430:
65:
Degeneracy results because there are more codons than encodable amino acids. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required (20 amino acids plus stop) and the next largest number of bases is three, then 4³
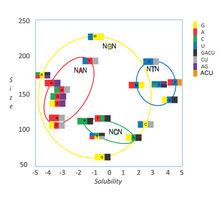
453:). For example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids, NCN yields amino acid residues that are small in size and moderate in hydropathy, and NAN encodes average size hydrophilic residues. These tendencies may result from the shared ancestry of the
107:
A position is said to be non-degenerate if any mutation at this position changes the amino acid. For example, all three positions of methionine's AUG are non-degenerate, because the only codon coding for methionine is AUG. The same goes for tryptophan's UGG.
46:
Degeneracy of the genetic code was identified by
Lagerkvist. For instance, codons GAA and GAG both specify glutamic acid and exhibit redundancy; but, neither specifies any other amino acid and thus are not ambiguous or demonstrate no ambiguity.
50:
The codons encoding one amino acid may differ in any of their three positions; however, more often than not, this difference is in the second or third position. For instance, the amino acid
418:
restricts this in practice in many organisms; twofold degenerate codons can withstand silence mutation rather than
Missense or Nonsense point mutations at the third position. Since
34:, exhibited as the multiplicity of three-base pair codon combinations that specify an amino acid. The degeneracy of the genetic code is what accounts for the existence of
441:
A practical consequence of redundancy is that some errors in the genetic code cause only a synonymous mutation, or an error that would not affect the protein because the
426:(purine to pyrimidine or vice versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.
85:
of four possible nucleotides (A, C, G, T) at this position specify the same amino acid. A nucleotide substitution at a 4-fold degenerate site is always a
748:
916:
547:
481:
501:
800:
780:
888:
638:
655:
683:"The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems"
1042:
986:
981:
1133:
1069:
1000:
741:
805:
1123:
414:. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although
58:
is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position); and the amino acid
1020:
454:
1128:
1064:
852:
765:
734:
1025:
842:
827:
96:
on some of the substitutions. An example (and the only) 3-fold degenerate site is the third position of an
1030:
955:
847:
93:
945:
930:
810:
450:
419:
136:
582:"Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon"
1052:
950:
868:
460:
These variable codes for amino acids are allowed because of modified bases in the first base of the
878:
86:
62:
is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second, or third position).
35:
1013:
896:
502:"The Information in DNA Determines Cellular Function via Translation | Learn Science at Scitable"
1097:
712:
634:
611:
543:
429:
702:
694:
601:
593:
465:
434:
415:
123:. Only two amino acids are specified by a single codon each. One of these is the amino-acid
1074:
911:
757:
127:, specified by the codon AUG, which also specifies the start of translation; the other is
422:
mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than
901:
873:
707:
682:
667:
606:
581:
411:
54:
is specified by GAA and GAG codons (difference in the third position); the amino acid
1117:
1102:
789:
51:
976:
906:
775:
423:
31:
698:
1079:
1008:
446:
442:
104:. In computation, this position is often treated as a twofold degenerate site.
1037:
656:"Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering"
563:
128:
124:
101:
97:
461:
716:
615:
1057:
1047:
971:
120:
597:
793:
469:
116:
55:
410:
These properties of the genetic code make it more fault-tolerant for
112:
59:
66:
gives 64 possible codons, meaning that some degeneracy must exist.
815:
726:
428:
27:
631:
Reaction centers of photosynthetic bacteria: Feldafing-II-Meeting
538:
Watson JD, Baker TA, Bell SP, Gann A, Levine M, Oosick R (2008).
730:
100:
codon. AUU, AUC, or AUA all encode isoleucine, but AUG encodes
785:
135:
Inverse table for the standard genetic code (compressed using
111:
There are three amino acids encoded by six different codons:
433:
Grouping of codons by amino acid residue molar volume and
633:. Vol. 6. Berlin: Springer-Verlag. pp. 209–18.
564:
Two out of three: An alternative method for codon reading
449:
is maintained by equivalent substitution of amino acids (
629:
Yang; et al. (1990). Michel-Beyerle, M. E. (ed.).
1090:
999:
964:
938:
929:
887:
861:
835:
826:
764:
464:of the tRNA, and the base-pair formed is called a
742:
8:
542:. San Francisco: Pearson/Benjamin Cummings.
74:The examples all refer to the standard code.
935:
897:Precursor mRNA (pre-mRNA / hnRNA)
832:
749:
735:
727:
706:
605:
133:
16:Redundancy of codons in the genetic code
493:
472:and the Non-Watson-Crick U-G basepair.
92:A less degenerate site would produce a
580:Lehmann, J; Libchaber, A (July 2008).
77:A position of a codon is said to be a
917:Histone acetylation and deacetylation
533:
531:
529:
527:
525:
523:
521:
482:Neutral theory of molecular evolution
7:
982:Ribosome-nascent chain complex (RNC)
681:Varani G, McClain WH (July 2000).
89:with no change on the amino acid.
14:
987:Post-translational modification
131:, specified by the codon UGG.
81:-fold degenerate site if only
1:
540:Molecular Biology of the Gene
468:. The modified bases include
299:UCU, UCC, UCA, UCG; AGU, AGC
200:CUU, CUC, CUA, CUG; UUA, UUG
189:CGU, CGC, CGA, CGG; AGA, AGG
654:Füllen G, Youvan DC (1994).
699:10.1093/embo-reports/kvd001
1150:
455:aminoacyl tRNA synthetases
152:
73:
457:related to these codons.
338:
241:
30:is the redundancy of the
1048:sequestration (P-bodies)
660:Complexity International
562:Lagerkvist, U. (1978.) "
1026:Gene regulatory network
1031:cis-regulatory element
438:
94:nonsynonymous mutation
451:conservative mutation
432:
1134:Protein biosynthesis
1053:alternative splicing
1043:Post-transcriptional
869:Transcription factor
36:synonymous mutations
977:Transfer RNA (tRNA)
598:10.1261/rna.1029808
376:GUU, GUC, GUA, GUG
347:GGU, GGC, GGA, GGG
330:CAA, CAG; GAA, GAG
319:ACU, ACC, ACA, ACG
279:CCU, CCC, CCA, CCG
250:AAU, AAC; GAU, GAC
169:GCU, GCC, GCA, GCG
140:
87:synonymous mutation
1124:Molecular genetics
1091:Influential people
1070:Post-translational
889:Post-transcription
439:
134:
1111:
1110:
995:
994:
925:
924:
801:Special transfers
549:978-0-8053-9592-1
403:
402:
1141:
936:
833:
751:
744:
737:
728:
721:
720:
710:
678:
672:
671:
666:. Archived from
651:
645:
644:
626:
620:
619:
609:
577:
571:
560:
554:
553:
535:
516:
515:
513:
512:
498:
466:wobble base pair
416:codon usage bias
141:
1149:
1148:
1144:
1143:
1142:
1140:
1139:
1138:
1129:Gene expression
1114:
1113:
1112:
1107:
1086:
1021:Transcriptional
991:
960:
921:
912:Polyadenylation
883:
857:
822:
816:Protein→Protein
767:
760:
758:Gene expression
755:
725:
724:
680:
679:
675:
653:
652:
648:
641:
628:
627:
623:
579:
578:
574:
561:
557:
550:
537:
536:
519:
510:
508:
500:
499:
495:
490:
478:
412:point mutations
408:
204:
193:
75:
72:
44:
17:
12:
11:
5:
1147:
1145:
1137:
1136:
1131:
1126:
1116:
1115:
1109:
1108:
1106:
1105:
1100:
1098:François Jacob
1094:
1092:
1088:
1087:
1085:
1084:
1083:
1082:
1077:
1067:
1062:
1061:
1060:
1055:
1050:
1040:
1035:
1034:
1033:
1028:
1018:
1017:
1016:
1005:
1003:
997:
996:
993:
992:
990:
989:
984:
979:
974:
968:
966:
962:
961:
959:
958:
953:
948:
942:
940:
933:
927:
926:
923:
922:
920:
919:
914:
909:
904:
899:
893:
891:
885:
884:
882:
881:
876:
874:RNA polymerase
871:
865:
863:
859:
858:
856:
855:
850:
845:
839:
837:
830:
824:
823:
821:
820:
819:
818:
813:
808:
798:
797:
796:
778:
772:
770:
762:
761:
756:
754:
753:
746:
739:
731:
723:
722:
673:
670:on 2011-03-15.
646:
639:
621:
572:
555:
548:
517:
506:www.nature.com
492:
491:
489:
486:
485:
484:
477:
474:
447:hydrophobicity
443:hydrophilicity
407:
404:
401:
400:
397:
396:UAA, UGA, UAG
394:
391:
388:
387:AUG, CUG, UUG
385:
381:
380:
377:
374:
371:
368:
365:
361:
360:
357:
354:
351:
348:
345:
341:
340:
337:
334:
331:
328:
327:Gln or Glu, Z
324:
323:
320:
317:
314:
311:
308:
304:
303:
300:
297:
294:
291:
288:
284:
283:
280:
277:
274:
271:
268:
264:
263:
260:
257:
254:
251:
248:
247:Asn or Asp, B
244:
243:
240:
237:
234:
231:
227:
226:
223:
220:
217:
214:
211:
207:
206:
201:
198:
195:
190:
187:
183:
182:
179:
178:AUU, AUC, AUA
176:
173:
170:
167:
163:
162:
159:
156:
153:
151:
148:
145:
137:IUPAC notation
71:
68:
43:
40:
15:
13:
10:
9:
6:
4:
3:
2:
1146:
1135:
1132:
1130:
1127:
1125:
1122:
1121:
1119:
1104:
1103:Jacques Monod
1101:
1099:
1096:
1095:
1093:
1089:
1081:
1078:
1076:
1073:
1072:
1071:
1068:
1066:
1065:Translational
1063:
1059:
1056:
1054:
1051:
1049:
1046:
1045:
1044:
1041:
1039:
1036:
1032:
1029:
1027:
1024:
1023:
1022:
1019:
1015:
1012:
1011:
1010:
1007:
1006:
1004:
1002:
998:
988:
985:
983:
980:
978:
975:
973:
970:
969:
967:
963:
957:
954:
952:
949:
947:
944:
943:
941:
937:
934:
932:
928:
918:
915:
913:
910:
908:
905:
903:
900:
898:
895:
894:
892:
890:
886:
880:
877:
875:
872:
870:
867:
866:
864:
860:
854:
851:
849:
846:
844:
841:
840:
838:
834:
831:
829:
828:Transcription
825:
817:
814:
812:
809:
807:
804:
803:
802:
799:
795:
791:
787:
784:
783:
782:
781:Central dogma
779:
777:
774:
773:
771:
769:
763:
759:
752:
747:
745:
740:
738:
733:
732:
729:
718:
714:
709:
704:
700:
696:
692:
688:
684:
677:
674:
669:
665:
661:
657:
650:
647:
642:
640:3-540-53420-2
636:
632:
625:
622:
617:
613:
608:
603:
599:
595:
592:(7): 1264–9.
591:
587:
583:
576:
573:
570:, 75:1759-62.
569:
565:
559:
556:
551:
545:
541:
534:
532:
530:
528:
526:
524:
522:
518:
507:
503:
497:
494:
487:
483:
480:
479:
475:
473:
471:
467:
463:
458:
456:
452:
448:
444:
436:
431:
427:
425:
421:
417:
413:
405:
398:
395:
392:
389:
386:
383:
382:
378:
375:
372:
369:
366:
363:
362:
358:
355:
352:
349:
346:
343:
342:
335:
332:
329:
326:
325:
321:
318:
315:
312:
309:
306:
305:
301:
298:
295:
292:
289:
286:
285:
281:
278:
275:
272:
269:
266:
265:
261:
258:
255:
252:
249:
246:
245:
238:
235:
232:
229:
228:
224:
221:
218:
215:
212:
209:
208:
203:CUN, UUR; or
202:
199:
196:
191:
188:
185:
184:
180:
177:
174:
171:
168:
165:
164:
160:
157:
154:
149:
146:
143:
142:
138:
132:
130:
126:
122:
118:
114:
109:
105:
103:
99:
95:
90:
88:
84:
80:
69:
67:
63:
61:
57:
53:
52:glutamic acid
48:
41:
39:
37:
33:
29:
25:
21:
1080:irreversible
965:Key elements
862:Key elements
776:Genetic code
766:Introduction
693:(1): 18–23.
690:
686:
676:
668:the original
663:
659:
649:
630:
624:
589:
585:
575:
567:
558:
539:
509:. Retrieved
505:
496:
459:
440:
424:transversion
409:
406:Implications
192:CGN, AGR; or
110:
106:
91:
82:
78:
76:
64:
49:
45:
32:genetic code
23:
19:
18:
931:Translation
768:to genetics
161:Compressed
150:Compressed
70:Terminology
1118:Categories
1075:reversible
1038:lac operon
1014:imprinting
1009:Epigenetic
1001:Regulation
956:Eukaryotic
902:5' capping
853:Eukaryotic
511:2021-07-14
488:References
435:hydropathy
420:transition
158:DNA codons
155:Amino acid
147:DNA codons
144:Amino acid
129:tryptophan
125:methionine
102:methionine
98:isoleucine
42:Background
24:redundancy
20:Degeneracy
946:Bacterial
843:Bacterial
462:anticodon
399:URA, UAR
367:CAU, CAC
356:UAU, UAC
310:GAA, GAG
302:UCN, AGY
290:CAA, CAG
270:UGU, UGC
259:UUU, UUC
233:GAU, GAC
222:AAA, AAG
213:AAU, AAC
205:CUY, YUR
194:CGY, MGR
1058:microRNA
972:Ribosome
951:Archaeal
907:Splicing
879:Promoter
848:Archaeal
792: →
788: →
717:11256617
687:EMBO Rep
616:18495942
476:See also
121:arginine
811:RNA→DNA
806:RNA→RNA
794:Protein
708:1083677
607:2441979
470:inosine
373:Val, V
364:His, H
353:Tyr, Y
344:Gly, G
336:Trp, W
316:Thr, T
307:Glu, E
296:Ser, S
287:Gln, Q
276:Pro, P
267:Cys, C
256:Phe, F
239:Met, M
230:Asp, D
219:Lys, K
210:Asn, N
197:Leu, L
186:Arg, R
175:Ile, I
166:Ala, A
117:leucine
56:leucine
715:
705:
637:
614:
604:
546:
384:START
119:, and
113:serine
60:serine
28:codons
939:Types
836:Types
393:STOP
713:PMID
635:ISBN
612:PMID
568:PNAS
544:ISBN
390:HUG
379:GUN
370:CAY
359:UAY
350:GGN
339:UGG
333:SAR
322:ACN
313:GAR
293:CAR
282:CCN
273:UGY
262:UUY
253:RAY
242:AUG
236:GAY
225:AAR
216:AAY
181:AUH
172:GCN
790:RNA
786:DNA
703:PMC
695:doi
602:PMC
594:doi
586:RNA
566:",
445:or
26:of
22:or
1120::
711:.
701:.
689:.
685:.
662:.
658:.
610:.
600:.
590:14
588:.
584:.
520:^
504:.
139:)
115:,
38:.
750:e
743:t
736:v
719:.
697::
691:1
664:1
643:.
618:.
596::
552:.
514:.
437:.
83:n
79:n
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.