672:17173615
631:17085598
570:15 April
546:18223035
528:25224208
489:17502617
430:17953486
376:26369910
330:11 April
273:12 April
195:See also
175:method.
169:in vitro
139:telomere
133:Top-down
110:in vitro
622:1859930
599:Bibcode
537:2254918
480:1885604
457:Bibcode
421:2041994
189:de novo
173:de novo
123:de novo
99:De novo
91:de novo
76:into a
670:
629:
619:
544:
534:
526:
487:
477:
428:
418:
374:
159:Plants
524:JSTOR
302:(PDF)
119:maize
74:genes
668:PMID
627:PMID
572:2012
542:PMID
485:PMID
426:PMID
372:PMID
332:2012
275:2012
44:and
658:doi
617:PMC
607:doi
595:103
532:PMC
516:doi
475:PMC
465:doi
453:104
416:PMC
406:doi
362:doi
245:doi
66:DNA
688::
666:.
652:.
648:.
625:.
615:.
605:.
593:.
589:.
540:.
530:.
522:.
512:20
510:.
506:.
483:.
473:.
463:.
451:.
447:.
424:.
414:.
400:.
396:.
384:^
370:.
358:14
356:.
352:.
340:^
308:.
304:.
283:^
225:^
80:.
56:.
40:,
24:A
674:.
660::
654:3
633:.
609::
601::
574:.
548:.
518::
491:.
467::
459::
432:.
408::
402:3
378:.
364::
334:.
310:2
277:.
251:.
247::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.