38:
1404:
1110:
870:
267:
9052:
803:
1494:
any point in the lifespan of the patient. The other two forms appear in infancy. Common symptoms of the lethal neonatal form and the severe infantile forms are liver failure, heart problems, seizures and death. The myopathic form is characterized by muscle pain and weakness following vigorous exercise. Treatment generally includes dietary modifications and carnitine supplements.
1241:
1493:
to be processed as a fuel source. The disease is caused by a defect in the gene CPT2. This deficiency will present in patients in one of three ways: lethal neonatal, severe infantile hepatocardiomuscular, and myopathic form. The myopathic is the least severe form of the deficiency and can manifest at
1257:
which has a broad targeting capacity. Due to this, alcohol sulfotransferase is also known by several other names including "hydroxysteroid sulfotransferase," "steroid sulfokinase," and "estrogen sulfotransferase." Decreases in its activity has been linked to human liver disease. This transferase acts
877:
Enzymes that transfer aldehyde or ketone groups and included in EC 2.2. This category consists of various transketolases and transaldolases. Transaldolase, the namesake of aldehyde transferases, is an important part of the pentose phosphate pathway. The reaction it catalyzes consists of a transfer
2509:
Wongtrakul J, Pongjaroenkit S, Leelapat P, Nachaiwieng W, Prapanthadara LA, Ketterman AJ (Mar 2010). "Expression and characterization of three new glutathione transferases, an epsilon (AcGSTE2-2), omega (AcGSTO1-1), and theta (AcGSTT1-1) from
Anopheles cracens (Diptera: Culicidae), a major Thai
1192:
is divided up in categories based on the type of group that accepts the transfer. Groups that are classified as phosphate acceptors include: alcohols, carboxy groups, nitrogenous groups, and phosphate groups. Further constituents of this subclass of transferases are various kinases. A prominent
1056:
EC 2.5 relates to enzymes that transfer alkyl or aryl groups, but does not include methyl groups. This is in contrast to functional groups that become alkyl groups when transferred, as those are included in EC 2.3. EC 2.5 currently only possesses one sub-class: Alkyl and aryl transferases.
1758:
The family of glutathione transferases (GST) is extremely diverse, and therefore can be used for a number of biotechnological purposes. Plants use glutathione transferases as a means to segregate toxic metals from the rest of the cell. These glutathione transferases can be used to create
1365:. The full name of A transferase is alpha 1-3-N-acetylgalactosaminyltransferase and its function in the cell is to add N-acetylgalactosamine to H-antigen, creating A-antigen. The full name of B transferase is alpha 1-3-galactosyltransferase, and its function in the cell is to add a
1290:-containing groups. This category only contains two transferases, and thus is one of the smallest categories of transferase. Selenocysteine synthase, which was first added to the classification system in 1999, converts seryl-tRNA(Sec UCA) into selenocysteyl-tRNA(Sec UCA).
1248:
Transfer of sulfur-containing groups is covered by EC 2.8 and is subdivided into the subcategories of sulfurtransferases, sulfotransferases, and CoA-transferases, as well as enzymes that transfer alkylthio groups. A specific group of sulfotransferases are those that use
301:
added to pigeon breast muscle. This observance was later verified by the discovery of its reaction mechanism by
Braunstein and Kritzmann in 1937. Their analysis showed that this reversible reaction could be applied to other tissues. This assertion was validated by
1394:
and the gene itself is over 18kb long. The alleles for A and B transferases are extremely similar. The resulting enzymes only differ in 4 amino acid residues. The differing residues are located at positions 176, 235, 266, and 268 in the enzymes.
1462:, which usually first manifests during infancy. Disease sufferers experience nausea, vomiting, inability to feed, and breathing difficulties. In extreme cases, ketoacidosis can lead to coma and death. The deficiency is caused by
234:
In the above reaction (where the dash represents a bond, not a minus sign), X would be the donor, and Y would be the acceptor. R denotes the functional group transferred as a result of transferase activity. The donor is often a
921:
Transfer of acyl groups or acyl groups that become alkyl groups during the process of being transferred are key aspects of EC 2.3. Further, this category also differentiates between amino-acyl and non-amino-acyl groups.
8613:
229:
2390:
Munch-Petersen A, Kalckar HM, Cutolo E, Smith EE (Dec 1953). "Uridyl transferases and the formation of uridine triphosphate; enzymic production of uridine triphosphate: uridine diphosphoglucose pyrophosphorolysis".
1559:. Acetylcholine is involved in many neuropsychic functions such as memory, attention, sleep and arousal. The enzyme is globular in shape and consists of a single amino acid chain. ChAT functions to transfer an
4042:
Wünschiers R, Jahn M, Jahn D, Schomburg I, Peifer S, Heinzle E, Burtscher H, Garbe J, Steen A, Schobert M, Oesterhelt D, Wachtveitl J, Chang A (2010). "Chapter 3: Metabolism". In Michal G, Schomburg D (eds.).
8986:
6975:
6970:
395:
of transferases are constructed in the form of "donor:acceptor grouptransferase." For example, methylamine:L-glutamate N-methyltransferase would be the standard naming convention for the transferase
8805:
373:. Initially, the exact mechanism of Pipe was unknown, due to a lack of information on its substrate. Research into Pipe's catalytic activity eliminated the likelihood of it being a heparan sulfate
5498:
Kumar B, Singh-Pareek SL, Sopory SK (2008). "Chapter 23: Glutathione
Homeostasis and Abiotic Stresses in Plants: Physiological, Biochemical and Molecular Approaches". In Kumar A, Sopory S (eds.).
4713:
Karadag N, Zenciroglu A, Eminoglu FT, Dilli D, Karagol BS, Kundak A, Dursun A, Hakan N, Okumus N (2013). "Literature review and outcome of classic galactosemia diagnosed in the neonatal period".
1382:: Type A (express A antigens), Type B (express B antigens), Type AB (express both A and B antigens) and Type O (express neither A nor B antigens). The gene for A and B transferases is located on
3478:
Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H, Christen P (Apr 1984). "Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure".
5021:"Long-term effects of selective immunolesions of cholinergic neurons of the nucleus basalis magnocellularis on the ascending cholinergic pathways in the rat: a model for Alzheimer's disease"
367:
Classification of transferases continues to this day, with new ones being discovered frequently. An example of this is Pipe, a sulfotransferase involved in the dorsal-ventral patterning of
8623:
9029:
1639:
also show a marked decrease in ChAT production. Though the specific cause of the reduced production is not clear, it is believed that the death of medium-sized motor neurons with spiny
8618:
270:
Biodegradation of dopamine via catechol-O-methyltransferase (along with other enzymes). The mechanism for dopamine degradation led to the Nobel Prize in
Physiology or Medicine in 1970.
7348:
7224:
6378:
1763:
to detect contaminants such as herbicides and insecticides. Glutathione transferases are also used in transgenic plants to increase resistance to both biotic and abiotic stress.
1541:. Currently, the only available treatment is early diagnosis followed by adherence to a diet devoid of lactose, and prescription of antibiotics for infections that may develop.
247:
Some of the most important discoveries relating to transferases occurred as early as the 1930s. Earliest discoveries of transferase activity occurred in other classifications of
7549:
1276:
1174:
1099:
1043:
967:
904:
433:
4477:"Analysis of common mutations in the galactose-1-phosphate uridyl transferase gene: new assays to increase the sensitivity and specificity of newborn screening for galactosemia"
8979:
7015:
1623:. Low levels of ChAT activity are an early indication of the disease and are detectable long before motor neurons begin to die. This can even be detected before the patient is
5425:
Maselli RA, Chen D, Mo D, Bowe C, Fenton G, Wollmann RL (Feb 2003). "Choline acetyltransferase mutations in myasthenic syndrome due to deficient acetylcholine resynthesis".
4791:
Braida D, Ponzoni L, Martucci R, Sparatore F, Gotti C, Sala M (May 2014). "Role of neuronal nicotinic acetylcholine receptors (nAChRs) on learning and memory in zebrafish".
8798:
8738:
8716:
7680:
1339:
1230:
855:
5989:
448:
However, other accepted names are more frequently used for transferases, and are often formed as "acceptor grouptransferase" or "donor grouptransferase." For example, a
6190:
8534:
1679:. SIDS infants also display fewer neurons capable of producing ChAT in the vagus system. These defects in the medulla could lead to an inability to control essential
8972:
7329:
4124:"ABO ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) [ Homo sapiens (human) ]"
7397:
6846:
8791:
6144:
1353:
system. Both A and B transferases are glycosyltransferases, meaning they transfer a sugar molecule onto an H-antigen. This allows H-antigen to synthesize the
263:. Prior to the realization that individual enzymes were capable of such a task, it was believed that two or more enzymes enacted functional group transfers.
7506:
6663:
6658:
6045:
1478:
488:
classification). These categories comprise over 450 different unique enzymes. In the EC numbering system, transferases have been given a classification of
8644:
7476:
6208:
6114:
314:
in 1937. This in turn would pave the way for the possibility that similar transfers were a primary means of producing most amino acids via amino transfer.
1250:
7760:
6464:
6371:
6309:
822:, and amido groups. Carbamoyltransferases, as an example, transfer a carbamoyl group from one molecule to another. Carbamoyl groups follow the formula NH
8870:
5278:
Mancama D, Mata I, Kerwin RW, Arranz MJ (Oct 2007). "Choline acetyltransferase variants and their influence in schizophrenia and olanzapine response".
8539:
7542:
7151:
7146:
1513:(GALT) has any number of mutations, leading to a deficiency in the amount of GALT produced. There are two forms of Galactosemia: classic and Duarte.
1510:
396:
7750:
6646:
1820:. These efforts are focused on sequencing the subunits of the rubber transferase enzyme complex in order to transfect these genes into other plants.
8706:
7622:
6088:
8860:
3567:"Biochemical characterization of the human cyclin-dependent protein kinase activating kinase. Identification of p35 as a novel regulatory subunit"
317:
Another such example of early transferase research and later reclassification involved the discovery of uridyl transferase. In 1953, the enzyme
4573:
Mahmood U, Imran M, Naik SI, Cheema HA, Saeed A, Arshad M, Mahmood S (Nov 2012). "Detection of common mutations in the GALT gene through ARMS".
7673:
7511:
6653:
6319:
6035:
5071:"Neural restrictive silencer factor and choline acetyltransferase expression in cerebral tissue of Alzheimer's Disease patients: A pilot study"
162:
8345:
6364:
5982:
4165:
3631:
2903:
2282:
1731:
353:
9024:
8953:
8855:
7535:
6641:
6273:
381:
7765:
5326:"Reduction in choline acetyltransferase immunoreactivity but not muscarinic-m2 receptor immunoreactivity in the brainstem of SIDS infants"
4885:"Mice with selective elimination of striatal acetylcholine release are lean, show altered energy homeostasis and changed sleep/wake cycle"
7810:
6813:
6345:
6065:
6061:
3818:"Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORα and RORγ and its potential relevance to human liver diseases"
3816:
Ou Z, Shi X, Gilroy RK, Kirisci L, Romkes M, Lynch C, Wang H, Xu M, Jiang M, Ren S, Gramignoli R, Strom SC, Huang M, Xie W (Jan 2013).
1812:
in a number of commercial uses. Efforts are being made to produce transgenic plants capable of synthesizing natural rubber, including
8479:
6839:
6781:
2079:
1743:
484:
Described primarily based on the type of biochemical group transferred, transferases can be divided into ten categories (based on the
8049:
7843:
7666:
7632:
7392:
6597:
6535:
5698:
5507:
4060:
2661:"Drosophila pipe protein activity in the ovary and the embryonic salivary gland does not require heparan sulfate glycosaminoglycans"
2002:"Pyruvate dehydrogenase kinase-4 contributes to the recirculation of gluconeogenic precursors during postexercise glycogen recovery"
1710:(inability to breathe) that can be fatal. ChAT deficiency is implicated in myasthenia syndromes where the transition problem occurs
1474:
460:
acceptor. In practice, many molecules are not referred to using this terminology due to more prevalent common names. For example,
8190:
7740:
7642:
7402:
6592:
6587:
6229:
5975:
1750:
end of an existing DNA molecule. Terminal transferase is one of the few DNA polymerases that can function without an RNA primer.
1307:
5528:
Chronopoulou EG, Labrou NE (2009). "Glutathione transferases: emerging multidisciplinary tools in red and green biotechnology".
129:. Transferases are also utilized during translation. In this case, an amino acid chain is the functional group transferred by a
37:
8907:
7423:
6530:
6304:
6185:
5202:
Smith R, Chung H, Rundquist S, Maat-Schieman ML, Colgan L, Englund E, Liu YJ, Roos RA, Faull RL, Brundin P, Li JY (Nov 2006).
4258:"ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase)"
7597:
7407:
6545:
6457:
6294:
6289:
6213:
6093:
4621:
4209:"ABO Blood Group (Transferase A, Alpha 1-3-N-Acetylgalactosaminyltransferase;Transferase B, Alpha 1-3-Galactosyltransferase)"
2230:
Braunstein AE, Kritzmann MG (1937). "Formation and
Breakdown of Amino-acids by Inter-molecular Transfer of the Amino Group".
1699:
827:
5854:
3173:
1591:. Patients with Alzheimer's disease show a 30 to 90% reduction in activity in several regions of the brain, including the
8843:
8763:
8758:
8318:
8136:
7587:
6832:
6710:
6690:
6604:
6070:
6030:
1934:"Model reactions for CoA transferase involving thiol transfer. Anhydride formation from thiol esters and carboxylic acids"
1612:
318:
9042:
8628:
8516:
8484:
8469:
6942:
6631:
6525:
6520:
6299:
6139:
1668:
496:
is not considered a functional group when it comes to transferase targets; instead, hydrogen transfer is included under
377:. Further research has shown that Pipe targets the ovarian structures for sulfation. Pipe is currently classified as a
345:
1403:
8151:
6927:
6793:
6567:
6314:
5565:"Expression patterns of glutathione transferase gene (GstI) in maize seedlings under juglone-induced oxidative stress"
5500:
Recent advances in plant biotechnology and its applications : Prof. Dr. Karl-Hermann
Neumann commemorative volume
1833:
330:
4278:
1412:
galactose-1-phosphate uridyltransferase. A deficiency of the human isoform of this transferase causes of galactosemia
5069:
González-Castañeda RE, Sánchez-González VJ, Flores-Soto M, Vázquez-Camacho G, Macías-Islas MA, Ortiz GG (Mar 2013).
4526:"Genetic basis of transferase-deficient galactosaemia in Ireland and the population history of the Irish Travellers"
4355:
3201:"Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome"
8934:
7755:
7367:
6803:
6636:
6510:
1703:
1133:
but it has since been reclassified as a subcategory of EC 2.1 (single-carbon transferring enzymes). In the case of
879:
5839:
9012:
8944:
8922:
8890:
8831:
8749:
8727:
8702:
8677:
8597:
8205:
8165:
8110:
8087:
8059:
8027:
7893:
7801:
7782:
7745:
7706:
7647:
7617:
7579:
7492:
7384:
7297:
6932:
6872:
6751:
6727:
6678:
6562:
6515:
6490:
6450:
6408:
6333:
6248:
6158:
6124:
6040:
6011:
5955:
5942:
5929:
5916:
5903:
5890:
5877:
2555:"Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity"
1857:
1853:
1764:
1549:
883:
476:
strand. In the EC system of classification, the accepted name for RNA polymerase is DNA-directed RNA polymerase.
442:
97:
Transferases are involved in myriad reactions in the cell. Three examples of these reactions are the activity of
7980:
5849:
9003:
8902:
8822:
8214:
8195:
8126:
7860:
7697:
7607:
7570:
6937:
6910:
6863:
6756:
6481:
6399:
6177:
6002:
5803:
5746:
4380:
1254:
485:
46:
2339:
Hird FJ, Rowsell EV (Sep 1950). "Additional transaminations by insoluble particle preparations of rat liver".
4945:"Choline acetyltransferase: the structure, distribution and pathologic changes in the central nervous system"
2442:
8072:
7427:
6990:
6798:
6705:
5751:
2468:
Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT (Nov 1996).
1636:
1454:
are created upon the breakdown of fats in the body and are an important energy source. Inability to utilize
1194:
465:
102:
771:
8865:
8526:
8299:
8209:
7718:
6905:
6766:
6761:
6626:
6234:
5243:
Karson CN, Casanova MF, Kleinman JE, Griffin WS (Mar 1993). "Choline acetyltransferase in schizophrenia".
1849:
1588:
1428:
1134:
118:
5644:
4333:
1659:, which is believed to correlate with the decreased cognitive functioning experienced by these patients.
1261:
1159:
1084:
1028:
952:
889:
775:
418:
9072:
8912:
8380:
8335:
8037:
8022:
7958:
7938:
7836:
7471:
6957:
6947:
6746:
6741:
6416:
6134:
5772:
5691:
5472:
1747:
1526:
1521:. Galactosemia renders infants unable to process the sugars in breast milk, which leads to vomiting and
1420:
1130:
1109:
935:
906:
449:
340:
Another example of historical significance relating to transferase is the discovery of the mechanism of
5844:
3029:
2759:"Mutational study of heparan sulfate 2-O-sulfotransferase and chondroitin sulfate 2-O-sulfotransferase"
7821:
5615:
3995:"Mutational analysis of Escherichia coli MoeA: two functional activities map to the active site cleft"
3147:
8880:
8294:
8289:
8169:
8032:
8012:
7921:
7735:
7339:
7166:
6900:
6808:
6695:
6682:
6582:
6129:
5378:
3406:
2713:
2615:
2400:
2348:
2239:
2104:
1841:
1829:
923:
910:
643:
322:
130:
4079:
Nishida C, Tomita T, Nishiyama M, Suzuki R, Hara M, Itoh Y, Ogawa H, Okumura K, Nishiyama C (2011).
3974:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3907:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3771:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3706:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3546:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3459:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
3010:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
2955:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
2926:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
2808:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
2604:"Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient"
1887:. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
1209:. Once combined, the CDK-cyclin complex is capable of enacting its function within the cell cycle.
8964:
8557:
8549:
8330:
7997:
7933:
7897:
7637:
7612:
7602:
7301:
7216:
7114:
6887:
6859:
6540:
6098:
5808:
5669:
4715:
2702:"Sulfation of eggshell components by Pipe defines dorsal-ventral polarity in the Drosophila embryo"
1837:
1514:
1045:
990:
831:
810:
EC 2.1 includes enzymes that transfer single-carbon groups. This category consists of transfers of
708:
639:
612:
604:
326:
303:
110:
4257:
3725:
Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC (Jun 2001).
3395:"C. elegans aging is modulated by hydrogen sulfide and the sulfhydrylase/cysteine synthase cysl-2"
1775:. Further, glutathione transferase genes have been investigated due to their ability to prevent
1324:
1215:
840:
8783:
8420:
8358:
8309:
8044:
7871:
7791:
7454:
7138:
6876:
5741:
5450:
5303:
5155:"Early presymptomatic cholinergic dysfunction in a murine model of amyotrophic lateral sclerosis"
5051:
4975:
4914:
4816:
4695:
4598:
4555:
3596:
2641:
2584:
2535:
2424:
2372:
2255:
2128:
1800:
1680:
1522:
1189:
1126:
691:
671:
608:
311:
294:
252:
90:(called the donor) to another (called the acceptor). They are involved in hundreds of different
5967:
411:
is the EC category grouping. This same action by the transferase can be illustrated as follows:
3623:
2977:
464:
is the modern common name for what was formerly known as RNA nucleotidyltransferase, a kind of
9016:
8835:
8435:
8425:
8415:
8340:
8324:
8283:
8269:
8264:
8002:
7965:
7627:
7496:
6922:
6824:
6771:
6500:
6200:
6015:
5596:
5545:
5503:
5442:
5404:
5347:
5295:
5260:
5225:
5184:
5135:
5100:
5043:
4967:
4906:
4865:
4808:
4773:
4732:
4687:
4652:
4590:
4547:
4506:
4424:
4161:
4102:
4056:
4024:
3949:
3882:
3847:
3764:
3746:
3627:
3588:
3495:
3434:
3323:
3230:
2899:
2850:
2780:
2739:
2682:
2633:
2576:
2527:
2491:
2416:
2364:
2321:
2278:
2212:
2177:
2120:
2075:
2031:
1955:
1656:
1533:, failure to grow, and mental impairment, among others. Buildup of a second toxic substance,
1058:
751:
727:
531:
527:
408:
374:
91:
5020:
3673:
3055:
8894:
8768:
8430:
8410:
8352:
8248:
8146:
8131:
8077:
7975:
7970:
7829:
7334:
7321:
6965:
6915:
6736:
6700:
5787:
5782:
5756:
5684:
5586:
5576:
5537:
5434:
5394:
5386:
5337:
5287:
5252:
5215:
5174:
5166:
5127:
5090:
5082:
5035:
4959:
4896:
4855:
4847:
4800:
4763:
4724:
4679:
4582:
4537:
4496:
4488:
4235:
4092:
4048:
4014:
4006:
3939:
3874:
3837:
3829:
3738:
3615:
3578:
3487:
3424:
3414:
3367:
3313:
3220:
3212:
3101:
2840:
2770:
2729:
2721:
2672:
2623:
2566:
2519:
2481:
2408:
2356:
2311:
2247:
2204:
2167:
2159:
2148:"A quantitative study of succinic acid in muscle: Glutamic and aspartic acids as precursors"
2112:
2067:
2021:
2013:
1945:
1809:
1776:
1739:
1553:
1070:
1010:
765:
731:
290:
75:
7658:
7206:
7201:
8875:
8691:
8681:
8503:
8474:
8100:
8007:
7992:
7353:
6776:
6477:
6442:
6252:
5834:
5818:
5731:
4993:
4752:"Human choline acetyltransferase gene maps to region 10q11-q22.2 by in situ hybridization"
2000:
Herbst EA, MacPherson RE, LeBlanc PJ, Roy BD, Jeoung NH, Harris RA, Peters SJ (Jan 2014).
1845:
1772:
1443:
1350:
1014:
1006:
1002:
745:
721:
685:
661:
633:
598:
580:
574:
546:
521:
392:
6356:
5645:"Development of Domestic Natural Rubber-Producing Industrial Crops Through Biotechnology"
4208:
3699:
3082:
2919:
2801:
156:
Mechanistically, an enzyme that catalyzed the following reaction would be a transferase:
7527:
5382:
3410:
2717:
2619:
2404:
2352:
2243:
2108:
1025:
and UDP-galactose. This occurs via the following pathway: UDP-β-D-galactose + D-glucose
9056:
8659:
8498:
8405:
8397:
8274:
8237:
5872:
5813:
5591:
5564:
5399:
5366:
5179:
5154:
5095:
5070:
5039:
4860:
4835:
4501:
4476:
4019:
3994:
3842:
3817:
3429:
3394:
3225:
3200:
2734:
2701:
2172:
2147:
2026:
2001:
1735:
1684:
1575:
cells and exists in two forms: soluble and membrane bound. The ChAT gene is located on
1506:
1198:
939:
869:
497:
461:
349:
274:
41:
4492:
3944:
3927:
3318:
2845:
2828:
2571:
2554:
2486:
2469:
2195:
Snell EE, Jenkins WT (December 1959). "The mechanism of the transamination reaction".
1950:
1933:
9066:
7928:
7358:
6895:
6426:
6421:
6162:
5777:
5736:
5390:
5342:
5325:
4963:
4768:
4751:
3878:
3790:
3616:
3491:
2869:
1715:
1596:
1592:
1576:
1556:
1518:
1315:
1206:
1142:
998:
931:
815:
556:
552:
361:
341:
334:
298:
51:
5454:
5086:
5055:
4979:
4944:
4918:
4699:
4602:
4559:
3967:
3600:
3106:
2645:
2588:
2539:
1974:
1880:
1651:
Patients with
Schizophrenia also exhibit decreased levels of ChAT, localized to the
1517:
is generally less severe than classic galactosemia and is caused by a deficiency of
1156:
The reaction, for example, follows the following order: L-aspartate +2-oxoglutarate
266:
8017:
7987:
7916:
7710:
6431:
6080:
5726:
5307:
5153:
Casas C, Herrando-Grabulosa M, Manzano R, Mancuso R, Osta R, Navarro X (Mar 2013).
4851:
4820:
3900:
3865:
Sekura RD, Marcus CJ, Lyon ES, Jakoby WB (May 1979). "Assay of sulfotransferases".
3452:
3003:
2376:
2259:
2132:
1861:
1780:
1768:
1672:
1600:
1560:
1502:
1490:
1459:
1383:
1374:
1354:
1188:-containing groups, it also includes nuclotidyl transferases as well. Sub-category
1122:
994:
667:
357:
307:
138:
79:
63:
4750:
Strauss WL, Kemper RR, Jayakar P, Kong CF, Hersh LB, Hilt DC, Rabin M (Feb 1991).
4524:
Murphy M, McHugh B, Tighe O, Mayne P, O'Neill C, Naughten E, Croke DT (Jul 1999).
3928:"Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence"
2428:
2006:
American
Journal of Physiology. Regulatory, Integrative and Comparative Physiology
1466:
in the gene OXCT1. Treatments mostly rely on controlling the diet of the patient.
321:
was shown to be a transferase, when it was found that it could reversibly produce
4728:
4182:
4154:
3539:
3419:
1603:. However, ChAT deficiency is not believed to be the main cause of this disease.
1244:
Ribbon diagram of a variant structure of estrogen sulfotransferase (PDB 1aqy EBI)
94:
throughout biology, and are integral to some of life's most important processes.
17:
8583:
8573:
8174:
7911:
6715:
5950:
5721:
5131:
4449:
3972:
School of
Biological & Chemical Sciences at Queen Mary, University of London
3905:
School of
Biological & Chemical Sciences at Queen Mary, University of London
3769:
School of Biological & Chemical Sciences at Queen Mary, University of London
3704:
School of Biological & Chemical Sciences at Queen Mary, University of London
3544:
School of Biological & Chemical Sciences at Queen Mary, University of London
3008:
School of Biological & Chemical Sciences at Queen Mary, University of London
2953:
School of Biological & Chemical Sciences at Queen Mary, University of London
2806:
School of Biological & Chemical Sciences at Queen Mary, University of London
1981:. Department of Computing Science and Biological Sciences, University of Alberta
1711:
1688:
1652:
1624:
1616:
1482:
1118:
469:
404:
400:
260:
9051:
5541:
4586:
3300:
Fitzgerald DK, Brodbeck U, Kiyosawa I, Mawal R, Colvin B, Ebner KE (Apr 1970).
2948:
2300:"Rudolf Schoenheimer and the concept of the dynamic state of body constituents"
2017:
1714:. These syndromes are characterized by the patients’ inability to resynthesize
1525:
within days of birth. Most symptoms of the disease are caused by a buildup of
8926:
8385:
8228:
8141:
7906:
7856:
7592:
4804:
4683:
4644:
4420:
4081:"B-transferase with a Pro234Ser substitution acquires AB-transferase activity"
4052:
2725:
2628:
2603:
1906:
1534:
1486:
1379:
1358:
1299:
1185:
1146:
802:
780:
704:
695:
369:
134:
126:
114:
98:
55:
3301:
3154:. The Biochemistry and Biophysics Program at Renssalaer Polytechnic Institute
2316:
2299:
8848:
8664:
8577:
8114:
7880:
6258:
5924:
5898:
4542:
4525:
3199:
Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V (May 2009).
3060:
ChEBI: The database and ontology of Chemical Entities of Biological Interest
2071:
1817:
1760:
1366:
1319:
938:. As an aminoacyltransferase, it catalyzes the transfer of a peptide to an
858:
835:
819:
286:
106:
5665:
5600:
5549:
5446:
5408:
5351:
5299:
5229:
5188:
5139:
5104:
5047:
4971:
4910:
4883:
Guzman MS, De Jaeger X, Drangova M, Prado MA, Gros R, Prado VF (Mar 2013).
4812:
4736:
4691:
4656:
4594:
4551:
4510:
4428:
4106:
4028:
3851:
3750:
3742:
3438:
3274:
3234:
2784:
2775:
2758:
2743:
2686:
2637:
2531:
2420:
2368:
2216:
2208:
2181:
2124:
2035:
5264:
5256:
4869:
4777:
3953:
3592:
3583:
3566:
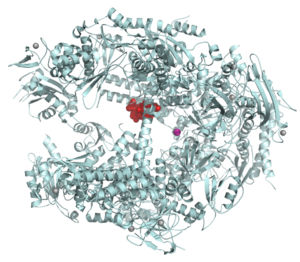
3499:
3327:
2854:
2580:
2495:
2325:
1959:
1240:
1129:
and other nitrogen group transferring enzymes. EC 2.6 previously included
224:{\displaystyle {\ce {X-R + Y}}\quad {\xrightarrow{}}\quad {\ce {X + Y-R}}}
149:
and its subsequent addition to the amino acid attached to the tRNA in the
8999:
8511:
8091:
7693:
6505:
6391:
5581:
5291:
5220:
5203:
5019:
Szigeti C, Bencsik N, Simonka AJ, Legradi A, Kasa P, Gulya K (May 2013).
3886:
3833:
1783:
1676:
1640:
1538:
1463:
1362:
1306:-containing groups. However, as of 2011, only one enzyme has been added:
1303:
1287:
1138:
1062:
986:
927:
784:
756:
712:
676:
617:
561:
493:
256:
236:
146:
122:
87:
83:
31:
3647:
8462:
8447:
8063:
7516:
7374:
7107:
7102:
7097:
7092:
7087:
7055:
6979:
6337:
5204:"Cholinergic neuronal defect without cell loss in Huntington's disease"
4123:
4097:
4080:
3341:
3248:
1813:
1568:
1564:
1455:
1451:
1427:. The most common result of a transferase deficiency is a buildup of a
1424:
1408:
1022:
1018:
625:
621:
435:
58:," RNA polymerases are classified as a form of nucleotidyl transferase.
5438:
4901:
4884:
4010:
3513:
3216:
3121:
2677:
2660:
2412:
2163:
1552:(also known as ChAT or CAT) is an important enzyme which produces the
1009:
to other molecules. An example of a prominent glycosyltransferase is
198:
8818:
8440:
7875:
7562:
7312:
7082:
7077:
7072:
7067:
7062:
7050:
7045:
7040:
7035:
7030:
7025:
7020:
7008:
7003:
6998:
6786:
6614:
6609:
6550:
6395:
6167:
6119:
5937:
5707:
5170:
5118:
Rowland LP, Shneider NA (May 2001). "Amyotrophic lateral sclerosis".
4475:
Dobrowolski SF, Banas RA, Suzow JG, Berkley M, Naylor EW (Feb 2003).
3393:
Qabazard B, Ahmed S, Li L, Arlt VM, Moore PK, Stürzenbaum SR (2013).
2360:
2251:
2116:
2062:
Boyce S, Tipton KF (2005). "Enzyme Classification and Nomenclature".
1795:
1530:
1447:
1391:
1202:
811:
736:
699:
565:
537:
453:
248:
191:
150:
142:
71:
4938:
4936:
4934:
4932:
4930:
4928:
4045:
Biochemical Pathways: an Atlas of Biochemistry and Molecular Biology
3726:
2523:
2095:
Morton RK (Jul 1953). "Transferase activity of hydrolytic enzymes".
1258:
via the following reaction: 3'-phosphoadenylyl sulfate + an alcohol
1212:
The reaction catalyzed by CDK is as follows: ATP + a target protein
1113:
Aspartate aminotransferase can act on several different amino acids
27:
Class of enzymes which transfer functional groups between molecules
8457:
8452:
8304:
7464:
7459:
7432:
7283:
7234:
7229:
6668:
6555:
5911:
5324:
Mallard C, Tolcos M, Leditschke J, Campbell P, Rees S (Mar 1999).
1707:
1620:
1572:
1439:
1402:
1239:
1150:
868:
801:
648:
589:
278:
265:
36:
8614:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
4836:"Cholinergic mechanisms in the rat somatosensory cerebral cortex"
2470:"A new enzyme superfamily - the phosphopantetheinyl transferases"
8368:
8363:
8259:
8254:
7953:
7948:
7943:
7770:
7728:
7723:
7566:
7278:
7273:
7268:
7263:
7258:
7253:
7246:
7241:
7196:
7191:
7186:
7181:
7176:
7171:
7161:
7156:
7128:
7123:
7118:
6572:
1615:
show a marked decrease in ChAT activity in motor neurons in the
1387:
1201:. As their name implies, CDKs are heavily dependent on specific
652:
585:
8968:
8787:
7825:
7662:
7531:
6976:
Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase
6971:
B-N-acetylglucosaminyl-glycopeptide b-1,4-galactosyltransferase
6828:
6446:
6360:
5971:
5680:
5420:
5418:
4311:
1125:(also known as "aminotransferase"), and a very small number of
707:-containing groups; subclasses are based on the acceptor (e.g.
5319:
5317:
3993:
Nichols JD, Xiang S, Schindelin H, Rajagopalan KV (Jan 2007).
3622:(8th ed.). Boston: McGraw-Hill/Higher Education. p.
3565:
Yee A, Wu L, Liu L, Kobayashi R, Xiong Y, Hall FL (Jan 1996).
473:
457:
1734:
are transferases that can be used to label DNA or to produce
1702:
is a family of diseases that are characterized by defects in
293:(also known as a "transaminase"), was first noted in 1930 by
2896:
To grasp the essence of life: a history of molecular biology
993:
is a subcategory of EC 2.4 transferases that is involved in
1607:
Amyotrophic lateral sclerosis (ALS or Lou Gehrig's disease)
1061:, for example, catalyzes the formation of acetic acids and
989:
groups, as well as those that transfer hexose and pentose.
5676:
4670:
Bosch AM (Aug 2006). "Classical galactosaemia revisited".
1794:
Currently the only available commercial source of natural
9030:
O-phospho-L-seryl-tRNASec:L-selenocysteinyl-tRNA synthase
3678:
Protein Data Bank in Europe Bringing Structure to Biology
1349:
The A and B transferases are the foundation of the human
5523:
5521:
5519:
4645:"Classic Galactosemia and Clinical Variant Galactosemia"
4187:
Human Blood: An Introduction to Its Components and Types
4147:
4145:
4143:
4141:
4118:
4116:
4074:
4072:
1587:
Decreased expression of ChAT is one of the hallmarks of
1265:
1163:
1088:
1032:
956:
893:
422:
5666:
Superfamilies of single-pass transmembrane transferases
5466:
5464:
4156:
Oxford Dictionary of Biochemistry and Molecular Biology
3302:"Alpha-lactalbumin and the lactose synthetase reaction"
2659:
Zhu X, Sen J, Stevens L, Goltz JS, Stein D (Sep 2005).
1298:
The category of EC 2.10 includes enzymes that transfer
7225:
Dolichyl-phosphate-mannose-protein mannosyltransferase
4047:(2nd ed.). Oxford: Wiley-Blackwell. p. 140.
9040:
8624:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
1529:
in the body. Common symptoms include liver failure,
1327:
1314:. The reaction it catalyzes is as follows: adenylyl-
1310:. This enzyme is a component of MoCo biosynthesis in
1264:
1218:
1162:
1087:
1031:
955:
892:
843:
421:
348:. This discovery was a large part of the reason for
165:
8629:
CDP-diacylglycerol—choline O-phosphatidyltransferase
5330:
Journal of Neuropathology and Experimental Neurology
4414:
4412:
4410:
4408:
4406:
4404:
4402:
4306:
4304:
4302:
4300:
4279:"Succinyl-CoA:3-ketoacid CoA transferase deficiency"
1489:
lacks the ability to transport fatty acids into the
981:
EC 2.4: glycosyl, hexosyl, and pentosyl transferases
9011:
8943:
8921:
8889:
8830:
8748:
8726:
8701:
8676:
8657:
8637:
8619:
CDP-diacylglycerol—serine O-phosphatidyltransferase
8606:
8596:
8566:
8548:
8525:
8497:
8396:
8236:
8227:
8204:
8164:
8109:
8086:
8058:
7892:
7868:
7800:
7781:
7705:
7578:
7491:
7447:
7416:
7383:
7320:
7311:
7296:
7215:
7137:
6989:
6956:
6886:
6871:
6726:
6677:
6489:
6407:
6332:
6282:
6266:
6247:
6222:
6199:
6176:
6157:
6107:
6079:
6054:
6023:
6010:
5863:
5827:
5796:
5765:
5714:
4312:"SUCCINYL-CoA:3-OXOACID CoA TRANSFERASE DEFICIENCY"
133:. The transfer involves the removal of the growing
5367:"New horizons for congenital myasthenic syndromes"
4153:
2829:"The enzymatic synthesis of N-methylglutamic acid"
1671:infants show decreased levels of ChAT in both the
1505:results from an inability to process galactose, a
1333:
1270:
1224:
1168:
1093:
1037:
961:
898:
882:(also known as G3P). The reaction is as follows:
849:
452:is a transferase that catalyzes the transfer of a
427:
223:
4616:
4614:
4612:
4189:. Behavioral Sciences Department, Palomar College
6191:3-methyl-2-oxobutanoate hydroxymethyltransferase
5365:Engel AG, Shen XM, Selcen D, Sine S (Dec 2012).
2553:Sen J, Goltz JS, Stevens L, Stein D (Nov 1998).
1738:. It accomplishes both of these tasks by adding
1278:adenosine 3',5'bisphosphate + an alkyl sulfate.
113:, which is part of the pathway that metabolizes
4356:"Succinyl-CoA 3-Oxoacid Transferase Deficiency"
3618:Human genetics : concepts and applications
2052:. Upper Saddle River, NJ: Pearson, 2013. Print.
1253:as a sulfate group donor. Within this group is
504:Classification of transferases into subclasses
7398:Hypoxanthine-guanine phosphoribosyltransferase
4421:"Carnitine Palmitoyltransferase II Deficiency"
3765:"EC 2.8 Transferring Sulfur-Containing Groups"
3700:"EC 2.8 Transferring Sulfur-Containing Groups"
3004:"EC 2.1.3: Carboxy- and Carbamoyltransferases"
2197:Journal of Cellular and Comparative Physiology
1643:leads to the lower levels of ChAT production.
1121:groups is EC 2.6. This includes enzymes like
806:Reaction involving aspartate transcarbamylase.
8980:
8799:
7837:
7674:
7543:
6840:
6458:
6372:
6145:Cyclopropane-fatty-acyl-phospholipid synthase
5983:
5692:
4381:"Carnitine plamitoyltransferase I deficiency"
4160:(Rev. ed.). Oxford: Oxford Univ. Press.
3727:"Structure and function of sulfotransferases"
2757:Xu D, Song D, Pedersen LC, Liu J (Mar 2007).
2443:"Physiology or Medicine 1970 - Press Release"
8:
7507:Beta-galactoside alpha-2,6-sialyltransferase
6664:2-acylglycerol-3-phosphate O-acyltransferase
6659:1-acylglycerol-3-phosphate O-acyltransferase
6046:Phosphatidylethanolamine N-methyltransferase
5639:
5637:
5502:. New Delhi: I.K. International Pub. House.
2796:
2794:
1767:are currently being explored as targets for
1537:, occurs in the lenses of the eyes, causing
1509:. This deficiency occurs when the gene for
1184:While EC 2.7 includes enzymes that transfer
8645:N-acetylglucosamine-1-phosphate transferase
8535:UTP—glucose-1-phosphate uridylyltransferase
7477:Indolylacetylinositol arabinosyltransferase
6209:Phosphoribosylglycinamide formyltransferase
6115:Phosphatidyl ethanolamine methyltransferase
5569:International Journal of Molecular Sciences
5280:American Journal of Medical Genetics Part B
4085:Bioscience, Biotechnology, and Biochemistry
2943:
2941:
1369:molecule to H-antigen, creating B-antigen.
500:, due to electron transfer considerations.
8987:
8973:
8965:
8806:
8792:
8784:
8673:
8603:
8233:
8224:
7889:
7844:
7830:
7822:
7761:Branched-chain amino acid aminotransferase
7681:
7667:
7659:
7550:
7536:
7528:
7317:
7308:
6883:
6847:
6833:
6825:
6465:
6451:
6443:
6379:
6365:
6357:
6310:3-hydroxymethylcephem carbamoyltransferase
6263:
6173:
6020:
5990:
5976:
5968:
5699:
5685:
5677:
5371:Annals of the New York Academy of Sciences
1836:or anchored to membranes through a single
1197:(or CDK), which comprises a sub-family of
878:of a dihydroxyacetone functional group to
8739:serine/threonine-specific protein kinases
8717:serine/threonine-specific protein kinases
8540:Galactose-1-phosphate uridylyltransferase
5590:
5580:
5398:
5341:
5219:
5178:
5094:
4900:
4859:
4767:
4541:
4500:
4096:
4018:
3943:
3841:
3582:
3428:
3418:
3317:
3224:
3205:Nature Structural & Molecular Biology
3105:
2898:. Dordrecht: Springer. pp. 198–199.
2844:
2827:Shaw WV, Tsai L, Stadtman ER (Feb 1966).
2774:
2733:
2700:Zhang Z, Stevens LM, Stein D (Jul 2009).
2676:
2627:
2570:
2485:
2315:
2171:
2025:
1949:
1511:galactose-1-phosphate uridylyltransferase
1326:
1263:
1217:
1161:
1117:The grouping consistent with transfer of
1086:
1030:
954:
891:
842:
420:
397:methylamine-glutamate N-methyltransferase
212:
202:
192:
186:
170:
166:
164:
7623:Farnesyl-diphosphate farnesyltransferase
4152:Datta SP, Smith GH, Campbell PN (2000).
3797:. Kyoto University Bioinformatics Center
2874:Chem1 General Chemistry Virtual Textbook
1108:
942:, following this reaction: peptidyl-tRNA
865:EC 2.2: aldehyde and ketone transferases
502:
54:(in red). Despite the use of the term "
9047:
3731:Archives of Biochemistry and Biophysics
3680:. The European Bioinformatics Institute
3374:. European Molecular Biology Laboratory
3180:. European Molecular Biology Laboratory
3062:. European Molecular Biology Laboratory
1872:
873:The reaction catalyzed by transaldolase
297:, after observing the disappearance of
74:that catalyse the transfer of specific
7512:Monosialoganglioside sialyltransferase
7349:NAD(P):arginine ADP-ribosyltransferase
7330:NAD:diphthamide ADP-ribosyltransferase
6654:Glycerol-3-phosphate O-acyltransferase
6320:N-acetylornithine carbamoyltransferase
6089:Betaine-homocysteine methyltransferase
6036:Phenylethanolamine N-methyltransferase
4672:Journal of Inherited Metabolic Disease
3652:ExPASy: Bioinformatics Resource Portal
3518:ExPASy: Bioinformatics Resource Portal
3346:ExPASy: Bioinformatics Resource Portal
3253:ExPASy: Bioinformatics Resource Portal
3126:ExPASy: Bioinformatics Resource Portal
2275:The Dynamic State of Body Constituents
1779:and have shown improved resistance in
1545:Choline acetyltransferase deficiencies
1286:EC 2.9 includes enzymes that transfer
1153:group from one molecule to the other.
985:EC 2.4 includes enzymes that transfer
8871:thiosulfate—dithiol sulfurtransferase
6647:Lecithin—cholesterol acyltransferase
5477:Biotechnology and Genetic Engineering
3083:"Aspartate Carbamyl Transferase from
2972:
2970:
1361:conjugates that are known as the A/B
354:Nobel Prize in Physiology or Medicine
7:
9025:L-seryl-tRNASec selenium transferase
8954:Coenzyme-B sulfoethylthiotransferase
8856:3-mercaptopyruvate sulfurtransferase
7751:Kynurenine—oxoglutarate transaminase
6642:Glyceronephosphate O-acyltransferase
6274:methylmalonyl-CoA carboxytransferase
4998:GeneCards: The Human Gene Compendium
4481:The Journal of Molecular Diagnostics
4213:GeneCards: The Human Gene Compendium
1885:Enzyme Nomenclature. Recommendations
1695:Congenital myasthenic syndrome (CMS)
1017:. Its primary action is to produce
382:heparan sulfate 2-O-sulfotransferase
8861:thiosulfate—thiol sulfurtransferase
6814:Sulfoacetaldehyde acetyltransferase
6506:Acetyl-Coenzyme A acetyltransferase
6346:Arginine:glycine amidinotransferase
6066:Acetylserotonin O-methyltransferase
6062:5-hydroxyindole-O-methyltransferase
5120:The New England Journal of Medicine
4361:. Climb National Information Centre
3932:The Journal of Biological Chemistry
3654:. Swiss Institute of Bioinformatics
3571:The Journal of Biological Chemistry
3520:. Swiss Institute of Bioinformatics
3348:. Swiss Institute of Bioinformatics
3306:The Journal of Biological Chemistry
3255:. Swiss Institute of Bioinformatics
3128:. Swiss Institute of Bioinformatics
2833:The Journal of Biological Chemistry
2763:The Journal of Biological Chemistry
1938:The Journal of Biological Chemistry
1663:Sudden infant death syndrome (SIDS)
1271:{\displaystyle \rightleftharpoons }
1169:{\displaystyle \rightleftharpoons }
1094:{\displaystyle \rightleftharpoons }
1052:EC 2.5: alkyl and aryl transferases
1038:{\displaystyle \rightleftharpoons }
962:{\displaystyle \rightleftharpoons }
899:{\displaystyle \rightleftharpoons }
428:{\displaystyle \rightleftharpoons }
6730:: converted into alkyl on transfer
5245:The American Journal of Psychiatry
5040:10.1016/j.brainresbull.2013.01.007
4530:European Journal of Human Genetics
3926:Forchhammer K, Böck A (Apr 1991).
1881:"EC 2.7.7 Nucleotidyltransferases"
1706:which leads to recurrent bouts of
798:EC 2.1: single carbon transferases
25:
8050:Glucose-1,6-bisphosphate synthase
7633:Alkylglycerone phosphate synthase
7393:Adenine phosphoribosyltransferase
6536:Chloramphenicol acetyltransferase
1828:Many transferases associate with
1808:). Natural rubber is superior to
1742:in the form of a template to the
1475:Carnitine palmitoyltransferase II
655:groups, other than methyl groups
9050:
8191:Ribose-phosphate diphosphokinase
7811:Pyridoxine 5'-phosphate synthase
7643:Geranylgeranyltransferase type 1
7403:Uracil phosphoribosyltransferase
6588:Carnitine O-palmitoyltransferase
6230:Glutamate formimidoyltransferase
5391:10.1111/j.1749-6632.2012.06803.x
5343:10.1097/00005072-199903000-00005
4964:10.1046/j.1440-1827.1999.00977.x
1932:Moore SA, Jencks WP (Sep 1982).
1824:Membrane-associated transferases
1481:) leads to an excess long chain
1308:molybdopterin molybdotransferase
1105:EC 2.6: nitrogenous transferases
1013:which is a dimer possessing two
285:) group from an amino acid to a
8908:Tyrosylprotein sulfotransferase
7766:Alanine—glyoxylate transaminase
7424:Purine nucleoside phosphorylase
6531:Beta-galactoside transacetylase
6305:Putrescine carbamoyltransferase
6186:Serine hydroxymethyltransferase
5530:Recent Patents on Biotechnology
5087:10.1590/S1415-47572013000100005
5000:. Weizmann Institute of Science
4215:. Weizmann Institute of Science
3152:Molecular Biochemistry II Notes
3107:10.3891/acta.chem.scand.10-0548
3081:Reichard P, Hanshoff G (1956).
3056:"carbamoyl group (CHEBI:23004)"
2602:Moussian B, Roth S (Nov 2005).
1979:Small Molecule Pathway Database
1667:Recent studies have shown that
1423:are at the root of many common
1180:EC 2.7: phosphorus transferases
201:
185:
7598:Methionine adenosyltransferase
7408:Amidophosphoribosyltransferase
6546:Serotonin N-acetyl transferase
6493:: other than amino-acyl groups
6295:Ornithine carbamoyltransferase
6290:Aspartate carbamoyltransferase
6214:Inosine monophosphate synthase
6094:Homocysteine methyltransferase
5075:Genetics and Molecular Biology
4852:10.1113/jphysiol.1972.sp009951
4456:. National Institute of Health
4387:. National Institute of Health
4285:. National Institute of Health
4234:Moran, Lawrence (2007-02-22).
2984:. Encyclopædia Britannica, Inc
2949:"EC2 Transferase Nomenclature"
2894:Hausmann R (3 December 2010).
1913:. National Institute of Health
1378:to have any of four different
1328:
1219:
930:that facilitates formation of
844:
830:such a transfer is written as
1:
8844:Thiosulfate sulfurtransferase
8764:Protein-histidine tele-kinase
8759:Protein-histidine pros-kinase
8638:Glycosyl-1-phosphotransferase
7588:Dimethylallyltranstransferase
6711:Keratinocyte transglutaminase
6691:Gamma-glutamyl transpeptidase
6605:Serine C-palmitoyltransferase
6071:Catechol-O-methyl transferase
6031:Histamine N-methyltransferase
4994:"Choline O-Acetyltransferase"
4493:10.1016/S1525-1578(10)60450-3
3945:10.1016/S0021-9258(18)38121-3
3319:10.1016/S0021-9258(18)63212-0
3275:"Keyword Glycosyltransferase"
2846:10.1016/S0021-9258(18)96855-9
2572:10.1016/s0092-8674(00)81615-3
2512:Journal of Medical Entomology
2487:10.1016/S1074-5521(96)90181-7
2064:Encyclopedia of Life Sciences
2050:Molecular Biology of the Gene
1951:10.1016/S0021-9258(18)33907-3
1848:. Some others are multi-span
1282:EC 2.9: selenium transferases
1149:, it reversibly transfers an
886:+ glyceraldehyde 3-phosphate
588:groups or groups that become
319:UDP-glucose pyrophosphorylase
8470:RNA-dependent RNA polymerase
6943:Ceramide glucosyltransferase
6632:Aminolevulinic acid synthase
6526:Acetyl-CoA C-acyltransferase
6521:Dihydrolipoyl transacetylase
6300:Oxamate carbamoyltransferase
6140:Thiopurine methyltransferase
5622:. University of Nevada, Reno
4769:10.1016/0888-7543(91)90273-H
4729:10.7754/clin.lab.2013.121235
3879:10.1016/0003-2697(79)90188-x
3648:"ENZYME Entry: EC 2.7.11.22"
3492:10.1016/0022-2836(84)90333-4
3480:Journal of Molecular Biology
3420:10.1371/journal.pone.0080135
2870:"Naming Chemical Substances"
2277:. Hafner Publishing Co Ltd.
1834:peripheral membrane proteins
1334:{\displaystyle \rightarrow }
1225:{\displaystyle \rightarrow }
1176:oxaloacetate + L-glutamate.
850:{\displaystyle \rightarrow }
346:catechol-O-methyltransferase
8377:RNA-directed DNA polymerase
8245:DNA-directed DNA polymerase
6794:2-hydroxyglutarate synthase
6315:Lysine carbamoyltransferase
6001:: one carbon transferases (
5479:. Colorado State University
5132:10.1056/NEJM200105313442207
3342:"ENZYME entry: EC 2.4.1.22"
3249:"ENZYME entry: EC 2.3.2.12"
3148:"Pentose Phosphate Pathway"
2146:Needham, Dorothy M (1930).
1563:from acetyl co-enzyme A to
1442:transferase deficiency (or
1440:Succinyl-CoA:3-ketoacid CoA
1341:molybdenum cofactor + AMP.
1294:EC 2.10: metal transferases
1236:EC 2.8: sulfur transferases
472:to the 3’ end of a growing
9089:
8935:Propionate CoA-transferase
8730:: protein-dual-specificity
7756:Ornithine aminotransferase
7368:Poly ADP ribose polymerase
6804:2-isopropylmalate synthase
6637:Beta-ketoacyl-ACP synthase
6511:N-Acetylglutamate synthase
5542:10.2174/187220809789389135
4587:10.1016/j.gene.2012.08.010
3178:Enzyme Structures Database
3174:"EC 2.2.1.2 Transaldolase"
2298:Guggenheim KY (Nov 1991).
2018:10.1152/ajpregu.00150.2013
1854:oligosaccharyltransferases
1704:neuromuscular transmission
1477:deficiency (also known as
1386:. The gene contains seven
880:glyceraldehyde 3-phosphate
415:methylamine + L-glutamate
29:
7746:Tyrosine aminotransferase
7648:Porphobilinogen deaminase
7618:Glutathione S-transferase
7385:Phosphoribosyltransferase
6752:Decylhomocitrate synthase
6563:Histone acetyltransferase
6516:Choline acetyltransferase
6125:Histone methyltransferase
6041:Amine N-methyltransferase
5855:Michaelis–Menten kinetics
4889:Journal of Neurochemistry
4840:The Journal of Physiology
4805:10.1007/s00213-013-3340-1
4684:10.1007/s10545-006-0382-0
4053:10.1002/9781118657072.ch3
3094:Acta Chemica Scandinavica
2924:IUBMB Enzyme Nomenclature
2726:10.1016/j.cub.2009.05.050
2629:10.1016/j.cub.2005.10.026
1858:glutathione S-transferase
1550:Choline acetyltransferase
1345:Role in histo-blood group
973:+ peptidyl aminoacyl-tRNA
917:EC 2.3: acyl transferases
884:sedoheptulose 7-phosphate
70:is any one of a class of
8903:Alcohol sulfotransferase
8707:protein-serine/threonine
8607:Phosphatidyltransferases
8196:Thiamine diphosphokinase
7608:Dihydropteroate synthase
6938:1,3-Beta-glucan synthase
6757:2-methylcitrate synthase
6178:Hydroxymethyltransferase
5747:Diffusion-limited enzyme
5208:Human Molecular Genetics
3457:IUBMB Enzyme Nomenclatur
2304:The Journal of Nutrition
1765:Glutathione transferases
1754:Glutathione transferases
1606:
1446:) leads to a buildup of
1255:alcohol sulfotransferase
1232:ADP + a phosphoprotein.
857:L-carbamoyl aspartate +
277:, or the transfer of an
117:, and the regulation of
47:Saccharomyces cerevisiae
30:Not to be confused with
7428:Thymidine phosphorylase
6799:3-propylmalate synthase
6706:Tissue transglutaminase
5028:Brain Research Bulletin
4952:Pathology International
4543:10.1038/sj.ejhg.5200327
4454:Genetics Home Reference
4419:Weiser, Thomas (1993).
4385:Genetics Home Reference
4283:Genetics Home Reference
3867:Analytical Biochemistry
3822:Molecular Endocrinology
2982:Encyclopædia Britannica
2474:Chemistry & Biology
2273:Schoenheimer R (1949).
2072:10.1038/npg.els.0003893
1975:"Tryptophan Metabolism"
1911:Genetics Home Reference
1840:, for example numerous
1769:anti-cancer medications
1630:
1582:
1195:cyclin-dependent kinase
592:groups during transfer
466:nucleotidyl transferase
194: transferase
8866:tRNA sulfurtransferase
8527:Nucleotidyltransferase
8210:nucleotidyltransferase
8137:Nucleoside-diphosphate
7719:Aspartate transaminase
7322:ADP-ribosyltransferase
6767:3-ethylmalate synthase
6762:2-ethylmalate synthase
6627:Acyltransferase like 2
6235:Aminomethyltransferase
5563:Sytykiewicz H (2011).
5473:"Terminal Transferase"
3743:10.1006/abbi.2001.2368
3514:"Enzyme entry:2.6.1.1"
3030:"carbamoyltransferase"
2776:10.1074/jbc.M608062200
2317:10.1093/jn/121.11.1701
2209:10.1002/jcp.1030540413
1852:, for example certain
1850:transmembrane proteins
1683:functions such as the
1655:of the brain and the
1458:leads to intermittent
1413:
1335:
1272:
1245:
1226:
1170:
1135:aspartate transaminase
1114:
1101:L-cysteine + acetate.
1095:
1039:
963:
900:
874:
851:
807:
763:
743:
719:
683:
659:
631:
596:
572:
544:
519:
429:
271:
225:
199:
121:(PDH), which converts
119:pyruvate dehydrogenase
59:
8913:Aryl sulfotransferase
8381:Reverse transcriptase
7785:: Oximinotransferases
7472:Arabinosyltransferase
6948:N-glycosyltransferase
6747:Citrate (Re)-synthase
6742:Decylcitrate synthase
6683:Aminoacyltransferases
6583:palmitoyltransferases
6417:Acetolactate synthase
6135:DNA methyltransferase
5840:Eadie–Hofstee diagram
5773:Allosteric regulation
5257:10.1176/ajp.150.3.454
4834:Stone TW (Sep 1972).
3584:10.1074/jbc.271.1.471
1771:due to their role in
1732:Terminal transferases
1727:Terminal transferases
1722:Uses in biotechnology
1527:galactose-1-phosphate
1406:
1336:
1273:
1243:
1227:
1171:
1112:
1096:
1069:-acetyl-L-serine and
1040:
964:
907:erythrose 4-phosphate
901:
872:
852:
805:
772:molybdenumtransferase
515:Group(s) transferred
450:DNA methyltransferase
430:
407:is the acceptor, and
269:
226:
187:
40:
8881:cysteine desulfurase
8170:diphosphotransferase
8152:Thiamine-diphosphate
7859:-containing groups (
7736:Alanine transaminase
7340:Pseudomonas exotoxin
6860:glycosyltransferases
6809:Homocitrate synthase
6696:Peptidyl transferase
6130:Thymidylate synthase
5850:Lineweaver–Burk plot
5582:10.3390/ijms12117982
5292:10.1002/ajmg.b.30468
3834:10.1210/me.2012-1145
3281:. UniProt Consortium
3122:"ENZYME class 2.2.1"
1842:glycosyltransferases
1830:biological membranes
1631:Huntington's disease
1325:
1262:
1216:
1160:
1085:
1077:-acetyl-L-serine + H
1029:
1005:through transfer of
953:
924:Peptidyl transferase
911:fructose 6-phosphate
890:
841:
644:chlorophyll synthase
443:N-methyl-L-glutamate
419:
163:
131:peptidyl transferase
92:biochemical pathways
9002:-containing group (
8821:-containing group (
8752:: protein-histidine
8670:; protein acceptor)
8558:mRNA capping enzyme
8550:Guanylyltransferase
7638:Farnesyltransferase
7613:Spermidine synthase
7603:Riboflavin synthase
7115:Hyaluronan synthase
6541:N-acetyltransferase
6099:Methionine synthase
5670:Membranome database
5383:2012NYASA1275...54E
4716:Clinical Laboratory
3411:2013PLoSO...880135Q
3034:The Free Dictionary
2802:"EC 2 Introduction"
2718:2009CBio...19.1200Z
2620:2005CBio...15.R887M
2405:1953Natur.172.1036M
2353:1950Natur.166..517H
2244:1937Natur.140R.503B
2109:1953Natur.172...65M
1838:transmembrane helix
1790:Rubber transferases
1653:mesopontine tegment
1589:Alzheimer's disease
1583:Alzheimer's disease
1515:Duarte galactosemia
1372:It is possible for
1137:, which can act on
1127:oximinotransferases
991:Glycosyltransferase
832:carbamoyl phosphate
818:, formyl, carboxy,
776:tungstentransferase
759:-containing groups
739:-containing groups
640:riboflavin synthase
620:groups, as well as
613:pentosyltransferase
605:glycosyltransferase
505:
304:Rudolf Schoenheimer
197:
111:N-acetyltransferase
9017:Selenotransferases
8836:Sulfurtransferases
8028:Phosphoinositide 3
7872:phosphotransferase
7792:Oximinotransferase
7455:Xylosyltransferase
6928:Debranching enzyme
6501:acetyltransferases
5809:Enzyme superfamily
5742:Enzyme promiscuity
5427:Muscle & Nerve
5221:10.1093/hmg/ddl252
5159:Brain and Behavior
4943:Oda Y (Nov 1999).
4793:Psychopharmacology
4183:"ABO Blood Groups"
4098:10.1271/bbb.110276
1806:Hevea brasiliensis
1414:
1331:
1268:
1246:
1222:
1190:phosphotransferase
1166:
1131:amidinotransferase
1115:
1091:
1035:
959:
896:
875:
847:
808:
692:phosphotransferase
672:oximinotransferase
609:hexosyltransferase
503:
425:
295:Dorothy M. Needham
272:
253:beta-galactosidase
221:
105:, which transfers
60:
9038:
9037:
8962:
8961:
8895:Sulfotransferases
8781:
8780:
8777:
8776:
8653:
8652:
8592:
8591:
8493:
8492:
8406:Template-directed
8160:
8159:
8127:Phosphomevalonate
7819:
7818:
7741:GABA transaminase
7656:
7655:
7628:Spermine synthase
7525:
7524:
7487:
7486:
7443:
7442:
7292:
7291:
6923:Glycogen synthase
6822:
6821:
6772:ATP citrate lyase
6440:
6439:
6354:
6353:
6328:
6327:
6243:
6242:
6201:Formyltransferase
6153:
6152:
5965:
5964:
5616:"What is Rubber?"
5439:10.1002/mus.10300
4902:10.1111/jnc.12128
4723:(9–10): 1139–46.
4643:Berry GT (2000).
4334:"SCOT deficiency"
4167:978-0-19-850673-7
4011:10.1021/bi061551q
3633:978-0-07-299539-8
3217:10.1038/nsmb.1577
2905:978-90-481-6205-5
2678:10.1242/dev.01962
2510:malaria vector".
2413:10.1038/1721036a0
2284:978-0-02-851800-8
2238:(3542): 503–504.
2164:10.1042/bj0240208
2048:Watson, James D.
1657:nucleus accumbens
1479:CPT-II deficiency
1470:CPT-II deficiency
1059:Cysteine synthase
790:
789:
752:selenotransferase
728:sulfurtransferase
532:formyltransferase
528:methyltransferase
409:methyltransferase
375:glycosaminoglycan
219:
211:
205:
195:
183:
177:
169:
76:functional groups
18:Formyltransferase
16:(Redirected from
9080:
9055:
9054:
9046:
8989:
8982:
8975:
8966:
8927:CoA-transferases
8808:
8801:
8794:
8785:
8769:Histidine kinase
8692:tyrosine kinases
8682:protein-tyrosine
8674:
8604:
8411:RNA polymerase I
8234:
8225:
8078:Aspartate kinase
8073:Phosphoglycerate
7890:
7846:
7839:
7832:
7823:
7683:
7676:
7669:
7660:
7552:
7545:
7538:
7529:
7335:Diphtheria toxin
7318:
7309:
6966:Lactose synthase
6933:Branching enzyme
6884:
6849:
6842:
6835:
6826:
6782:HMG-CoA synthase
6737:Citrate synthase
6701:Transglutaminase
6478:acyltransferases
6467:
6460:
6453:
6444:
6381:
6374:
6367:
6358:
6264:
6174:
6021:
5992:
5985:
5978:
5969:
5845:Hanes–Woolf plot
5788:Enzyme activator
5783:Enzyme inhibitor
5757:Enzyme catalysis
5701:
5694:
5687:
5678:
5672:
5663:
5657:
5656:
5654:
5652:
5641:
5632:
5631:
5629:
5627:
5611:
5605:
5604:
5594:
5584:
5560:
5554:
5553:
5525:
5514:
5513:
5495:
5489:
5488:
5486:
5484:
5468:
5459:
5458:
5422:
5413:
5412:
5402:
5362:
5356:
5355:
5345:
5321:
5312:
5311:
5275:
5269:
5268:
5240:
5234:
5233:
5223:
5199:
5193:
5192:
5182:
5171:10.1002/brb3.104
5150:
5144:
5143:
5126:(22): 1688–700.
5115:
5109:
5108:
5098:
5066:
5060:
5059:
5025:
5016:
5010:
5009:
5007:
5005:
4990:
4984:
4983:
4949:
4940:
4923:
4922:
4904:
4880:
4874:
4873:
4863:
4831:
4825:
4824:
4788:
4782:
4781:
4771:
4747:
4741:
4740:
4710:
4704:
4703:
4667:
4661:
4660:
4640:
4634:
4633:
4631:
4629:
4618:
4607:
4606:
4570:
4564:
4563:
4545:
4521:
4515:
4514:
4504:
4472:
4466:
4465:
4463:
4461:
4446:
4440:
4439:
4437:
4435:
4416:
4397:
4396:
4394:
4392:
4377:
4371:
4370:
4368:
4366:
4360:
4352:
4346:
4345:
4343:
4341:
4330:
4324:
4323:
4321:
4319:
4308:
4295:
4294:
4292:
4290:
4275:
4269:
4268:
4266:
4264:
4253:
4247:
4246:
4244:
4242:
4236:"Human ABO Gene"
4231:
4225:
4224:
4222:
4220:
4205:
4199:
4198:
4196:
4194:
4178:
4172:
4171:
4159:
4149:
4136:
4135:
4133:
4131:
4120:
4111:
4110:
4100:
4076:
4067:
4066:
4039:
4033:
4032:
4022:
3990:
3984:
3983:
3981:
3979:
3964:
3958:
3957:
3947:
3923:
3917:
3916:
3914:
3912:
3897:
3891:
3890:
3862:
3856:
3855:
3845:
3813:
3807:
3806:
3804:
3802:
3791:"Enzyme 2.8.2.2"
3787:
3781:
3780:
3778:
3776:
3761:
3755:
3754:
3722:
3716:
3715:
3713:
3711:
3696:
3690:
3689:
3687:
3685:
3670:
3664:
3663:
3661:
3659:
3644:
3638:
3637:
3621:
3614:Lewis R (2008).
3611:
3605:
3604:
3586:
3562:
3556:
3555:
3553:
3551:
3536:
3530:
3529:
3527:
3525:
3510:
3504:
3503:
3475:
3469:
3468:
3466:
3464:
3449:
3443:
3442:
3432:
3422:
3390:
3384:
3383:
3381:
3379:
3364:
3358:
3357:
3355:
3353:
3338:
3332:
3331:
3321:
3297:
3291:
3290:
3288:
3286:
3271:
3265:
3264:
3262:
3260:
3245:
3239:
3238:
3228:
3196:
3190:
3189:
3187:
3185:
3170:
3164:
3163:
3161:
3159:
3144:
3138:
3137:
3135:
3133:
3118:
3112:
3111:
3109:
3091:
3085:Escherichia coli
3078:
3072:
3071:
3069:
3067:
3052:
3046:
3045:
3043:
3041:
3026:
3020:
3019:
3017:
3015:
3000:
2994:
2993:
2991:
2989:
2974:
2965:
2964:
2962:
2960:
2945:
2936:
2935:
2933:
2931:
2916:
2910:
2909:
2891:
2885:
2884:
2882:
2880:
2865:
2859:
2858:
2848:
2824:
2818:
2817:
2815:
2813:
2798:
2789:
2788:
2778:
2754:
2748:
2747:
2737:
2697:
2691:
2690:
2680:
2656:
2650:
2649:
2631:
2599:
2593:
2592:
2574:
2550:
2544:
2543:
2506:
2500:
2499:
2489:
2465:
2459:
2458:
2456:
2454:
2449:. Nobel Media AB
2439:
2433:
2432:
2399:(4388): 1036–7.
2387:
2381:
2380:
2361:10.1038/166517a0
2336:
2330:
2329:
2319:
2295:
2289:
2288:
2270:
2264:
2263:
2252:10.1038/140503b0
2227:
2221:
2220:
2192:
2186:
2185:
2175:
2143:
2137:
2136:
2117:10.1038/172065a0
2092:
2086:
2085:
2059:
2053:
2046:
2040:
2039:
2029:
1997:
1991:
1990:
1988:
1986:
1970:
1964:
1963:
1953:
1944:(18): 10882–92.
1929:
1923:
1922:
1920:
1918:
1903:
1897:
1896:
1894:
1892:
1877:
1810:synthetic rubber
1777:oxidative damage
1740:deoxynucleotides
1554:neurotransmitter
1429:cellular product
1340:
1338:
1337:
1332:
1312:Escherichia coli
1277:
1275:
1274:
1269:
1231:
1229:
1228:
1223:
1175:
1173:
1172:
1167:
1100:
1098:
1097:
1092:
1071:hydrogen sulfide
1044:
1042:
1041:
1036:
1015:protein subunits
1011:lactose synthase
968:
966:
965:
960:
946:+ aminoacyl-tRNA
905:
903:
902:
897:
856:
854:
853:
848:
732:sulfotransferase
506:
434:
432:
431:
426:
393:Systematic names
358:Sir Bernard Katz
291:aminotransferase
259:, and acid/base
230:
228:
227:
222:
220:
217:
216:
209:
203:
200:
196:
193:
184:
181:
175:
174:
167:
141:molecule in the
109:, the action of
86:group) from one
50:complexed with
21:
9088:
9087:
9083:
9082:
9081:
9079:
9078:
9077:
9063:
9062:
9061:
9049:
9041:
9039:
9034:
9007:
8993:
8963:
8958:
8939:
8917:
8885:
8876:biotin synthase
8826:
8812:
8782:
8773:
8744:
8722:
8697:
8668:
8662:
8658:2.7.10-2.7.13:
8649:
8633:
8600:: miscellaneous
8588:
8562:
8544:
8521:
8504:exoribonuclease
8501:
8489:
8475:Polyadenylation
8392:
8218:
8212:
8200:
8182:
8178:
8172:
8156:
8118:
8105:
8082:
8054:
7884:
7878:
7870:
7864:
7850:
7820:
7815:
7796:
7777:
7701:
7687:
7657:
7652:
7574:
7556:
7526:
7521:
7498:
7483:
7439:
7412:
7379:
7354:Pertussis toxin
7303:
7288:
7211:
7133:
6985:
6952:
6878:
6867:
6853:
6823:
6818:
6777:Malate synthase
6722:
6673:
6485:
6471:
6441:
6436:
6403:
6385:
6355:
6350:
6324:
6278:
6256:
6239:
6218:
6195:
6166:
6149:
6103:
6075:
6050:
6006:
5996:
5966:
5961:
5873:Oxidoreductases
5859:
5835:Enzyme kinetics
5823:
5819:List of enzymes
5792:
5761:
5732:Catalytic triad
5710:
5705:
5675:
5664:
5660:
5650:
5648:
5643:
5642:
5635:
5625:
5623:
5613:
5612:
5608:
5575:(11): 7982–95.
5562:
5561:
5557:
5527:
5526:
5517:
5510:
5497:
5496:
5492:
5482:
5480:
5470:
5469:
5462:
5424:
5423:
5416:
5364:
5363:
5359:
5323:
5322:
5315:
5277:
5276:
5272:
5242:
5241:
5237:
5214:(21): 3119–31.
5201:
5200:
5196:
5152:
5151:
5147:
5117:
5116:
5112:
5068:
5067:
5063:
5023:
5018:
5017:
5013:
5003:
5001:
4992:
4991:
4987:
4947:
4942:
4941:
4926:
4882:
4881:
4877:
4833:
4832:
4828:
4790:
4789:
4785:
4749:
4748:
4744:
4712:
4711:
4707:
4669:
4668:
4664:
4642:
4641:
4637:
4627:
4625:
4620:
4619:
4610:
4572:
4571:
4567:
4523:
4522:
4518:
4474:
4473:
4469:
4459:
4457:
4448:
4447:
4443:
4433:
4431:
4418:
4417:
4400:
4390:
4388:
4379:
4378:
4374:
4364:
4362:
4358:
4354:
4353:
4349:
4339:
4337:
4332:
4331:
4327:
4317:
4315:
4310:
4309:
4298:
4288:
4286:
4277:
4276:
4272:
4262:
4260:
4256:Kidd, Kenneth.
4255:
4254:
4250:
4240:
4238:
4233:
4232:
4228:
4218:
4216:
4207:
4206:
4202:
4192:
4190:
4180:
4179:
4175:
4168:
4151:
4150:
4139:
4129:
4127:
4122:
4121:
4114:
4078:
4077:
4070:
4063:
4041:
4040:
4036:
3992:
3991:
3987:
3977:
3975:
3966:
3965:
3961:
3925:
3924:
3920:
3910:
3908:
3899:
3898:
3894:
3864:
3863:
3859:
3815:
3814:
3810:
3800:
3798:
3789:
3788:
3784:
3774:
3772:
3763:
3762:
3758:
3724:
3723:
3719:
3709:
3707:
3698:
3697:
3693:
3683:
3681:
3672:
3671:
3667:
3657:
3655:
3646:
3645:
3641:
3634:
3613:
3612:
3608:
3564:
3563:
3559:
3549:
3547:
3538:
3537:
3533:
3523:
3521:
3512:
3511:
3507:
3477:
3476:
3472:
3462:
3460:
3451:
3450:
3446:
3392:
3391:
3387:
3377:
3375:
3366:
3365:
3361:
3351:
3349:
3340:
3339:
3335:
3299:
3298:
3294:
3284:
3282:
3273:
3272:
3268:
3258:
3256:
3247:
3246:
3242:
3198:
3197:
3193:
3183:
3181:
3172:
3171:
3167:
3157:
3155:
3146:
3145:
3141:
3131:
3129:
3120:
3119:
3115:
3089:
3080:
3079:
3075:
3065:
3063:
3054:
3053:
3049:
3039:
3037:
3028:
3027:
3023:
3013:
3011:
3002:
3001:
2997:
2987:
2985:
2976:
2975:
2968:
2958:
2956:
2947:
2946:
2939:
2929:
2927:
2918:
2917:
2913:
2906:
2893:
2892:
2888:
2878:
2876:
2867:
2866:
2862:
2826:
2825:
2821:
2811:
2809:
2800:
2799:
2792:
2769:(11): 8356–67.
2756:
2755:
2751:
2706:Current Biology
2699:
2698:
2694:
2671:(17): 3813–22.
2658:
2657:
2653:
2614:(21): R887–99.
2608:Current Biology
2601:
2600:
2596:
2552:
2551:
2547:
2524:10.1603/me09132
2508:
2507:
2503:
2467:
2466:
2462:
2452:
2450:
2441:
2440:
2436:
2389:
2388:
2384:
2347:(4221): 517–8.
2338:
2337:
2333:
2297:
2296:
2292:
2285:
2272:
2271:
2267:
2229:
2228:
2224:
2203:(S1): 161–177.
2194:
2193:
2189:
2145:
2144:
2140:
2094:
2093:
2089:
2082:
2061:
2060:
2056:
2047:
2043:
1999:
1998:
1994:
1984:
1982:
1972:
1971:
1967:
1931:
1930:
1926:
1916:
1914:
1905:
1904:
1900:
1890:
1888:
1879:
1878:
1874:
1870:
1846:Golgi apparatus
1826:
1792:
1773:drug resistance
1756:
1736:plasmid vectors
1729:
1724:
1712:presynaptically
1697:
1665:
1649:
1633:
1609:
1585:
1547:
1500:
1472:
1444:SCOT deficiency
1437:
1435:SCOT deficiency
1401:
1351:ABO blood group
1347:
1323:
1322:
1296:
1284:
1260:
1259:
1238:
1214:
1213:
1199:protein kinases
1182:
1158:
1157:
1107:
1083:
1082:
1080:
1076:
1068:
1054:
1027:
1026:
1007:monosaccharides
1003:polysaccharides
983:
976:
972:
951:
950:
949:
945:
919:
888:
887:
867:
839:
838:
825:
800:
795:
581:acyltransferase
498:oxidoreductases
482:
468:that transfers
439:
417:
416:
390:
333:and an organic
284:
245:
161:
160:
137:chain from the
35:
28:
23:
22:
15:
12:
11:
5:
9086:
9084:
9076:
9075:
9065:
9064:
9060:
9059:
9036:
9035:
9033:
9032:
9027:
9021:
9019:
9009:
9008:
8994:
8992:
8991:
8984:
8977:
8969:
8960:
8959:
8957:
8956:
8950:
8948:
8941:
8940:
8938:
8937:
8931:
8929:
8919:
8918:
8916:
8915:
8910:
8905:
8899:
8897:
8887:
8886:
8884:
8883:
8878:
8873:
8868:
8863:
8858:
8853:
8852:
8851:
8840:
8838:
8828:
8827:
8813:
8811:
8810:
8803:
8796:
8788:
8779:
8778:
8775:
8774:
8772:
8771:
8766:
8761:
8755:
8753:
8746:
8745:
8743:
8742:
8733:
8731:
8724:
8723:
8721:
8720:
8711:
8709:
8699:
8698:
8696:
8695:
8686:
8684:
8671:
8666:
8660:protein kinase
8655:
8654:
8651:
8650:
8648:
8647:
8641:
8639:
8635:
8634:
8632:
8631:
8626:
8621:
8616:
8610:
8608:
8601:
8594:
8593:
8590:
8589:
8587:
8586:
8581:
8570:
8568:
8564:
8563:
8561:
8560:
8554:
8552:
8546:
8545:
8543:
8542:
8537:
8531:
8529:
8523:
8522:
8520:
8519:
8514:
8508:
8506:
8499:Phosphorolytic
8495:
8494:
8491:
8490:
8488:
8487:
8482:
8477:
8472:
8467:
8466:
8465:
8460:
8455:
8445:
8444:
8443:
8433:
8428:
8423:
8418:
8413:
8408:
8402:
8400:
8398:RNA polymerase
8394:
8393:
8391:
8390:
8389:
8388:
8378:
8374:
8373:
8372:
8371:
8366:
8361:
8350:
8349:
8348:
8343:
8338:
8333:
8322:
8316:
8315:
8314:
8307:
8302:
8297:
8292:
8281:
8280:
8279:
8272:
8267:
8262:
8257:
8246:
8242:
8240:
8238:DNA polymerase
8231:
8222:
8216:
8202:
8201:
8199:
8198:
8193:
8187:
8185:
8180:
8176:
8162:
8161:
8158:
8157:
8155:
8154:
8149:
8144:
8139:
8134:
8129:
8123:
8121:
8116:
8107:
8106:
8104:
8103:
8097:
8095:
8084:
8083:
8081:
8080:
8075:
8069:
8067:
8056:
8055:
8053:
8052:
8047:
8042:
8041:
8040:
8035:
8025:
8023:Diacylglycerol
8020:
8015:
8010:
8005:
8000:
7995:
7990:
7985:
7984:
7983:
7973:
7968:
7963:
7962:
7961:
7956:
7951:
7946:
7941:
7934:Phosphofructo-
7931:
7926:
7925:
7924:
7914:
7909:
7903:
7901:
7887:
7882:
7866:
7865:
7851:
7849:
7848:
7841:
7834:
7826:
7817:
7816:
7814:
7813:
7807:
7805:
7798:
7797:
7795:
7794:
7788:
7786:
7779:
7778:
7776:
7775:
7774:
7773:
7763:
7758:
7753:
7748:
7743:
7738:
7733:
7732:
7731:
7726:
7715:
7713:
7703:
7702:
7688:
7686:
7685:
7678:
7671:
7663:
7654:
7653:
7651:
7650:
7645:
7640:
7635:
7630:
7625:
7620:
7615:
7610:
7605:
7600:
7595:
7590:
7584:
7582:
7576:
7575:
7557:
7555:
7554:
7547:
7540:
7532:
7523:
7522:
7520:
7519:
7514:
7509:
7503:
7501:
7489:
7488:
7485:
7484:
7482:
7481:
7480:
7479:
7469:
7468:
7467:
7462:
7451:
7449:
7445:
7444:
7441:
7440:
7438:
7437:
7436:
7435:
7420:
7418:
7414:
7413:
7411:
7410:
7405:
7400:
7395:
7389:
7387:
7381:
7380:
7378:
7377:
7371:
7370:
7364:
7363:
7362:
7361:
7356:
7345:
7344:
7343:
7342:
7337:
7326:
7324:
7315:
7306:
7294:
7293:
7290:
7289:
7287:
7286:
7281:
7276:
7271:
7266:
7261:
7256:
7250:
7249:
7244:
7239:
7238:
7237:
7232:
7221:
7219:
7213:
7212:
7210:
7209:
7204:
7199:
7194:
7189:
7184:
7179:
7174:
7169:
7164:
7159:
7154:
7149:
7143:
7141:
7135:
7134:
7132:
7131:
7126:
7121:
7111:
7110:
7105:
7100:
7095:
7090:
7085:
7080:
7075:
7070:
7065:
7059:
7058:
7053:
7048:
7043:
7038:
7033:
7028:
7023:
7018:
7012:
7011:
7006:
7001:
6995:
6993:
6987:
6986:
6984:
6983:
6973:
6968:
6962:
6960:
6954:
6953:
6951:
6950:
6945:
6940:
6935:
6930:
6925:
6920:
6919:
6918:
6913:
6908:
6903:
6892:
6890:
6881:
6869:
6868:
6854:
6852:
6851:
6844:
6837:
6829:
6820:
6819:
6817:
6816:
6811:
6806:
6801:
6796:
6791:
6790:
6789:
6779:
6774:
6769:
6764:
6759:
6754:
6749:
6744:
6739:
6733:
6731:
6724:
6723:
6721:
6720:
6719:
6718:
6713:
6708:
6698:
6693:
6687:
6685:
6675:
6674:
6672:
6671:
6666:
6661:
6656:
6650:
6649:
6644:
6639:
6634:
6629:
6620:
6619:
6618:
6617:
6612:
6602:
6601:
6600:
6595:
6578:
6577:
6576:
6575:
6570:
6560:
6559:
6558:
6553:
6548:
6538:
6533:
6528:
6523:
6518:
6513:
6508:
6496:
6494:
6487:
6486:
6472:
6470:
6469:
6462:
6455:
6447:
6438:
6437:
6435:
6434:
6429:
6424:
6419:
6413:
6411:
6405:
6404:
6386:
6384:
6383:
6376:
6369:
6361:
6352:
6351:
6349:
6348:
6342:
6340:
6330:
6329:
6326:
6325:
6323:
6322:
6317:
6312:
6307:
6302:
6297:
6292:
6286:
6284:
6280:
6279:
6277:
6276:
6270:
6268:
6261:
6245:
6244:
6241:
6240:
6238:
6237:
6232:
6226:
6224:
6220:
6219:
6217:
6216:
6211:
6205:
6203:
6197:
6196:
6194:
6193:
6188:
6182:
6180:
6171:
6155:
6154:
6151:
6150:
6148:
6147:
6142:
6137:
6132:
6127:
6122:
6117:
6111:
6109:
6105:
6104:
6102:
6101:
6096:
6091:
6085:
6083:
6077:
6076:
6074:
6073:
6068:
6058:
6056:
6052:
6051:
6049:
6048:
6043:
6038:
6033:
6027:
6025:
6018:
6008:
6007:
5997:
5995:
5994:
5987:
5980:
5972:
5963:
5962:
5960:
5959:
5946:
5933:
5920:
5907:
5894:
5881:
5867:
5865:
5861:
5860:
5858:
5857:
5852:
5847:
5842:
5837:
5831:
5829:
5825:
5824:
5822:
5821:
5816:
5811:
5806:
5800:
5798:
5797:Classification
5794:
5793:
5791:
5790:
5785:
5780:
5775:
5769:
5767:
5763:
5762:
5760:
5759:
5754:
5749:
5744:
5739:
5734:
5729:
5724:
5718:
5716:
5712:
5711:
5706:
5704:
5703:
5696:
5689:
5681:
5674:
5673:
5658:
5633:
5606:
5555:
5515:
5508:
5490:
5460:
5414:
5357:
5313:
5270:
5235:
5194:
5145:
5110:
5061:
5011:
4985:
4958:(11): 921–37.
4924:
4875:
4826:
4799:(9): 1975–85.
4783:
4742:
4705:
4662:
4635:
4622:"Galactosemia"
4608:
4565:
4516:
4467:
4450:"Galactosemia"
4441:
4398:
4372:
4347:
4325:
4296:
4270:
4248:
4226:
4200:
4173:
4166:
4137:
4112:
4068:
4061:
4034:
3985:
3959:
3938:(10): 6324–8.
3918:
3892:
3857:
3808:
3782:
3756:
3717:
3691:
3674:"1aqy Summary"
3665:
3639:
3632:
3606:
3557:
3531:
3505:
3486:(3): 497–525.
3470:
3444:
3405:(11): e80135.
3385:
3359:
3333:
3292:
3266:
3240:
3191:
3165:
3139:
3113:
3073:
3047:
3021:
2995:
2966:
2937:
2911:
2904:
2886:
2860:
2819:
2790:
2749:
2712:(14): 1200–5.
2692:
2651:
2594:
2545:
2501:
2480:(11): 923–36.
2460:
2447:Nobelprize.org
2434:
2382:
2331:
2310:(11): 1701–4.
2290:
2283:
2265:
2222:
2187:
2138:
2103:(4367): 65–8.
2087:
2081:978-0470016176
2080:
2054:
2041:
1992:
1965:
1924:
1898:
1871:
1869:
1866:
1856:or microsomal
1825:
1822:
1791:
1788:
1755:
1752:
1728:
1725:
1723:
1720:
1696:
1693:
1685:cardiovascular
1664:
1661:
1648:
1645:
1635:Patients with
1632:
1629:
1611:Patients with
1608:
1605:
1584:
1581:
1546:
1543:
1499:
1496:
1471:
1468:
1436:
1433:
1400:
1397:
1346:
1343:
1330:
1295:
1292:
1283:
1280:
1267:
1237:
1234:
1221:
1205:molecules for
1181:
1178:
1165:
1106:
1103:
1090:
1078:
1074:
1066:
1053:
1050:
1034:
982:
979:
974:
970:
958:
947:
943:
940:aminoacyl-tRNA
918:
915:
895:
866:
863:
846:
823:
799:
796:
794:
791:
788:
787:
778:
769:
761:
760:
754:
749:
741:
740:
734:
725:
717:
716:
702:
689:
681:
680:
674:
665:
657:
656:
646:
637:
629:
628:
615:
602:
594:
593:
583:
578:
570:
569:
559:
550:
542:
541:
534:
525:
517:
516:
513:
510:
481:
480:Classification
478:
462:RNA polymerase
446:
445:
437:
424:
403:is the donor,
389:
386:
350:Julius Axelrod
282:
275:Transamination
244:
241:
232:
231:
215:
208:
190:
180:
173:
42:RNA polymerase
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
9085:
9074:
9071:
9070:
9068:
9058:
9053:
9048:
9044:
9031:
9028:
9026:
9023:
9022:
9020:
9018:
9014:
9010:
9005:
9001:
8997:
8990:
8985:
8983:
8978:
8976:
8971:
8970:
8967:
8955:
8952:
8951:
8949:
8946:
8942:
8936:
8933:
8932:
8930:
8928:
8924:
8920:
8914:
8911:
8909:
8906:
8904:
8901:
8900:
8898:
8896:
8892:
8888:
8882:
8879:
8877:
8874:
8872:
8869:
8867:
8864:
8862:
8859:
8857:
8854:
8850:
8847:
8846:
8845:
8842:
8841:
8839:
8837:
8833:
8829:
8824:
8820:
8816:
8809:
8804:
8802:
8797:
8795:
8790:
8789:
8786:
8770:
8767:
8765:
8762:
8760:
8757:
8756:
8754:
8751:
8747:
8741:
8740:
8735:
8734:
8732:
8729:
8725:
8719:
8718:
8713:
8712:
8710:
8708:
8704:
8700:
8694:
8693:
8688:
8687:
8685:
8683:
8679:
8675:
8672:
8669:
8661:
8656:
8646:
8643:
8642:
8640:
8636:
8630:
8627:
8625:
8622:
8620:
8617:
8615:
8612:
8611:
8609:
8605:
8602:
8599:
8595:
8585:
8582:
8579:
8575:
8572:
8571:
8569:
8565:
8559:
8556:
8555:
8553:
8551:
8547:
8541:
8538:
8536:
8533:
8532:
8530:
8528:
8524:
8518:
8515:
8513:
8510:
8509:
8507:
8505:
8500:
8496:
8486:
8483:
8481:
8478:
8476:
8473:
8471:
8468:
8464:
8461:
8459:
8456:
8454:
8451:
8450:
8449:
8446:
8442:
8439:
8438:
8437:
8434:
8432:
8429:
8427:
8424:
8422:
8419:
8417:
8414:
8412:
8409:
8407:
8404:
8403:
8401:
8399:
8395:
8387:
8384:
8383:
8382:
8379:
8376:
8375:
8370:
8367:
8365:
8362:
8360:
8357:
8356:
8354:
8351:
8347:
8344:
8342:
8339:
8337:
8334:
8332:
8329:
8328:
8326:
8323:
8320:
8317:
8313:
8312:
8308:
8306:
8303:
8301:
8298:
8296:
8293:
8291:
8288:
8287:
8285:
8282:
8278:
8277:
8273:
8271:
8268:
8266:
8263:
8261:
8258:
8256:
8253:
8252:
8250:
8247:
8244:
8243:
8241:
8239:
8235:
8232:
8230:
8226:
8223:
8220:
8211:
8207:
8203:
8197:
8194:
8192:
8189:
8188:
8186:
8183:
8171:
8167:
8163:
8153:
8150:
8148:
8145:
8143:
8140:
8138:
8135:
8133:
8130:
8128:
8125:
8124:
8122:
8119:
8112:
8108:
8102:
8099:
8098:
8096:
8093:
8089:
8085:
8079:
8076:
8074:
8071:
8070:
8068:
8065:
8061:
8057:
8051:
8048:
8046:
8043:
8039:
8038:Class II PI 3
8036:
8034:
8031:
8030:
8029:
8026:
8024:
8021:
8019:
8016:
8014:
8013:Deoxycytidine
8011:
8009:
8006:
8004:
8001:
7999:
7996:
7994:
7991:
7989:
7986:
7982:
7981:ADP-thymidine
7979:
7978:
7977:
7974:
7972:
7969:
7967:
7964:
7960:
7957:
7955:
7952:
7950:
7947:
7945:
7942:
7940:
7937:
7936:
7935:
7932:
7930:
7927:
7923:
7920:
7919:
7918:
7915:
7913:
7910:
7908:
7905:
7904:
7902:
7899:
7895:
7891:
7888:
7885:
7877:
7873:
7867:
7862:
7858:
7854:
7847:
7842:
7840:
7835:
7833:
7828:
7827:
7824:
7812:
7809:
7808:
7806:
7803:
7799:
7793:
7790:
7789:
7787:
7784:
7780:
7772:
7769:
7768:
7767:
7764:
7762:
7759:
7757:
7754:
7752:
7749:
7747:
7744:
7742:
7739:
7737:
7734:
7730:
7727:
7725:
7722:
7721:
7720:
7717:
7716:
7714:
7712:
7711:Transaminases
7708:
7704:
7699:
7695:
7691:
7684:
7679:
7677:
7672:
7670:
7665:
7664:
7661:
7649:
7646:
7644:
7641:
7639:
7636:
7634:
7631:
7629:
7626:
7624:
7621:
7619:
7616:
7614:
7611:
7609:
7606:
7604:
7601:
7599:
7596:
7594:
7591:
7589:
7586:
7585:
7583:
7581:
7577:
7572:
7568:
7564:
7560:
7553:
7548:
7546:
7541:
7539:
7534:
7533:
7530:
7518:
7515:
7513:
7510:
7508:
7505:
7504:
7502:
7500:
7494:
7490:
7478:
7475:
7474:
7473:
7470:
7466:
7463:
7461:
7458:
7457:
7456:
7453:
7452:
7450:
7446:
7434:
7431:
7430:
7429:
7425:
7422:
7421:
7419:
7415:
7409:
7406:
7404:
7401:
7399:
7396:
7394:
7391:
7390:
7388:
7386:
7382:
7376:
7373:
7372:
7369:
7366:
7365:
7360:
7359:Cholera toxin
7357:
7355:
7352:
7351:
7350:
7347:
7346:
7341:
7338:
7336:
7333:
7332:
7331:
7328:
7327:
7325:
7323:
7319:
7316:
7314:
7310:
7307:
7305:
7299:
7295:
7285:
7282:
7280:
7277:
7275:
7272:
7270:
7267:
7265:
7262:
7260:
7257:
7255:
7252:
7251:
7248:
7245:
7243:
7240:
7236:
7233:
7231:
7228:
7227:
7226:
7223:
7222:
7220:
7218:
7214:
7208:
7205:
7203:
7200:
7198:
7195:
7193:
7190:
7188:
7185:
7183:
7180:
7178:
7175:
7173:
7170:
7168:
7165:
7163:
7160:
7158:
7155:
7153:
7150:
7148:
7145:
7144:
7142:
7140:
7136:
7130:
7127:
7125:
7122:
7120:
7116:
7113:
7112:
7109:
7106:
7104:
7101:
7099:
7096:
7094:
7091:
7089:
7086:
7084:
7081:
7079:
7076:
7074:
7071:
7069:
7066:
7064:
7061:
7060:
7057:
7054:
7052:
7049:
7047:
7044:
7042:
7039:
7037:
7034:
7032:
7029:
7027:
7024:
7022:
7019:
7017:
7014:
7013:
7010:
7007:
7005:
7002:
7000:
6997:
6996:
6994:
6992:
6991:Glucuronosyl-
6988:
6981:
6977:
6974:
6972:
6969:
6967:
6964:
6963:
6961:
6959:
6955:
6949:
6946:
6944:
6941:
6939:
6936:
6934:
6931:
6929:
6926:
6924:
6921:
6917:
6914:
6912:
6909:
6907:
6904:
6902:
6899:
6898:
6897:
6896:Phosphorylase
6894:
6893:
6891:
6889:
6885:
6882:
6880:
6874:
6870:
6865:
6861:
6857:
6850:
6845:
6843:
6838:
6836:
6831:
6830:
6827:
6815:
6812:
6810:
6807:
6805:
6802:
6800:
6797:
6795:
6792:
6788:
6785:
6784:
6783:
6780:
6778:
6775:
6773:
6770:
6768:
6765:
6763:
6760:
6758:
6755:
6753:
6750:
6748:
6745:
6743:
6740:
6738:
6735:
6734:
6732:
6729:
6725:
6717:
6714:
6712:
6709:
6707:
6704:
6703:
6702:
6699:
6697:
6694:
6692:
6689:
6688:
6686:
6684:
6680:
6676:
6670:
6667:
6665:
6662:
6660:
6657:
6655:
6652:
6651:
6648:
6645:
6643:
6640:
6638:
6635:
6633:
6630:
6628:
6625:
6622:
6621:
6616:
6613:
6611:
6608:
6607:
6606:
6603:
6599:
6596:
6594:
6591:
6590:
6589:
6586:
6584:
6580:
6579:
6574:
6571:
6569:
6566:
6565:
6564:
6561:
6557:
6554:
6552:
6549:
6547:
6544:
6543:
6542:
6539:
6537:
6534:
6532:
6529:
6527:
6524:
6522:
6519:
6517:
6514:
6512:
6509:
6507:
6504:
6502:
6498:
6497:
6495:
6492:
6488:
6483:
6479:
6475:
6468:
6463:
6461:
6456:
6454:
6449:
6448:
6445:
6433:
6430:
6428:
6427:Transketolase
6425:
6423:
6422:Transaldolase
6420:
6418:
6415:
6414:
6412:
6410:
6406:
6401:
6397:
6393:
6389:
6382:
6377:
6375:
6370:
6368:
6363:
6362:
6359:
6347:
6344:
6343:
6341:
6339:
6335:
6331:
6321:
6318:
6316:
6313:
6311:
6308:
6306:
6303:
6301:
6298:
6296:
6293:
6291:
6288:
6287:
6285:
6281:
6275:
6272:
6271:
6269:
6265:
6262:
6260:
6254:
6250:
6246:
6236:
6233:
6231:
6228:
6227:
6225:
6221:
6215:
6212:
6210:
6207:
6206:
6204:
6202:
6198:
6192:
6189:
6187:
6184:
6183:
6181:
6179:
6175:
6172:
6170:- and Related
6169:
6164:
6163:Hydroxymethyl
6160:
6156:
6146:
6143:
6141:
6138:
6136:
6133:
6131:
6128:
6126:
6123:
6121:
6118:
6116:
6113:
6112:
6110:
6106:
6100:
6097:
6095:
6092:
6090:
6087:
6086:
6084:
6082:
6078:
6072:
6069:
6067:
6063:
6060:
6059:
6057:
6053:
6047:
6044:
6042:
6039:
6037:
6034:
6032:
6029:
6028:
6026:
6022:
6019:
6017:
6013:
6009:
6004:
6000:
5993:
5988:
5986:
5981:
5979:
5974:
5973:
5970:
5957:
5953:
5952:
5947:
5944:
5940:
5939:
5934:
5931:
5927:
5926:
5921:
5918:
5914:
5913:
5908:
5905:
5901:
5900:
5895:
5892:
5888:
5887:
5882:
5879:
5875:
5874:
5869:
5868:
5866:
5862:
5856:
5853:
5851:
5848:
5846:
5843:
5841:
5838:
5836:
5833:
5832:
5830:
5826:
5820:
5817:
5815:
5814:Enzyme family
5812:
5810:
5807:
5805:
5802:
5801:
5799:
5795:
5789:
5786:
5784:
5781:
5779:
5778:Cooperativity
5776:
5774:
5771:
5770:
5768:
5764:
5758:
5755:
5753:
5750:
5748:
5745:
5743:
5740:
5738:
5737:Oxyanion hole
5735:
5733:
5730:
5728:
5725:
5723:
5720:
5719:
5717:
5713:
5709:
5702:
5697:
5695:
5690:
5688:
5683:
5682:
5679:
5671:
5667:
5662:
5659:
5646:
5640:
5638:
5634:
5621:
5617:
5610:
5607:
5602:
5598:
5593:
5588:
5583:
5578:
5574:
5570:
5566:
5559:
5556:
5551:
5547:
5543:
5539:
5536:(3): 211–23.
5535:
5531:
5524:
5522:
5520:
5516:
5511:
5509:9788189866099
5505:
5501:
5494:
5491:
5478:
5474:
5467:
5465:
5461:
5456:
5452:
5448:
5444:
5440:
5436:
5432:
5428:
5421:
5419:
5415:
5410:
5406:
5401:
5396:
5392:
5388:
5384:
5380:
5376:
5372:
5368:
5361:
5358:
5353:
5349:
5344:
5339:
5336:(3): 255–64.
5335:
5331:
5327:
5320:
5318:
5314:
5309:
5305:
5301:
5297:
5293:
5289:
5286:(7): 849–53.
5285:
5281:
5274:
5271:
5266:
5262:
5258:
5254:
5250:
5246:
5239:
5236:
5231:
5227:
5222:
5217:
5213:
5209:
5205:
5198:
5195:
5190:
5186:
5181:
5176:
5172:
5168:
5165:(2): 145–58.
5164:
5160:
5156:
5149:
5146:
5141:
5137:
5133:
5129:
5125:
5121:
5114:
5111:
5106:
5102:
5097:
5092:
5088:
5084:
5080:
5076:
5072:
5065:
5062:
5057:
5053:
5049:
5045:
5041:
5037:
5033:
5029:
5022:
5015:
5012:
4999:
4995:
4989:
4986:
4981:
4977:
4973:
4969:
4965:
4961:
4957:
4953:
4946:
4939:
4937:
4935:
4933:
4931:
4929:
4925:
4920:
4916:
4912:
4908:
4903:
4898:
4895:(5): 658–69.
4894:
4890:
4886:
4879:
4876:
4871:
4867:
4862:
4857:
4853:
4849:
4846:(2): 485–99.
4845:
4841:
4837:
4830:
4827:
4822:
4818:
4814:
4810:
4806:
4802:
4798:
4794:
4787:
4784:
4779:
4775:
4770:
4765:
4761:
4757:
4753:
4746:
4743:
4738:
4734:
4730:
4726:
4722:
4718:
4717:
4709:
4706:
4701:
4697:
4693:
4689:
4685:
4681:
4678:(4): 516–25.
4677:
4673:
4666:
4663:
4658:
4654:
4650:
4646:
4639:
4636:
4623:
4617:
4615:
4613:
4609:
4604:
4600:
4596:
4592:
4588:
4584:
4580:
4576:
4569:
4566:
4561:
4557:
4553:
4549:
4544:
4539:
4536:(5): 549–54.
4535:
4531:
4527:
4520:
4517:
4512:
4508:
4503:
4498:
4494:
4490:
4486:
4482:
4478:
4471:
4468:
4455:
4451:
4445:
4442:
4430:
4426:
4422:
4415:
4413:
4411:
4409:
4407:
4405:
4403:
4399:
4386:
4382:
4376:
4373:
4357:
4351:
4348:
4335:
4329:
4326:
4313:
4307:
4305:
4303:
4301:
4297:
4284:
4280:
4274:
4271:
4259:
4252:
4249:
4237:
4230:
4227:
4214:
4210:
4204:
4201:
4188:
4184:
4177:
4174:
4169:
4163:
4158:
4157:
4148:
4146:
4144:
4142:
4138:
4125:
4119:
4117:
4113:
4108:
4104:
4099:
4094:
4091:(8): 1570–5.
4090:
4086:
4082:
4075:
4073:
4069:
4064:
4062:9780470146842
4058:
4054:
4050:
4046:
4038:
4035:
4030:
4026:
4021:
4016:
4012:
4008:
4004:
4000:
3996:
3989:
3986:
3973:
3969:
3963:
3960:
3955:
3951:
3946:
3941:
3937:
3933:
3929:
3922:
3919:
3906:
3902:
3896:
3893:
3888:
3884:
3880:
3876:
3872:
3868:
3861:
3858:
3853:
3849:
3844:
3839:
3835:
3831:
3828:(1): 106–15.
3827:
3823:
3819:
3812:
3809:
3796:
3792:
3786:
3783:
3770:
3766:
3760:
3757:
3752:
3748:
3744:
3740:
3737:(2): 149–57.
3736:
3732:
3728:
3721:
3718:
3705:
3701:
3695:
3692:
3679:
3675:
3669:
3666:
3653:
3649:
3643:
3640:
3635:
3629:
3625:
3620:
3619:
3610:
3607:
3602:
3598:
3594:
3590:
3585:
3580:
3576:
3572:
3568:
3561:
3558:
3545:
3541:
3535:
3532:
3519:
3515:
3509:
3506:
3501:
3497:
3493:
3489:
3485:
3481:
3474:
3471:
3458:
3454:
3448:
3445:
3440:
3436:
3431:
3426:
3421:
3416:
3412:
3408:
3404:
3400:
3396:
3389:
3386:
3373:
3369:
3363:
3360:
3347:
3343:
3337:
3334:
3329:
3325:
3320:
3315:
3312:(8): 2103–8.
3311:
3307:
3303:
3296:
3293:
3280:
3276:
3270:
3267:
3254:
3250:
3244:
3241:
3236:
3232:
3227:
3222:
3218:
3214:
3211:(5): 528–33.
3210:
3206:
3202:
3195:
3192:
3179:
3175:
3169:
3166:
3153:
3149:
3143:
3140:
3127:
3123:
3117:
3114:
3108:
3103:
3099:
3095:
3088:
3086:
3077:
3074:
3061:
3057:
3051:
3048:
3036:. Farlex, Inc
3035:
3031:
3025:
3022:
3009:
3005:
2999:
2996:
2983:
2979:
2978:"Transferase"
2973:
2971:
2967:
2954:
2950:
2944:
2942:
2938:
2925:
2921:
2915:
2912:
2907:
2901:
2897:
2890:
2887:
2875:
2871:
2864:
2861:
2856:
2852:
2847:
2842:
2839:(4): 935–45.
2838:
2834:
2830:
2823:
2820:
2807:
2803:
2797:
2795:
2791:
2786:
2782:
2777:
2772:
2768:
2764:
2760:
2753:
2750:
2745:
2741:
2736:
2731:
2727:
2723:
2719:
2715:
2711:
2707:
2703:
2696:
2693:
2688:
2684:
2679:
2674:
2670:
2666:
2662:
2655:
2652:
2647:
2643:
2639:
2635:
2630:
2625:
2621:
2617:
2613:
2609:
2605:
2598:
2595:
2590:
2586:
2582:
2578:
2573:
2568:
2565:(4): 471–81.
2564:
2560:
2556:
2549:
2546:
2541:
2537:
2533:
2529:
2525:
2521:
2518:(2): 162–71.
2517:
2513:
2505:
2502:
2497:
2493:
2488:
2483:
2479:
2475:
2471:
2464:
2461:
2448:
2444:
2438:
2435:
2430:
2426:
2422:
2418:
2414:
2410:
2406:
2402:
2398:
2394:
2386:
2383:
2378:
2374:
2370:
2366:
2362:
2358:
2354:
2350:
2346:
2342:
2335:
2332:
2327:
2323:
2318:
2313:
2309:
2305:
2301:
2294:
2291:
2286:
2280:
2276:
2269:
2266:
2261:
2257:
2253:
2249:
2245:
2241:
2237:
2233:
2226:
2223:
2218:
2214:
2210:
2206:
2202:
2198:
2191:
2188:
2183:
2179:
2174:
2169:
2165:
2161:
2158:(1): 208–27.
2157:
2153:
2149:
2142:
2139:
2134:
2130:
2126:
2122:
2118:
2114:
2110:
2106:
2102:
2098:
2091:
2088:
2083:
2077:
2073:
2069:
2065:
2058:
2055:
2051:
2045:
2042:
2037:
2033:
2028:
2023:
2019:
2015:
2012:(2): R102–7.
2011:
2007:
2003:
1996:
1993:
1980:
1976:
1969:
1966:
1961:
1957:
1952:
1947:
1943:
1939:
1935:
1928:
1925:
1912:
1908:
1907:"Transferase"
1902:
1899:
1886:
1882:
1876:
1873:
1867:
1865:
1863:
1859:
1855:
1851:
1847:
1843:
1839:
1835:
1831:
1823:
1821:
1819:
1815:
1811:
1807:
1803:
1802:
1797:
1789:
1787:
1785:
1782:
1778:
1774:
1770:
1766:
1762:
1753:
1751:
1749:
1745:
1741:
1737:
1733:
1726:
1721:
1719:
1717:
1716:acetylcholine
1713:
1709:
1705:
1701:
1694:
1692:
1690:
1686:
1682:
1678:
1674:
1670:
1662:
1660:
1658:
1654:
1647:Schizophrenia
1646:
1644:
1642:
1638:
1628:
1626:
1622:
1618:
1614:
1604:
1602:
1598:
1597:parietal lobe
1594:
1593:temporal lobe
1590:
1580:
1578:
1577:chromosome 10
1574:
1570:
1566:
1562:
1558:
1557:acetylcholine
1555:
1551:
1544:
1542:
1540:
1536:
1532:
1528:
1524:
1520:
1519:galactokinase
1516:
1512:
1508:
1504:
1497:
1495:
1492:
1488:
1484:
1480:
1476:
1469:
1467:
1465:
1461:
1457:
1453:
1449:
1445:
1441:
1434:
1432:
1430:
1426:
1422:
1417:
1411:
1410:
1405:
1398:
1396:
1393:
1389:
1385:
1381:
1377:
1376:
1370:
1368:
1364:
1360:
1356:
1352:
1344:
1342:
1321:
1317:
1316:molybdopterin
1313:
1309:
1305:
1301:
1293:
1291:
1289:
1281:
1279:
1256:
1252:
1242:
1235:
1233:
1210:
1208:
1204:
1200:
1196:
1191:
1187:
1179:
1177:
1154:
1152:
1148:
1144:
1143:phenylalanine
1140:
1136:
1132:
1128:
1124:
1120:
1111:
1104:
1102:
1072:
1064:
1060:
1051:
1049:
1047:
1024:
1020:
1016:
1012:
1008:
1004:
1000:
999:disaccharides
996:
992:
988:
980:
978:
941:
937:
933:
932:peptide bonds
929:
925:
916:
914:
912:
908:
885:
881:
871:
864:
862:
860:
837:
833:
829:
821:
817:
816:hydroxymethyl
813:
804:
797:
792:
786:
782:
779:
777:
773:
770:
768:
767:
762:
758:
755:
753:
750:
748:
747:
742:
738:
735:
733:
729:
726:
724:
723:
718:
714:
710:
706:
703:
701:
697:
693:
690:
688:
687:
682:
678:
675:
673:
669:
666:
664:
663:
658:
654:
650:
647:
645:
641:
638:
636:
635:
630:
627:
623:
619:
616:
614:
610:
606:
603:
601:
600:
595:
591:
587:
584:
582:
579:
577:
576:
571:
567:
563:
560:
558:
557:transaldolase
554:
553:transketolase
551:
549:
548:
543:
539:
535:
533:
529:
526:
524:
523:
518:
514:
511:
508:
507:
501:
499:
495:
491:
487:
479:
477:
475:
471:
467:
463:
459:
455:
451:
444:
440:
414:
413:
412:
410:
406:
402:
398:
394:
387:
385:
383:
380:
376:
372:
371:
365:
363:
362:Ulf von Euler
359:
356:(shared with
355:
351:
347:
344:breakdown by
343:
342:catecholamine
338:
336:
335:pyrophosphate
332:
328:
324:
320:
315:
313:
309:
308:radioisotopes
306:'s work with
305:
300:
299:glutamic acid
296:
292:
288:
280:
276:
268:
264:
262:
258:
254:
250:
242:
240:
238:
213:
206:
188:
178:
171:
159:
158:
157:
154:
152:
148:
144:
140:
136:
132:
128:
124:
120:
116:
112:
108:
104:
100:
95:
93:
89:
85:
81:
77:
73:
69:
65:
57:
53:
49:
48:
43:
39:
33:
19:
9073:Transferases
8996:Transferases
8995:
8815:Transferases
8814:
8736:
8714:
8689:
8310:
8275:
8033:Class I PI 3
7998:Pantothenate
7869:2.7.1-2.7.4:
7853:Transferases
7852:
7689:
7593:Thiaminase I
7559:Transferases
7558:
7499:transferases
7304:transferases
6879:transferases
6856:Transferases
6855:
6623:
6581:
6499:
6474:Transferases
6473:
6432:DXP synthase
6388:Transferases
6387:
6081:Homocysteine
5998:
5951:Translocases
5948:
5935:
5922:
5909:
5896:
5886:Transferases
5885:
5883:
5870:
5727:Binding site
5661:
5649:. Retrieved
5624:. Retrieved
5619:
5614:Shintani D.
5609:
5572:
5568:
5558:
5533:
5529:
5499:
5493:
5481:. Retrieved
5476:
5433:(2): 180–7.
5430:
5426:
5377:(1): 54–62.
5374:
5370:
5360:
5333:
5329:
5283:
5279:
5273:
5251:(3): 454–9.
5248:
5244:
5238:
5211:
5207:
5197:
5162:
5158:
5148:
5123:
5119:
5113:
5081:(1): 28–36.
5078:
5074:
5064:
5031:
5027:
5014:
5002:. Retrieved
4997:
4988:
4955:
4951:
4892:
4888:
4878:
4843:
4839:
4829:
4796:
4792:
4786:
4762:(2): 396–8.
4759:
4755:
4745:
4720:
4714:
4708:
4675:
4671:
4665:
4649:GeneReviews
4648:
4638:
4626:. Retrieved
4581:(2): 291–4.
4578:
4574:
4568:
4533:
4529:
4519:
4484:
4480:
4470:
4458:. Retrieved
4453:
4444:
4432:. Retrieved
4389:. Retrieved
4384:
4375:
4363:. Retrieved
4350:
4338:. Retrieved
4328:
4316:. Retrieved
4287:. Retrieved
4282:
4273:
4261:. Retrieved
4251:
4239:. Retrieved
4229:
4217:. Retrieved
4212:
4203:
4191:. Retrieved
4186:
4176:
4155:
4128:. Retrieved
4088:
4084:
4044:
4037:
4005:(1): 78–86.
4002:
3999:Biochemistry
3998:
3988:
3976:. Retrieved
3971:
3962:
3935:
3931:
3921:
3909:. Retrieved
3904:
3895:
3870:
3866:
3860:
3825:
3821:
3811:
3799:. Retrieved
3794:
3785:
3773:. Retrieved
3768:
3759:
3734:
3730:
3720:
3708:. Retrieved
3703:
3694:
3682:. Retrieved
3677:
3668:
3656:. Retrieved
3651:
3642:
3617:
3609:
3577:(1): 471–7.
3574:
3570:
3560:
3548:. Retrieved
3543:
3534:
3522:. Retrieved
3517:
3508:
3483:
3479:
3473:
3461:. Retrieved
3456:
3447:
3402:
3398:
3388:
3376:. Retrieved
3371:
3362:
3350:. Retrieved
3345:
3336:
3309:
3305:
3295:
3283:. Retrieved
3278:
3269:
3257:. Retrieved
3252:
3243:
3208:
3204:
3194:
3182:. Retrieved
3177:
3168:
3156:. Retrieved
3151:
3142:
3130:. Retrieved
3125:
3116:
3097:
3093:
3084:
3076:
3064:. Retrieved
3059:
3050:
3038:. Retrieved
3033:
3024:
3012:. Retrieved
3007:
2998:
2986:. Retrieved
2981:
2957:. Retrieved
2952:
2928:. Retrieved
2923:
2920:"EC 2.7.7.6"
2914:
2895:
2889:
2877:. Retrieved
2873:
2863:
2836:
2832:
2822:
2810:. Retrieved
2805:
2766:
2762:
2752:
2709:
2705:
2695:
2668:
2664:
2654:
2611:
2607:
2597:
2562:
2558:
2548:
2515:
2511:
2504:
2477:
2473:
2463:
2451:. Retrieved
2446:
2437:
2396:
2392:
2385:
2344:
2340:
2334:
2307:
2303:
2293:
2274:
2268:
2235:
2231:
2225:
2200:
2196:
2190:
2155:
2151:
2141:
2100:
2096:
2090:
2063:
2057:
2049:
2044:
2009:
2005:
1995:
1983:. Retrieved
1978:
1968:
1941:
1937:
1927:
1915:. Retrieved
1910:
1901:
1889:. Retrieved
1884:
1875:
1862:MAPEG family
1827:
1805:
1799:
1793:
1757:
1730:
1698:
1673:hypothalamus
1666:
1650:
1637:Huntington's
1634:
1610:
1601:frontal lobe
1586:
1561:acetyl group
1548:
1507:simple sugar
1503:Galactosemia
1501:
1498:Galactosemia
1491:mitochondria
1473:
1460:ketoacidosis
1438:
1421:deficiencies
1419:Transferase
1418:
1415:
1407:
1399:Deficiencies
1384:chromosome 9
1375:Homo sapiens
1373:
1371:
1355:glycoprotein
1348:
1311:
1297:
1285:
1247:
1211:
1183:
1155:
1123:transaminase
1116:
1055:
995:biosynthesis
984:
920:
876:
809:
764:
744:
720:
684:
668:transaminase
660:
632:
597:
573:
545:
520:
489:
483:
447:
391:
388:Nomenclature
378:
368:
366:
339:
316:
273:
251:, including
246:
233:
155:
107:thiol esters
96:
67:
64:biochemistry
61:
45:
8947:: Alkylthio
8584:Transposase
8574:Recombinase
8219:-nucleoside
8045:Sphingosine
7694:nitrogenous
7690:Transferase
6958:Galactosyl-
6716:Factor XIII
5999:Transferase
5722:Active site
5651:23 November
5626:23 November
5483:10 November
4628:22 November
4487:(1): 42–7.
4434:22 November
4365:22 November
4340:22 November
4318:22 November
3978:11 December
3968:"EC 2.10.1"
3911:11 December
3873:(1): 82–6.
3801:11 December
3795:Kegg: DBGET
3775:11 December
3710:11 December
3684:11 December
3524:28 November
3463:28 November
3378:26 November
3352:26 November
3285:26 November
3259:26 November
3184:25 November
3158:25 November
3132:25 November
3100:: 548–566.
3066:25 November
3040:25 November
3014:25 November
2930:12 November
2879:13 November
2665:Development
1973:Wishart D.
1689:respiratory
1625:symptomatic
1617:spinal cord
1483:fatty acids
1380:blood types
1119:nitrogenous
1048:+ lactose.
936:translation
677:nitrogenous
470:nucleotides
456:group to a
405:L-glutamate
401:methylamine
331:UDP-glucose
261:phosphatase
103:transferase
68:transferase
8386:Telomerase
8229:Polymerase
8003:Mevalonate
7966:Riboflavin
7857:phosphorus
6911:Cellobiose
5925:Isomerases
5899:Hydrolases
5766:Regulation
5620:Elastomics
5471:Bowen, R.
5004:5 December
4460:4 November
4391:4 November
4289:4 November
4263:2 December
4241:2 December
4219:2 December
4193:2 December
4181:O'Neil D.
4130:2 December
3901:"EC 2.9.1"
3658:4 December
3550:4 December
3453:"EC 2.6.2"
2959:4 November
2812:5 November
2453:5 November
1985:4 November
1917:4 November
1868:References
1781:transgenic
1761:biosensors
1744:downstream
1535:galactitol
1359:glycolipid
1300:molybdenum
1207:activation
1193:kinase is
1186:phosphorus
1147:tryptophan
781:molybdenum
705:phosphorus
696:polymerase
379:Drosophila
370:Drosophila
135:amino acid
127:acetyl CoA
115:tryptophan
99:coenzyme A
56:polymerase
52:α-Amanitin
8849:Rhodanese
8578:Integrase
8502:3' to 5'
8147:Guanylate
8142:Uridylate
8132:Adenylate
7976:Thymidine
7971:Shikimate
7302:Pentosyl-
7217:Mannosyl-
6888:Glucosyl-
6283:Carbamoyl
6259:Carbamoyl
5804:EC number
2868:Lower S.
2152:Biochem J
1891:4 October
1818:sunflower
1784:cultigens
1691:systems.
1681:autonomic
1641:dendrites
1539:cataracts
1485:, as the
1425:illnesses
1367:galactose
1329:→
1320:molybdate
1266:⇌
1220:→
1164:⇌
1089:⇌
1033:⇌
957:⇌
894:⇌
859:phosphate
845:→
836:aspartate
820:carbamoyl
509:EC number
486:EC Number
423:⇌
287:keto acid
214:−
172:−
9067:Category
9000:selenium
8512:RNase PH
8120:acceptor
8101:Creatine
8094:acceptor
8066:acceptor
8008:Pyruvate
7993:Glycerol
7954:Platelet
7929:Galacto-
7900:acceptor
7696:groups (
7139:Fucosyl-
6906:Glycogen
6877:Hexosyl-
6568:P300/CBP
6392:aldehyde
5828:Kinetics
5752:Cofactor
5715:Activity
5601:22174645
5550:19747150
5455:10373463
5447:12548525
5409:23278578
5352:10197817
5300:17503482
5230:16987871
5189:23531559
5140:11386269
5105:23569405
5056:22103097
5048:23357177
5034:: 9–16.
4980:23621617
4972:10594838
4919:22798872
4911:23240572
4813:24311357
4756:Genomics
4737:24273939
4700:16382462
4692:16838075
4657:20301691
4603:11479049
4595:22963887
4560:22402528
4552:10439960
4511:12552079
4429:20301431
4107:21821934
4029:17198377
3852:23211525
3751:11396917
3601:20348897
3540:"EC 2.7"
3439:24260346
3399:PLOS ONE
3368:"EC 2.5"
3235:19363482
2785:17227754
2744:19540119
2687:16049108
2646:15984116
2638:16271864
2589:27722532
2540:23558834
2532:20380296
2421:13111246
2369:14780123
2217:13832270
2182:16744345
2125:13072573
2036:24305065
1677:striatum
1675:and the
1599:and the
1569:synapses
1523:anorexia
1464:mutation
1390:and six
1363:antigens
1304:tungsten
1288:selenium
1139:tyrosine
1063:cysteine
987:glycosyl
928:ribozyme
785:tungsten
757:selenium
715:, etc.)
713:carboxyl
626:pentoses
618:glycosyl
562:aldehyde
512:Examples
494:Hydrogen
399:, where
352:’s 1970
257:protease
237:coenzyme
189:→
147:ribosome
123:pyruvate
88:molecule
84:glycosyl
78:(e.g. a
32:Esterase
9057:Biology
8463:PrimPol
8448:Primase
7922:Hepatic
7917:Fructo-
7804:: Other
7517:ST8SIA4
7375:Sirtuin
7108:UGT2B28
7103:UGT2B17
7098:UGT2B15
7093:UGT2B11
7088:UGT2B10
7056:UGT1A10
6980:C1GALT1
6338:Amidine
6267:Carboxy
6253:Carboxy
6016:Methyl-
5938:Ligases
5708:Enzymes
5592:3233451
5400:3546605
5379:Bibcode
5308:6882521
5265:8434662
5180:3607155
5096:3615522
4870:5074408
4861:1331117
4821:8707545
4778:1840566
4502:1907369
4423:. NIH.
4020:1868504
3954:2007585
3843:3545217
3593:8550604
3500:6143829
3430:3832670
3407:Bibcode
3328:5440844
3279:UniProt
3226:2679717
2988:28 July
2855:5905132
2735:2733793
2714:Bibcode
2616:Bibcode
2581:9827800
2496:8939709
2401:Bibcode
2377:4215187
2349:Bibcode
2326:1941176
2260:4009655
2240:Bibcode
2173:1254374
2133:4180213
2105:Bibcode
2027:3921314
1960:6955307
1814:tobacco
1804:plant (
1798:is the
1746:end or
1567:in the
1565:choline
1456:ketones
1452:Ketones
1448:ketones
1409:E. coli
1392:introns
1023:glucose
1019:lactose
934:during
826:CO. In
766:EC 2.10
709:alcohol
679:groups
622:hexoses
568:groups
540:groups
536:single-
312:tracers
249:enzymes
243:History
145:of the
72:enzymes
9043:Portal
8819:sulfur
8750:2.7.13
8728:2.7.12
8703:2.7.11
8678:2.7.10
8517:PNPase
8485:PNPase
8441:POLRMT
8436:ssRNAP
7949:Muscle
7912:Gluco-
7876:kinase
7802:2.6.99
7497:Sialyl
7493:2.4.99
7313:Ribose
7152:POFUT2
7147:POFUT1
7083:UGT2B7
7078:UGT2B4
7073:UGT2A3
7068:UGT2A2
7063:UGT2A1
7051:UGT1A9
7046:UGT1A8
7041:UGT1A7
7036:UGT1A6
7031:UGT1A5
7026:UGT1A4
7021:UGT1A3
7016:UGT1A1
7009:B3GAT3
7004:B3GAT2
6999:B3GAT1
6901:Starch
6787:HMGCS2
6624:other:
6615:SPTLC2
6610:SPTLC1
6551:HGSNAT
6396:ketone
6168:Formyl
6120:DNMT3B
5912:Lyases
5647:. USDA
5599:
5589:
5548:
5506:
5453:
5445:
5407:
5397:
5350:
5306:
5298:
5263:
5228:
5187:
5177:
5138:
5103:
5093:
5054:
5046:
4978:
4970:
4917:
4909:
4868:
4858:
4819:
4811:
4776:
4735:
4698:
4690:
4655:
4624:. NORD
4601:
4593:
4558:
4550:
4509:
4499:
4427:
4314:. OMIM
4164:
4126:. NCBI
4105:
4059:
4027:
4017:
3952:
3887:495970
3885:
3850:
3840:
3749:
3630:
3599:
3591:
3498:
3437:
3427:
3372:IntEnz
3326:
3233:
3223:
2902:
2853:
2783:
2742:
2732:
2685:
2644:
2636:
2587:
2579:
2538:
2530:
2494:
2429:452922
2427:
2419:
2393:Nature
2375:
2367:
2341:Nature
2324:
2281:
2258:
2232:Nature
2215:
2180:
2170:
2131:
2123:
2097:Nature
2078:
2034:
2024:
1958:
1796:rubber
1595:, the
1531:sepsis
1203:cyclin
1145:, and
1065:from O
828:ATCase
812:methyl
746:EC 2.9
737:sulfur
722:EC 2.8
700:kinase
698:, and
686:EC 2.7
670:, and
662:EC 2.6
634:EC 2.5
611:, and
599:EC 2.4
575:EC 2.3
566:ketone
547:EC 2.2
538:carbon
522:EC 2.1
454:methyl
289:by an
281:(or NH
151:P-site
143:A-site
101:(CoA)
80:methyl
9013:2.9.1
8945:2.8.4
8923:2.8.3
8891:2.8.2
8832:2.8.1
8598:2.7.8
8567:Other
8206:2.7.7
8166:2.7.6
8111:2.7.4
8088:2.7.3
8060:2.7.2
7944:Liver
7907:Hexo-
7894:2.7.1
7783:2.6.3
7707:2.6.1
7580:2.5.1
7563:alkyl
7465:XYLT2
7460:XYLT1
7448:Other
7433:ECGF1
7417:Other
7298:2.4.2
7284:ALG12
7235:POMT2
7230:POMT1
7207:FUT11
7202:FUT10
6873:2.4.1
6728:2.3.3
6679:2.3.2
6669:ABHD5
6556:ARD1A
6491:2.3.1
6409:2.2.1
6334:2.1.4
6249:2.1.3
6223:Other
6159:2.1.2
6108:Other
6012:2.1.1
5864:Types
5451:S2CID
5304:S2CID
5052:S2CID
5024:(PDF)
4976:S2CID
4948:(PDF)
4915:S2CID
4817:S2CID
4696:S2CID
4599:S2CID
4556:S2CID
4359:(PDF)
4336:. NIH
3597:S2CID
3090:(PDF)
2642:S2CID
2585:S2CID
2536:S2CID
2425:S2CID
2373:S2CID
2256:S2CID
2129:S2CID
1860:from
1801:Hevea
1708:apnea
1621:brain
1573:nerve
1388:exons
1151:amino
1021:from
926:is a
649:alkyl
590:alkyl
329:from
279:amine
44:from
9006:2.9)
8825:2.8)
8737:see
8715:see
8690:see
8064:COOH
7863:2.7)
7771:AGXT
7729:GOT2
7724:GOT1
7700:2.6)
7573:2.5)
7567:aryl
7565:and
7279:ALG9
7274:ALG8
7269:ALG6
7264:ALG3
7259:ALG2
7254:ALG1
7247:DPM3
7242:DPM1
7197:FUT9
7192:FUT8
7187:FUT7
7182:FUT6
7177:FUT5
7172:FUT4
7167:FUT3
7162:FUT2
7157:FUT1
7129:HAS3
7124:HAS2
7119:HAS1
6916:Myo-
6866:2.4)
6598:CPT2
6593:CPT1
6573:NAT2
6484:2.3)
6402:2.2)
6257:and
6005:2.1)
5956:list
5949:EC7
5943:list
5936:EC6
5930:list
5923:EC5
5917:list
5910:EC4
5904:list
5897:EC3
5891:list
5884:EC2
5878:list
5871:EC1
5653:2013
5628:2013
5597:PMID
5546:PMID
5504:ISBN
5485:2013
5443:PMID
5405:PMID
5375:1275
5348:PMID
5296:PMID
5284:144B
5261:PMID
5226:PMID
5185:PMID
5136:PMID
5101:PMID
5044:PMID
5006:2013
4968:PMID
4907:PMID
4866:PMID
4809:PMID
4774:PMID
4733:PMID
4688:PMID
4653:PMID
4630:2013
4591:PMID
4575:Gene
4548:PMID
4507:PMID
4462:2013
4436:2013
4425:PMID
4393:2013
4367:2013
4342:2013
4320:2013
4291:2013
4265:2013
4243:2013
4221:2013
4195:2013
4162:ISBN
4132:2013
4103:PMID
4057:ISBN
4025:PMID
3980:2013
3950:PMID
3913:2013
3883:PMID
3848:PMID
3803:2013
3777:2013
3747:PMID
3712:2013
3686:2013
3660:2013
3628:ISBN
3589:PMID
3552:2013
3526:2013
3496:PMID
3465:2013
3435:PMID
3380:2013
3354:2013
3324:PMID
3287:2013
3261:2013
3231:PMID
3186:2013
3160:2013
3134:2013
3068:2013
3042:2013
3016:2013
2990:2016
2961:2013
2932:2013
2900:ISBN
2881:2013
2851:PMID
2814:2013
2781:PMID
2740:PMID
2683:PMID
2634:PMID
2577:PMID
2559:Cell
2528:PMID
2492:PMID
2455:2013
2417:PMID
2365:PMID
2322:PMID
2279:ISBN
2213:PMID
2178:PMID
2121:PMID
2076:ISBN
2032:PMID
1987:2013
1956:PMID
1919:2013
1893:2020
1816:and
1687:and
1669:SIDS
1619:and
1487:body
1357:and
1251:PAPS
1001:and
969:tRNA
834:+ L-
793:Role
774:and
730:and
653:aryl
642:and
624:and
586:acyl
555:and
530:and
360:and
325:and
139:tRNA
66:, a
8480:PAP
8421:III
8355:/Y
8346:TDT
8327:/X
8319:III
8311:Pfu
8286:/B
8276:Taq
8251:/A
8018:PFP
7988:NAD
5668:in
5587:PMC
5577:doi
5538:doi
5435:doi
5395:PMC
5387:doi
5338:doi
5288:doi
5253:doi
5249:150
5216:doi
5175:PMC
5167:doi
5128:doi
5124:344
5091:PMC
5083:doi
5036:doi
4960:doi
4897:doi
4893:124
4856:PMC
4848:doi
4844:225
4801:doi
4797:231
4764:doi
4725:doi
4680:doi
4583:doi
4579:509
4538:doi
4497:PMC
4489:doi
4093:doi
4049:doi
4015:PMC
4007:doi
3940:doi
3936:266
3875:doi
3838:PMC
3830:doi
3739:doi
3735:390
3579:doi
3575:271
3488:doi
3484:174
3425:PMC
3415:doi
3314:doi
3310:245
3221:PMC
3213:doi
3102:doi
2841:doi
2837:241
2771:doi
2767:282
2730:PMC
2722:doi
2673:doi
2669:132
2624:doi
2567:doi
2520:doi
2482:doi
2409:doi
2397:172
2357:doi
2345:166
2312:doi
2308:121
2248:doi
2236:140
2205:doi
2168:PMC
2160:doi
2113:doi
2101:172
2068:doi
2022:PMC
2014:doi
2010:306
1946:doi
1942:257
1844:in
1832:as
1700:CMS
1613:ALS
1571:of
1302:or
1073:: O
1046:UDP
997:of
783:or
651:or
564:or
490:EC2
474:RNA
458:DNA
364:).
327:G1P
323:UTP
310:as
125:to
82:or
62:In
9069::
9015::
9004:EC
8998::
8925::
8893::
8834::
8823:EC
8817::
8705::
8680::
8665:PO
8426:IV
8416:II
8325:IV
8321:/C
8284:II
8270:T7
8215:PO
8208::
8168::
8115:PO
8113::
8090::
8062::
7898:OH
7896::
7881:PO
7861:EC
7855::
7709::
7698:EC
7692::
7571:EC
7561::
7495::
7426::
7300::
7117::
6875::
6864:EC
6858::
6681::
6482:EC
6476::
6400:EC
6390::
6336::
6251::
6165:-,
6161::
6055:O-
6024:N-
6014::
6003:EC
5636:^
5618:.
5595:.
5585:.
5573:12
5571:.
5567:.
5544:.
5532:.
5518:^
5475:.
5463:^
5449:.
5441:.
5431:27
5429:.
5417:^
5403:.
5393:.
5385:.
5373:.
5369:.
5346:.
5334:58
5332:.
5328:.
5316:^
5302:.
5294:.
5282:.
5259:.
5247:.
5224:.
5212:15
5210:.
5206:.
5183:.
5173:.
5161:.
5157:.
5134:.
5122:.
5099:.
5089:.
5079:36
5077:.
5073:.
5050:.
5042:.
5032:94
5030:.
5026:.
4996:.
4974:.
4966:.
4956:49
4954:.
4950:.
4927:^
4913:.
4905:.
4891:.
4887:.
4864:.
4854:.
4842:.
4838:.
4815:.
4807:.
4795:.
4772:.
4758:.
4754:.
4731:.
4721:59
4719:.
4694:.
4686:.
4676:29
4674:.
4651:.
4647:.
4611:^
4597:.
4589:.
4577:.
4554:.
4546:.
4532:.
4528:.
4505:.
4495:.
4483:.
4479:.
4452:.
4401:^
4383:.
4299:^
4281:.
4211:.
4185:.
4140:^
4115:^
4101:.
4089:75
4087:.
4083:.
4071:^
4055:.
4023:.
4013:.
4003:46
4001:.
3997:.
3970:.
3948:.
3934:.
3930:.
3903:.
3881:.
3871:95
3869:.
3846:.
3836:.
3826:27
3824:.
3820:.
3793:.
3767:.
3745:.
3733:.
3729:.
3702:.
3676:.
3650:.
3626:.
3624:32
3595:.
3587:.
3573:.
3569:.
3542:.
3516:.
3494:.
3482:.
3455:.
3433:.
3423:.
3413:.
3401:.
3397:.
3370:.
3344:.
3322:.
3308:.
3304:.
3277:.
3251:.
3229:.
3219:.
3209:16
3207:.
3203:.
3176:.
3150:.
3124:.
3098:10
3096:.
3092:.
3058:.
3032:.
3006:.
2980:.
2969:^
2951:.
2940:^
2922:.
2872:.
2849:.
2835:.
2831:.
2804:.
2793:^
2779:.
2765:.
2761:.
2738:.
2728:.
2720:.
2710:19
2708:.
2704:.
2681:.
2667:.
2663:.
2640:.
2632:.
2622:.
2612:15
2610:.
2606:.
2583:.
2575:.
2563:95
2561:.
2557:.
2534:.
2526:.
2516:47
2514:.
2490:.
2476:.
2472:.
2445:.
2423:.
2415:.
2407:.
2395:.
2371:.
2363:.
2355:.
2343:.
2320:.
2306:.
2302:.
2254:.
2246:.
2234:.
2211:.
2201:54
2199:.
2176:.
2166:.
2156:24
2154:.
2150:.
2127:.
2119:.
2111:.
2099:.
2074:.
2066:.
2030:.
2020:.
2008:.
2004:.
1977:.
1954:.
1940:.
1936:.
1909:.
1883:.
1864:.
1786:.
1748:3'
1718:.
1627:.
1579:.
1450:.
1431:.
1416:.
1318:+
1141:,
1081:S
977:.
913:.
909:+
861:.
814:,
711:,
694:,
607:,
492:.
441:+
436:NH
384:.
337:.
255:,
239:.
153:.
9045::
8988:e
8981:t
8974:v
8807:e
8800:t
8793:v
8667:4
8663:(
8580:)
8576:(
8458:2
8453:1
8431:V
8369:κ
8364:ι
8359:η
8353:V
8341:μ
8336:λ
8331:β
8305:ζ
8300:ε
8295:δ
8290:α
8265:ν
8260:θ
8255:γ
8249:I
8221:)
8217:4
8213:(
8184:)
8181:7
8179:O
8177:2
8175:P
8173:(
8117:4
8092:N
7959:2
7939:1
7886:)
7883:4
7879:(
7874:/
7845:e
7838:t
7831:v
7682:e
7675:t
7668:v
7569:(
7551:e
7544:t
7537:v
6982:)
6978:(
6862:(
6848:e
6841:t
6834:v
6585::
6503::
6480:(
6466:e
6459:t
6452:v
6398:(
6394:-
6380:e
6373:t
6366:v
6255:-
6064:/
5991:e
5984:t
5977:v
5958:)
5954:(
5945:)
5941:(
5932:)
5928:(
5919:)
5915:(
5906:)
5902:(
5893:)
5889:(
5880:)
5876:(
5700:e
5693:t
5686:v
5655:.
5630:.
5603:.
5579::
5552:.
5540::
5534:3
5512:.
5487:.
5457:.
5437::
5411:.
5389::
5381::
5354:.
5340::
5310:.
5290::
5267:.
5255::
5232:.
5218::
5191:.
5169::
5163:3
5142:.
5130::
5107:.
5085::
5058:.
5038::
5008:.
4982:.
4962::
4921:.
4899::
4872:.
4850::
4823:.
4803::
4780:.
4766::
4760:9
4739:.
4727::
4702:.
4682::
4659:.
4632:.
4605:.
4585::
4562:.
4540::
4534:7
4513:.
4491::
4485:5
4464:.
4438:.
4395:.
4369:.
4344:.
4322:.
4293:.
4267:.
4245:.
4223:.
4197:.
4170:.
4134:.
4109:.
4095::
4065:.
4051::
4031:.
4009::
3982:.
3956:.
3942::
3915:.
3889:.
3877::
3854:.
3832::
3805:.
3779:.
3753:.
3741::
3714:.
3688:.
3662:.
3636:.
3603:.
3581::
3554:.
3528:.
3502:.
3490::
3467:.
3441:.
3417::
3409::
3403:8
3382:.
3356:.
3330:.
3316::
3289:.
3263:.
3237:.
3215::
3188:.
3162:.
3136:.
3110:.
3104::
3087:"
3070:.
3044:.
3018:.
2992:.
2963:.
2934:.
2908:.
2883:.
2857:.
2843::
2816:.
2787:.
2773::
2746:.
2724::
2716::
2689:.
2675::
2648:.
2626::
2618::
2591:.
2569::
2542:.
2522::
2498:.
2484::
2478:3
2457:.
2431:.
2411::
2403::
2379:.
2359::
2351::
2328:.
2314::
2287:.
2262:.
2250::
2242::
2219:.
2207::
2184:.
2162::
2135:.
2115::
2107::
2084:.
2070::
2038:.
2016::
1989:.
1962:.
1948::
1921:.
1895:.
1079:2
1075:3
1067:3
975:B
971:A
948:B
944:A
824:2
438:3
283:2
218:R
210:Y
207:+
204:X
182:Y
179:+
176:R
168:X
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.