310:. Proteins encoded by aberrantly spliced pre-mRNAs are functionally different and contribute to the characteristic anomalies exhibited by cancer cells, including their ability to proliferate, invade and undergo angiogenesis, and metastasis. Minigenes help researchers identify genetic mutations in cancer that result in splicing errors and determine the downstream effects those splicing errors have on gene expression. Using knowledge obtained from studies employing minigenes, oncologists have proposed tests designed to detect products of abnormal gene expression for diagnostic purposes. Additionally, the prospect of using minigenes as a
278:. Tau protein isoforms are created by alternative splicing of exons 2, 3 and 10. The regulation of tau splicing is specific to stage of development, physiology and location. Errors in tau splicing can occur in both exons and introns and, depending on the error, result in changes to protein structure or loss of function. Aggregation of these abnormal tau proteins correlates directly with pathogenesis and disease progression. Minigenes have been used by several researchers to help understand the regulatory components responsible for mRNA splicing of the TAU gene.
150:" at 3' sense and anti-sense strands (Step 2). These "sticky-ends" can be easily incorporated into a TOPO Vector by ligation into a commercially available source which has ligase already attached to it at the sight of incorporation (Step 3). The subsequent TOPO Vectors can be transfected into E.coli cells (Step 4). After incubation, total RNA can be extracted from the bacterial colonies and analyzed using
213:. Minigenes have been applied to the study of a diverse array of genetic diseases due to the aforementioned abundance of alternatively spliced genes and the specificity and variation observed in splicing regulation. The following are examples of minigene use in various diseases. While it is not an exhaustive list, it does provide a better understanding of how minigenes are utilized.
76:
167:
121:
final size of the fragment, which is in turn a reflection of the complexity of the minigene itself. The number of foreign DNA elements (exon and introns) inserted into the constitutive exons and introns of a given fragment varies with the type of experiment and the information being sought. A typical experiment might involve
89:
Minigenes were first described as the somatic assembly of DNA segments and consisted of DNA regions known to encode the protein and the flanking regions required to express the protein. The term was first used in a paper in 1977 to describe the cloning of two minigenes that were designed to express a
241:
called isolated growth hormone deficiency (IGHD), a disease that results in growth failure. IGHD type II is an autosomal dominant form caused by a mutation in the intervening sequence (IVS) adjacent to exon 3 of the gene encoding growth hormone 1, the GH-1 gene. This mutated form of IVS3 causes exon
93:
RNA splicing was discovered in the late 1970s through the study of adenoviruses that invade mammals and replicate inside them. Researchers identified RNA molecules that contained sequences from noncontiguous parts of the virus’s genome. This discovery led to the conclusion that regulatory mechanisms
120:
and widespread use of computers, several good programs now exist for the identification of cis-acting control regions that affect the splicing outcomes of a gene and advanced programs can even consider splicing outcomes in various tissue types. Differences in minigenes are usually reflected in the
154:
to quantify ratios of exon inclusion/exclusion (step 5). The minigene can be transfected into different cell types with various splicing factors to test trans-acting elements (Step 6). The expressed genes or the proteins they encode can be analyzed to evaluate splicing components and their effects
186:
and identify potential targets of therapeutic intervention in these diseases, explicating the splicing elements involved is essential. Determining the complete set of components involved in splicing presents many challenges due to the abundance of alternative splicing, which occurs in most human
1047:
Anfossi, M; Vuono, R; Maletta, R; Virdee, K; Mirabelli, M; Colao, R; Puccio, G; Bernardi, L; Frangipane, F; Gallo, M; Geracitano, S; Tomaino, C; Curcio, SA; Zannino, G; Lamenza, F; Duyckaerts, C; Spillantini, MG; Losso, MA; Bruni, AC (2011). "Compound heterozygosity of 2 novel MAPT mutations in
79:
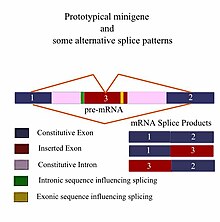
pre-mRNA splice patterns showing constitutive exons and introns and the inserted fragment. Orange lines show alternative splicing outcomes as dictated by the exonic sequences and intronic sequences (yellow and green bands) that influence splicing. These sequences may be splicing enhancers or
1134:
Adler, AS; McCleland, ML; Yee, S; Yaylaoglu, M; Hussain, S; Cosino, E; Quinones, G; Modrusan, Z; Seshagiri, S; Torres, E; Chopra, VS; Haley, B; Zhang, Z; Blackwood, EM; Singh, M; Junttila, M; Stephan, JP; Liu, J; Pau, G; Fearon, ER; Jiang, Z; Firestein, R (May 2014).
94:
existed which affected mature RNA and the genes it expresses. Using minigenes as a splice reporting vector to explore the effects of RNA splicing regulation naturally followed and remains the major use of minigenes to date.
141:
Once a suitable genomic fragment is chosen (Step 1), the exons and introns of the fragment can be inserted and amplified, along with the flanking constitutive exons and introns of the original gene, by
129:
which replace the wild-type gene and have been cloned into the same flanking sequences as the original fragment. These types of experiments help to determine the effect of various mutations on
629:
Grodecká, Lucie; Lockerová, Pavla; Ravčuková, Barbora; Buratti, Emanuele; Baralle, Francisco E.; Dušek, Ladislav; Freiberger, Tomáš; Spilianakis, Charalampos Babis (21 February 2014).
246:
within an intron splice enhancer (ISE) embedded in IVS3 was to blame for the skipping of E3. Moreover, it was determined that the function of the ISE is influenced by a nearby
110:
gene, i.e., the length of the fragment must include all upstream and downstream sequences which can affect its splicing. Therefore, most minigene designs begin with a thorough
1299:"Functional analysis of a large set of BRCA2 exon 7 variants highlights the predictive value of hexamer scores in detecting alterations of exonic splicing regulatory elements"
499:
Poonian, MS; McComas, WW; Nussbaum, AL (1977). "Chemical synthesis of two deoxyribododecanucleotides for the attachment of restriction termini to an artificial minigene".
1189:"HnRNP A1/A2 and SF2/ASF Regulate Alternative Splicing of Interferon Regulatory Factor-3 and Affect Immunomodulatory Functions in Human Non-Small Cell Lung Cancer Cells"
247:
286:
Cancer is a complex, heterogeneous disease that can be hereditary or the result of environmental stimuli. Minigenes are used to help oncologists understand the roles
290:
splicing plays in different cancer types. Of particular interest are cancer specific genetic mutations that disrupt normal splicing events, including those affecting
242:
3 to be skipped in the mRNA product. The mRNA (-E3) encodes a truncated form of hGH that then inhibits normal hGH secretion. Minigenes were used to determine that a
739:
191:. Splicing is distinctly conducted from cell type to cell type and across different stages of cellular development. Therefore, it is critical that any
299:
475:
225:. These effects on hormones have been identified as the cause of many endocrine disorders including thyroid-related pathological conditions,
374:
Stoss, O; Stoilov, P; Hartmann, AM; Nayler, O; Stamm, S (Dec 1999). "The in vivo minigene approach to analyze tissue-specific splicing".
56:
vectors) and act as a probe to determine which factors are important in splicing outcomes. They can be constructed to test the way both
1453:
102:
In order to provide a good minigene model, the gene fragment should have all of the necessary elements to ensure it exhibits the same
28:
gene fragment. This is a minigene in its most basic sense. More complex minigenes can be constructed containing multiple exons and
1091:
Rajan, P.; Elliott, DJ; Robson, CN; Leung, HY (Aug 2009). "Alternative splicing and biological heterogeneity in prostate cancer".
998:
Rodriguez-Martin, Teresa; Karen
Anthony; Mariano A. Garcia-Blanco; S. Gary Mansfield; Brian H. Anderton; Jean-Marc Gallo (2009).
688:
Barash, Yoseph; Vaquero-Garcia, Jorge; González-Vallinas, Juan; Xiong, Hui; Gao, Weijun; Lee, Leo J.; Frey, Brendan J. (2013).
946:
Kar, Amar; Fushimi, Kazuo; Zhou, Xiaohong; Ray, Payal; Shi, Chen; Chen, Xiaoping; Liu, Zhiren; Chen, She; Wu, Jane Y. (2011).
1483:
1473:
1463:
234:
237:. One specific example of a splicing error causing an endocrine disease that has been studied using minigenes is a type of
1246:
Acedo, Alberto; David J Sanz; Mercedes Durán; Mar
Infante; Lucía Pérez-Cabornero; Cristina Miner; Eladio A Velasco (2012).
230:
160:
1504:
554:
1550:
1346:
Daniotti, Jose L.; Aldo A. Vilcaes; Vanina Torres
Demichelis; Fernando M. Ruggiero; Macarena Rodriguez-Walker (2013).
766:"Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes"
1545:
1530:
156:
49:
948:"RNA Helicase p68 (DDX5) Regulates tau Exon 10 Splicing by Modulating a Stem-Loop Structure at the 5′ Splice Site"
1495:
827:
764:
Lim, Kian; Huat; Ferraris, Luciana; Filloux, Madeleine E.; Raphael, Benjamin J.; Fairbrother, William G. (2011).
238:
143:
61:
221:
RNA splicing errors can have drastic effects on how proteins function, including the hormones secreted by the
1000:"Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing"
1540:
1448:"Alternative pre-mRNA Splicing: Theory and Protocols", by Stefan Stamm, Chris Smith and Reinhard Lührmann
271:
267:
125:
minigenes which are expected to express genes normally in a comparison run against genetically engineered
116:
analysis of the requirements of the experiment before any "wet" lab work is conducted. With the advent of
57:
1478:"Alternative Splicing and Disease (Progress in Molecular and Subcellular Biology)" by Philippe Jeanteur
420:
Cooper, Thomas A. (December 2005). "Use of minigene systems to dissect alternative splicing elements".
1200:
777:
642:
311:
103:
1137:"An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth"
295:
1248:"Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes"
1328:
1116:
1073:
1348:"Glycosylation of Glycolipids in Cancer: Basis for Development of Novel Therapeutic Approaches"
1479:
1469:
1459:
1449:
1428:
1379:
1320:
1279:
1228:
1166:
1108:
1065:
1029:
977:
925:
876:
805:
721:
670:
611:
516:
481:
471:
437:
391:
263:
1555:
1418:
1410:
1397:
Aurisicchio, L; Fridman, A; Bagchi, A; Scarselli, E; La Monica, N; Ciliberto, G (Jan 2014).
1369:
1359:
1310:
1269:
1259:
1218:
1208:
1156:
1148:
1100:
1057:
1019:
1011:
967:
959:
915:
907:
866:
795:
785:
711:
701:
660:
650:
601:
593:
580:
Divina, Petr; Kvitkovicova, Andrea; Buratti, Emanuele; Vorechovsky, Igor (14 January 2009).
508:
463:
429:
383:
222:
1458:"Molecular Diagnostics, Second edition", by Ed. by George P. Patrinos and Whilhelm Ansorge
582:"Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping"
1508:
1297:
Di
Giacomo, D.; Gaildrat, P; Abuli, A; Abdat, J; Frébourg, T; Tosi, M; Martins, A (2013).
323:
69:
1061:
458:
Desviat, LR; Pérez, B; Ugarte, M (2012). "Minigenes to
Confirm Exon Skipping Mutations".
1204:
781:
646:
48:
biochemically assessed experiments. Specifically, minigenes are used as splice reporter
1535:
1423:
1398:
1374:
1347:
1274:
1247:
1223:
1188:
1161:
1136:
1024:
999:
972:
947:
920:
895:
800:
765:
716:
689:
665:
631:"Exon First Nucleotide Mutations in Splicing: Evaluation of In Silico Prediction Tools"
630:
606:
581:
348:
287:
243:
117:
387:
24:
and the control regions necessary for the gene to express itself in the same way as a
1524:
512:
53:
1332:
1120:
871:
854:
250:, revealing that this particular splicing error is caused by a trans-acting factor.
1077:
855:"Splicing variants impact in thyroid normal physiology and pathological conditions"
343:
328:
183:
179:
33:
182:
errors have been estimated to occur in a third of genetic diseases. To understand
1213:
655:
467:
433:
291:
259:
147:
706:
1501:
558:
303:
1364:
1104:
790:
690:"AVISPA: a web tool for the prediction and analysis of alternative splicing"
275:
166:
122:
112:
107:
25:
1432:
1383:
1324:
1283:
1232:
1187:
Guo, Rong; Yong Li; Jinying Ning; Dan Sun; Lianjun Lin; Xinmin Liu (2013).
1170:
1152:
1112:
1069:
1033:
981:
929:
880:
809:
725:
674:
615:
485:
441:
395:
911:
831:
597:
1015:
963:
520:
193:
130:
65:
44:
80:
silencers, polypyrimidine tract binding protein sites or other elements.
1414:
1315:
1298:
226:
38:
29:
338:
209:-regulatory elements and other regulators of pre-mature RNA splicing
197:
or bioinformatic assumptions about splicing regulation are confirmed
151:
126:
1264:
75:
534:
Clancy, S (2008). "RNA splicing: introns, exons and spliceosome".
307:
165:
74:
333:
21:
32:. Minigenes provide a valuable tool for researchers evaluating
187:
genes, and the specificity in which splicing is carried out
1399:"A novel minigene scaffold for therapeutic cancer vaccines"
1514:
993:
991:
146:. The primers for PCR can be chosen so that they leave "
1517:. Large database for retrieving information on genes.
1496:
Stefan Stamm's web page at the
University of Kentucky
462:. Methods Mol. Biol. Vol. 867. pp. 37–47.
1511:. A Good site for theoretical analysis of splicing.
300:heterogeneous nuclear ribonucleoparticules (hnRNP)
896:"Genetics of isolated growth hormone deficiency"
1182:
1180:
770:Proceedings of the National Academy of Sciences
941:
939:
8:
170:A typical cycle for constructing a minigene.
20:is a minimal gene fragment that includes an
1502:Christopher Burge's Lab at M.I.T. website
1422:
1373:
1363:
1314:
1273:
1263:
1222:
1212:
1160:
1023:
971:
919:
870:
799:
789:
715:
705:
664:
654:
605:
453:
451:
821:
819:
415:
413:
411:
409:
407:
405:
376:Brain Research. Brain Research Protocols
1468:"DNA Vaccines" edited by Hildegun Ertl
369:
367:
365:
363:
359:
853:Rosaria de Miranda, Elizabete (2009).
1498:. Good overview of minigene research.
7:
1062:10.1016/j.neurobiolaging.2010.12.013
201:. Minigenes are used to elucidate
155:via a variety of methods including
740:"Steps in producing a TOPO Vector"
586:European Journal of Human Genetics
304:serine/arginine-rich (SR) proteins
14:
308:small ribonucleoproteins (snRNP)
872:10.1590/S0004-27302009000600003
235:congenital adrenal hyperplasia
1:
900:J Clin Res Pediatr Endocrinol
388:10.1016/s1385-299x(99)00043-4
231:hyperinsulinemic hypoglycemia
161:size-exclusion chromatography
1214:10.1371/journal.pone.0062729
656:10.1371/journal.pone.0089570
513:10.1016/0378-1119(77)90040-3
859:Arq Bras Endocrinol Metabol
468:10.1007/978-1-61779-767-5_3
434:10.1016/j.ymeth.2005.07.015
1572:
1048:frontotemporal dementia".
707:10.1186/gb-2013-14-10-r114
274:diseases as well as other
264:neurodegenerative diseases
254:Neurodegenerative diseases
72:) affect gene expression.
239:growth hormone deficiency
104:alternative splicing (AS)
62:trans-regulatory elements
1365:10.3389/fonc.2013.00306
1105:10.1038/nrurol.2009.125
791:10.1073/pnas.1101135108
248:transposable AC element
58:cis-regulatory elements
1252:Breast Cancer Research
1153:10.1101/gad.237206.113
205:-regulatory elements,
171:
81:
1056:(4): 757.e1–757.e11.
912:10.4274/jcrpe.v2i2.52
598:10.1038/ejhg.2008.257
169:
78:
964:10.1128/MCB.01149-10
555:"Burge Lab Software"
553:Burge, Christopher.
312:cancer immunotherapy
296:RNA-binding proteins
1515:UCSC Genome Browser
1205:2013PLoSO...862729G
894:Mullis, PE (2010).
782:2011PNAS..10811093H
647:2014PLoSO...989570G
314:is being explored.
262:is associated with
1551:Molecular genetics
1507:2019-03-30 at the
1415:10.4161/onci.27529
1316:10.1002/humu.22428
1016:10.1093/hmg/ddp264
834:on 9 December 2013
217:Endocrine diseases
172:
127:allelic variations
82:
60:(RNA effects) and
1546:Synthetic biology
1531:Molecular biology
1010:(17): 3266–3273.
477:978-1-61779-766-8
1563:
1437:
1436:
1426:
1394:
1388:
1387:
1377:
1367:
1343:
1337:
1336:
1318:
1294:
1288:
1287:
1277:
1267:
1243:
1237:
1236:
1226:
1216:
1184:
1175:
1174:
1164:
1131:
1125:
1124:
1088:
1082:
1081:
1044:
1038:
1037:
1027:
995:
986:
985:
975:
958:(9): 1812–1821.
943:
934:
933:
923:
891:
885:
884:
874:
850:
844:
843:
841:
839:
830:. Archived from
828:"Stamms-lab.net"
823:
814:
813:
803:
793:
761:
755:
754:
752:
750:
736:
730:
729:
719:
709:
685:
679:
678:
668:
658:
626:
620:
619:
609:
577:
571:
570:
568:
566:
561:on 30 March 2019
557:. Archived from
550:
544:
543:
536:Nature Education
531:
525:
524:
496:
490:
489:
455:
446:
445:
417:
400:
399:
371:
258:Accumulation of
223:endocrine system
106:patterns as the
70:splicing factors
1571:
1570:
1566:
1565:
1564:
1562:
1561:
1560:
1521:
1520:
1509:Wayback Machine
1492:
1445:
1443:Further reading
1440:
1396:
1395:
1391:
1345:
1344:
1340:
1309:(11): 1547–57.
1296:
1295:
1291:
1265:10.1186/bcr3202
1245:
1244:
1240:
1186:
1185:
1178:
1147:(10): 1068–84.
1133:
1132:
1128:
1090:
1089:
1085:
1050:Neurobiol Aging
1046:
1045:
1041:
997:
996:
989:
952:Mol. Cell. Biol
945:
944:
937:
893:
892:
888:
852:
851:
847:
837:
835:
826:Stamm, Stefan.
825:
824:
817:
776:(27): 11093–6.
763:
762:
758:
748:
746:
738:
737:
733:
687:
686:
682:
628:
627:
623:
579:
578:
574:
564:
562:
552:
551:
547:
533:
532:
528:
507:(5–6): 357–72.
498:
497:
493:
478:
457:
456:
449:
419:
418:
403:
373:
372:
361:
357:
324:Recombinant DNA
320:
294:components and
284:
256:
219:
177:
139:
100:
87:
12:
11:
5:
1569:
1567:
1559:
1558:
1553:
1548:
1543:
1538:
1533:
1523:
1522:
1519:
1518:
1512:
1499:
1491:
1490:External links
1488:
1487:
1486:
1476:
1466:
1456:
1454:978-3527326068
1444:
1441:
1439:
1438:
1403:Oncoimmunology
1389:
1338:
1289:
1238:
1176:
1126:
1099:(8): 454–460.
1083:
1039:
987:
935:
886:
865:(6): 709–714.
845:
815:
756:
731:
694:Genome Biology
680:
621:
592:(6): 759–765.
572:
545:
526:
491:
476:
447:
428:(4): 331–340.
401:
358:
356:
353:
352:
351:
349:Cloning vector
346:
341:
336:
331:
326:
319:
316:
283:
280:
255:
252:
244:point mutation
218:
215:
176:
173:
138:
135:
118:Bioinformatics
99:
96:
86:
83:
36:patterns both
13:
10:
9:
6:
4:
3:
2:
1568:
1557:
1554:
1552:
1549:
1547:
1544:
1542:
1541:Biotechnology
1539:
1537:
1534:
1532:
1529:
1528:
1526:
1516:
1513:
1510:
1506:
1503:
1500:
1497:
1494:
1493:
1489:
1485:
1481:
1477:
1475:
1471:
1467:
1465:
1461:
1457:
1455:
1451:
1447:
1446:
1442:
1434:
1430:
1425:
1420:
1416:
1412:
1409:(1): e27529.
1408:
1404:
1400:
1393:
1390:
1385:
1381:
1376:
1371:
1366:
1361:
1357:
1353:
1349:
1342:
1339:
1334:
1330:
1326:
1322:
1317:
1312:
1308:
1304:
1300:
1293:
1290:
1285:
1281:
1276:
1271:
1266:
1261:
1257:
1253:
1249:
1242:
1239:
1234:
1230:
1225:
1220:
1215:
1210:
1206:
1202:
1199:(4): e62729.
1198:
1194:
1190:
1183:
1181:
1177:
1172:
1168:
1163:
1158:
1154:
1150:
1146:
1142:
1138:
1130:
1127:
1122:
1118:
1114:
1110:
1106:
1102:
1098:
1094:
1087:
1084:
1079:
1075:
1071:
1067:
1063:
1059:
1055:
1051:
1043:
1040:
1035:
1031:
1026:
1021:
1017:
1013:
1009:
1005:
1004:Hum Mol Genet
1001:
994:
992:
988:
983:
979:
974:
969:
965:
961:
957:
953:
949:
942:
940:
936:
931:
927:
922:
917:
913:
909:
905:
901:
897:
890:
887:
882:
878:
873:
868:
864:
860:
856:
849:
846:
833:
829:
822:
820:
816:
811:
807:
802:
797:
792:
787:
783:
779:
775:
771:
767:
760:
757:
745:
744:Life sciences
741:
735:
732:
727:
723:
718:
713:
708:
703:
699:
695:
691:
684:
681:
676:
672:
667:
662:
657:
652:
648:
644:
641:(2): e89570.
640:
636:
632:
625:
622:
617:
613:
608:
603:
599:
595:
591:
587:
583:
576:
573:
560:
556:
549:
546:
541:
537:
530:
527:
522:
518:
514:
510:
506:
502:
495:
492:
487:
483:
479:
473:
469:
465:
461:
460:Exon Skipping
454:
452:
448:
443:
439:
435:
431:
427:
423:
416:
414:
412:
410:
408:
406:
402:
397:
393:
389:
385:
382:(3): 383–94.
381:
377:
370:
368:
366:
364:
360:
354:
350:
347:
345:
342:
340:
337:
335:
332:
330:
327:
325:
322:
321:
317:
315:
313:
309:
305:
301:
297:
293:
289:
281:
279:
277:
273:
269:
265:
261:
253:
251:
249:
245:
240:
236:
232:
228:
224:
216:
214:
212:
208:
204:
200:
196:
195:
190:
185:
181:
174:
168:
164:
162:
158:
157:hybridization
153:
149:
145:
136:
134:
132:
128:
124:
119:
115:
114:
109:
105:
97:
95:
91:
84:
77:
73:
71:
67:
63:
59:
55:
54:exon-trapping
52:(also called
51:
47:
46:
41:
40:
35:
31:
27:
23:
19:
1406:
1402:
1392:
1355:
1351:
1341:
1306:
1302:
1292:
1255:
1251:
1241:
1196:
1192:
1144:
1140:
1129:
1096:
1093:Nat Rev Urol
1092:
1086:
1053:
1049:
1042:
1007:
1003:
955:
951:
906:(2): 52–62.
903:
899:
889:
862:
858:
848:
836:. Retrieved
832:the original
773:
769:
759:
747:. Retrieved
743:
734:
700:(10): R114.
697:
693:
683:
638:
634:
624:
589:
585:
575:
563:. Retrieved
559:the original
548:
539:
535:
529:
504:
500:
494:
459:
425:
421:
379:
375:
344:Transfection
329:RNA Splicing
285:
257:
220:
210:
206:
202:
198:
192:
188:
184:pathogenesis
180:RNA splicing
178:
140:
137:Construction
111:
101:
92:
88:
64:(associated
43:
37:
17:
15:
1352:Front Oncol
292:spliceosome
276:tauopathies
272:Parkinson's
268:Alzheimer's
260:tau protein
148:sticky ends
1525:Categories
1484:3540344489
1474:1461349257
1464:0123745373
1303:Hum. Mutat
1258:(3): R87.
355:References
266:including
133:splicing.
1141:Genes Dev
123:wild type
113:in silico
108:wild type
90:peptide.
30:intron(s)
26:wild type
1505:Archived
1433:24790791
1384:24392350
1333:22874730
1325:23983145
1284:22632462
1233:23658645
1193:PLOS ONE
1171:24788092
1121:30664940
1113:19657379
1070:21295377
1034:19498037
982:21343338
930:21274339
881:19893912
838:26 March
810:21685335
726:24156756
675:24586880
635:PLOS ONE
616:19142208
486:22454053
442:16314262
396:10592349
318:See also
298:such as
288:pre-mRNA
194:in vitro
131:pre-mRNA
66:proteins
45:in vitro
34:splicing
18:minigene
1556:Cloning
1424:4002591
1375:3867695
1358:: 306.
1275:3446350
1224:3639176
1201:Bibcode
1162:4035536
1078:5176440
1025:2722988
973:3133221
921:3014602
801:3131313
778:Bibcode
717:4014802
666:3931810
643:Bibcode
607:2947103
422:Methods
227:rickets
211:in vivo
199:in vivo
189:in vivo
85:History
50:vectors
39:in vivo
1482:
1472:
1462:
1452:
1431:
1421:
1382:
1372:
1331:
1323:
1282:
1272:
1231:
1221:
1169:
1159:
1119:
1111:
1076:
1068:
1032:
1022:
980:
970:
928:
918:
879:
808:
798:
724:
714:
673:
663:
614:
604:
521:590743
519:
484:
474:
440:
394:
339:Intron
282:Cancer
152:RT-PCR
1536:Genes
1329:S2CID
1117:S2CID
1074:S2CID
749:7 May
565:7 May
542:(31).
207:trans
98:Types
1480:ISBN
1470:ISBN
1460:ISBN
1450:ISBN
1429:PMID
1380:PMID
1321:PMID
1280:PMID
1229:PMID
1167:PMID
1109:PMID
1066:PMID
1030:PMID
978:PMID
926:PMID
877:PMID
840:2014
806:PMID
751:2014
722:PMID
671:PMID
612:PMID
567:2014
517:PMID
501:Gene
482:PMID
472:ISBN
438:PMID
392:PMID
334:Exon
306:and
270:and
233:and
175:Uses
42:and
22:exon
1419:PMC
1411:doi
1370:PMC
1360:doi
1311:doi
1270:PMC
1260:doi
1219:PMC
1209:doi
1157:PMC
1149:doi
1101:doi
1058:doi
1020:PMC
1012:doi
968:PMC
960:doi
916:PMC
908:doi
867:doi
796:PMC
786:doi
774:108
712:PMC
702:doi
661:PMC
651:doi
602:PMC
594:doi
509:doi
464:doi
430:doi
384:doi
203:cis
159:or
144:PCR
1527::
1427:.
1417:.
1405:.
1401:.
1378:.
1368:.
1354:.
1350:.
1327:.
1319:.
1307:34
1305:.
1301:.
1278:.
1268:.
1256:14
1254:.
1250:.
1227:.
1217:.
1207:.
1195:.
1191:.
1179:^
1165:.
1155:.
1145:28
1143:.
1139:.
1115:.
1107:.
1095:.
1072:.
1064:.
1054:32
1052:.
1028:.
1018:.
1008:18
1006:.
1002:.
990:^
976:.
966:.
956:31
954:.
950:.
938:^
924:.
914:.
902:.
898:.
875:.
863:53
861:.
857:.
818:^
804:.
794:.
784:.
772:.
768:.
742:.
720:.
710:.
698:14
696:.
692:.
669:.
659:.
649:.
637:.
633:.
610:.
600:.
590:17
588:.
584:.
538:.
515:.
503:.
480:.
470:.
450:^
436:.
426:37
424:.
404:^
390:.
378:.
362:^
302:,
229:,
163:.
16:A
1435:.
1413::
1407:3
1386:.
1362::
1356:3
1335:.
1313::
1286:.
1262::
1235:.
1211::
1203::
1197:8
1173:.
1151::
1123:.
1103::
1097:6
1080:.
1060::
1036:.
1014::
984:.
962::
932:.
910::
904:2
883:.
869::
842:.
812:.
788::
780::
753:.
728:.
704::
677:.
653::
645::
639:9
618:.
596::
569:.
540:1
523:.
511::
505:1
488:.
466::
444:.
432::
398:.
386::
380:4
68:/
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.