31:
4327:
204:
940:
676:(EC 2.7.1.11) catalyses the reverse conversion of fructose 6-phosphate to fructose-1,6-bisphosphate, but this is not just the reverse reaction, because the co-substrates are different (and so thermodynamic requirements are not violated). The two enzymes each catalyse the conversion in one direction only, and are regulated by metabolites such as
1689:
the gluconeogenesis pathway because it is rate-limiting and controls the incorporation of all three-carbon substrates into glucose but is not involved in glycogen breakdown and is removed from mitochondrial steps in the pathway. This means that altering its activity can have a large effect on gluconeogenesis while reducing the risk of
1673:
954:
492:
1688:
causes many serious problems, and treatments often focus on lowering blood sugar levels. Gluconeogenesis in the liver is a major cause of glucose overproduction in these patients, and so inhibition of gluconeogenesis is a reasonable way to treat type 2 diabetes. FBPase is a good enzyme to target in
1696:
Drug candidates have been developed that mimic the inhibitory activity of AMP on FBPase. Efforts were made to mimic the allosteric inhibitory effects of AMP while making the drug as structurally different from it as possible. Second-generation FBPase inhibitors have now been developed and have had
1663:
shows similar changes in activity to FBPase at colder temperatures, such as an upward shift in optimum pH at colder temperatures. This adaptation allows enzymes such as FBPase and fructose-1,6-bisphosphate aldolase to track intracellular pH changes in hibernating animals and match their activity
1639:
In addition to hibernation, there is evidence that FBPase activity varies significantly between warm and cold seasons even for animals that do not hibernate. In rabbits exposed to cold temperatures, FBPase activity decreased throughout the duration of cold exposure, increasing when temperatures
680:
so that high activity of one of them is accompanied by low activity of the other. More specifically, fructose 2,6-bisphosphate allosterically inhibits fructose 1,6-bisphosphatase, but activates phosphofructokinase-I. Fructose 1,6-bisphosphatase is involved in many different
2589:
Dang Q, Van Poelje PD, Erion MD (2012). "Chapter 11: The
Discovery and Development of MB07803, a Second-Generation Fructose-1,6-bisphosphatase Inhibitor with Improved Pharmacokinetic Properties, as a Potential Treatment of Type 2 Diabetes". In Jones RM (ed.).
1654:
Fructose 1,6-bisphosphate aldolase is another temperature dependent enzyme that plays an important role in the regulation of glycolysis and gluconeogenesis during hibernation. Its main role is in glycolysis instead of gluconeogenesis, but its
2674:
van Poelje PD, Potter SC, Chandramouli VC, Landau BR, Dang Q, Erion MD (June 2006). "Inhibition of fructose 1,6-bisphosphatase reduces excessive endogenous glucose production and attenuates hyperglycemia in Zucker diabetic fatty rats".
1604:
for its substrate FBP at 5 °C than at 37 °C. However, in a euthermic bat this decrease was only 25%, demonstrating the difference in temperature sensitivity between hibernating and euthermic bats. When sensitivity to
1523:
1061:
1664:
ranges to these shifts. Aldolase also complements the activity of FBPase in anoxic conditions (discussed above) by increasing glycolytic output while FBPase inhibition decreases gluconeogenesis activity.
1599:
resting metabolic rate. FBPase is modified in hibernating animals to be much more temperature sensitive than it is in euthermic animals. FBPase in the liver of a hibernating bat showed a 75% decrease in
1450:
988:
2317:
MacDonald JA, Storey KB (December 2002). "Purification and characterization of fructose bisphosphate aldolase from the ground squirrel, Spermophilus lateralis: enzyme role in mammalian hibernation".
2352:
Ekdahl KN, Ekman P (February 1984). "The effect of fructose 2,6-bisphosphate and AMP on the activity of phosphorylated and unphosphorylated fructose-1,6-bisphosphatase from rat liver".
1525:
1063:
1957:
Stec B, Yang H, Johnson KA, Chen L, Roberts MF (Nov 2000). "MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase".
2623:
591:
450:
328:
1632:, and this is another reason for reduced FBPase activity in hibernating animals. The substrate of FBPase, fructose 1,6-bisphosphate, has also been shown to activate
66:
2859:
867:, forming a four-layer alpha-beta-beta-alpha sandwich, unlike the more usual five-layered alpha-beta-alpha-beta-alpha arrangement. The arrangement of the
2718:
Kaur R, Dahiya L, Kumar M (December 2017). "Fructose-1,6-bisphosphatase inhibitors: A new valid approach for management of type 2 diabetes mellitus".
1473:
1470:
1011:
1008:
2993:
1482:
1020:
3401:
1511:
1049:
3648:
2651:
2607:
2501:
939:
539:
276:
4347:
3562:
2275:
Brooks SP, Storey KB (January 1992). "Mechanisms of glycolytic control during hibernation in the ground squirrel
Spermophilus lateralis".
1490:
1028:
1847:"Definition of a metal-dependent/Li-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure"
3663:
3657:
3458:
3208:
1454:
992:
90:
1810:
Zhang Y, Liang JY, Lipscomb WN (Feb 1993). "Structural similarities between fructose-1,6-bisphosphatase and inositol monophosphatase".
1501:
1039:
3506:
1706:
1906:"Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of
1492:
1478:
1462:
1030:
1016:
1000:
4046:
3617:
3406:
3058:
2769:
1498:
1457:
1036:
995:
1522:
1485:
1060:
1023:
2852:
1513:
1512:
1496:
1489:
1486:
1051:
1050:
1034:
1027:
1024:
953:
78:
1520:
1519:
1517:
1475:
1453:
1058:
1057:
1055:
1013:
991:
3724:
3612:
1636:
in glycolysis, linking increased glycolysis to decreased gluconeogenesis when FBPase activity is decreased during hibernation.
1474:
1456:
1012:
994:
1775:
Marcus F, Gontero B, Harrsch PB, Rittenhouse J (Mar 1986). "Amino acid sequence homology among fructose-1,6-bisphosphatases".
1466:
1465:
1004:
1003:
71:
3533:
3443:
3396:
2962:
1617:
were examined, FBPase from hibernating bats was much more sensitive to inhibitors at low temperature than in euthermic bats.
1524:
1472:
1062:
1010:
4202:
611:
470:
348:
3955:
3896:
2927:
2837:
1591:
processes to facilitate entry into hibernation, maintenance, arousal from hibernation, and adjustments to allow long-term
1479:
1461:
1017:
999:
4317:
2225:
Heldmaier G, Ortmann S, Elvert R (August 2004). "Natural hypometabolism during hibernation and daily torpor in mammals".
4000:
3567:
3557:
1493:
1458:
1031:
996:
1521:
1488:
1484:
1477:
1059:
1026:
1022:
1015:
3960:
3852:
2845:
1499:
1491:
1455:
1037:
1029:
993:
1504:
1042:
1494:
1483:
1467:
1032:
1021:
1005:
3546:
3542:
3538:
3454:
3287:
3175:
1460:
998:
4187:
2642:
Arch JR (2011). "Thermogenesis and
Related Metabolic Targets in Anti-Diabetic Therapy". In Schwanstecher M (ed.).
4357:
4303:
4290:
4277:
4264:
4251:
4238:
4225:
3808:
3754:
3714:
3673:
3577:
3416:
3384:
3270:
3234:
2980:
1614:
1526:
1481:
1064:
1019:
945:
677:
635:
4197:
1672:
1510:
1471:
1048:
1009:
4151:
4094:
3462:
3311:
3225:
2789:
2049:"The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea"
1681:
1656:
1518:
1502:
1500:
1495:
1464:
1459:
1056:
1040:
1038:
1033:
1002:
997:
825:
778:
131:
1516:
1515:
1514:
1509:
1508:
1507:
1497:
1487:
1480:
1476:
1469:
1468:
1463:
1054:
1053:
1052:
1047:
1046:
1045:
1035:
1025:
1018:
1014:
1007:
1006:
1001:
599:
458:
336:
1506:
1505:
1503:
1044:
1043:
1041:
4099:
3331:
3201:
3003:
2998:
2872:
2187:
Storey KB (December 1997). "Metabolic regulation in mammalian hibernation: enzyme and protein adaptations".
1610:
914:
1740:
Marcus F, Harrsch PB (May 1990). "Amino acid sequence of spinach chloroplast fructose-1,6-bisphosphatase".
3887:
3526:
3131:
30:
83:
4120:
4039:
3822:
3719:
3511:
3472:
3336:
3258:
3151:
3082:
2932:
2921:
2785:
1651:
digestion decreases gluconeogenesis relative to glycolysis during cold periods, similar to hibernation.
1606:
845:
4192:
2494:
Metabolic
Interconversion of Enzymes 1973 Third International Symposium held in Seattle, June 5-8, 1973
595:
454:
332:
4005:
3840:
3835:
3768:
3428:
3253:
3106:
3053:
2534:
1858:
1640:
became warmer again. The mechanism of this FBPase inhibition is thought to be digestion of FBPase by
959:
639:
552:
289:
148:
4156:
3860:
3830:
3634:
3629:
3553:
3489:
3275:
918:
853:
673:
4089:
3993:
3845:
3326:
3316:
3194:
2700:
2617:
2377:
2292:
2250:
1982:
1620:
During hibernation, respiration also dramatically decreases, resulting in conditions of relative
925:
lacking this enzyme are apparently still able to grow on gluconeogenic growth substrates such as
896:
2131:"Identification and expression of the Bacillus subtilis fructose-1, 6-bisphosphatase gene (fbp)"
2756:
Berg JM, Tymoczko JL, Stryer L (2002). "Glycolysis and
Gluconeogenesis". In Susan Moran (ed.).
1596:
4352:
3794:
3739:
3707:
3582:
3301:
2765:
2735:
2692:
2647:
2603:
2562:
2519:"Characterization of fructose-1,6-bisphosphate aldolase during anoxia in the tolerant turtle,
2497:
2467:
2418:
2369:
2334:
2242:
2204:
2160:
2111:
2070:
2026:
1974:
1939:
1886:
1827:
1792:
1757:
1621:
909:
782:
758:
586:
445:
323:
36:
2397:"Allosteric Inhibition of Rat Liver Fructose 1,6-Diphosphatase by Adenosine 5'-Monophosphate"
797:
pathways, including inositol signalling, gluconeogenesis, sulphate assimilation and possibly
4135:
4130:
4104:
4032:
3870:
3450:
3355:
3350:
3306:
3044:
2727:
2684:
2595:
2552:
2542:
2457:
2449:
2408:
2361:
2326:
2284:
2234:
2196:
2150:
2142:
2101:
2060:
2016:
1966:
1929:
1921:
1876:
1866:
1819:
1784:
1749:
840:
143:
578:
437:
315:
152:
136:
4182:
4166:
4079:
3972:
3786:
3702:
3697:
3692:
3605:
3600:
3360:
3238:
3126:
3013:
2880:
1716:
1647:, which are released at higher levels during colder periods. Inhibition of FBPase through
1633:
1625:
857:
714:
643:
172:
1449:
987:
35:
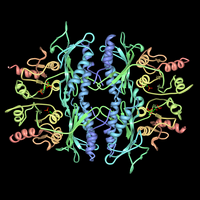
Fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex. Rendered from PDB
2538:
1862:
208:
crystal structure of rabbit liver fructose-1,6-bisphosphatase at 2.3 angstrom resolution
4331:
4220:
4161:
3945:
3940:
3935:
3321:
3296:
3292:
3265:
3248:
2557:
2518:
2462:
2437:
1545:
726:
2413:
2330:
2200:
2155:
2130:
2106:
2089:
1934:
1905:
1566:
1549:
1536:
4341:
4125:
4084:
3686:
3595:
3345:
3097:
2758:
2365:
2146:
1925:
1881:
1846:
1788:
1753:
1685:
892:
766:
730:
544:
281:
2704:
2381:
2254:
1659:
is the same as FBPase's, so its activity affects that of FBPase in gluconeogenesis.
1595:. During hibernation, an animal's metabolic rate may decrease to around 1/25 of its
520:
386:
232:
4074:
3813:
3744:
3467:
3388:
2296:
1986:
1690:
832:
790:
647:
574:
433:
311:
399:
245:
2833:
2819:
2805:
2731:
2547:
2090:"Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis"
1693:
and other potential side effects from altering other enzymes in gluconeogenesis.
532:
411:
257:
107:
4298:
4233:
4069:
3988:
3759:
3653:
3516:
3482:
3420:
3186:
3069:
2917:
2599:
1648:
1584:
864:
182:
4326:
2238:
1851:
Proceedings of the
National Academy of Sciences of the United States of America
3965:
3073:
2974:
2970:
2966:
2901:
2876:
2868:
2065:
2048:
1721:
1629:
1588:
972:
Click on genes, proteins and metabolites below to link to respective articles.
871:
813:
801:
794:
738:
682:
114:
4272:
4246:
3878:
3677:
3590:
3217:
2396:
1871:
887:
868:
836:
754:
698:
694:
651:
2739:
2696:
2566:
2422:
2338:
2246:
2074:
2030:
2021:
2004:
1978:
1943:
1823:
2471:
2373:
2208:
2164:
1890:
1831:
1796:
1761:
548:
285:
203:
3923:
3918:
3913:
3734:
3369:
3221:
3117:
2829:
2815:
2801:
2592:
New
Therapeutic Strategies for Type 2 Diabetes: Small Molecule Approaches
2129:
Fujita Y, Yoshida K, Miwa Y, Yanai N, Nagakawa E, Kasahara Y (Aug 1998).
2115:
1711:
1660:
1644:
1641:
1592:
930:
904:
849:
817:
774:
722:
686:
527:
406:
252:
119:
3930:
3908:
3903:
3521:
3093:
3008:
2438:"Control of glycolysis and gluconeogenesis in rat kidney cortex slices"
2288:
2005:"A novel candidate for the true fructose-1,6-bisphosphatase in archaea"
1697:
good results in clinical trials with non-human mammals and now humans.
900:
875:
821:
798:
786:
746:
742:
734:
702:
269:
264:
2688:
2453:
2003:
Rashid N, Imanaka H, Kanai T, Fukui T, Atomi H, Imanaka T (Aug 2002).
721:
was noted to be identical to that of inositol-1-phosphatase (IMPase).
4285:
4055:
3950:
3883:
3776:
3374:
3282:
3168:
3164:
3160:
3156:
2883:
926:
922:
770:
750:
606:
465:
343:
102:
4259:
3892:
3643:
3639:
1970:
1671:
861:
824:
so far, though a new group of FBPases (FBPase IV) which also show
3624:
3501:
3494:
3438:
3433:
3021:
3017:
2944:
2940:
2936:
2825:
2811:
2797:
762:
725:
polyphosphate 1-phosphatase (IPPase), IMPase and FBPase share a
718:
568:
515:
491:
427:
393:
381:
305:
239:
227:
95:
4028:
3190:
2841:
2913:
2909:
2905:
879:
690:
2523:: an assessment of enzyme activity, expression and structure"
1680:
Fructose 1,6-bisphosphatase is also a key player in treating
1448:
986:
2646:(1st ed.). Berlin, Heidelberg: Springer. p. 203.
2492:
Fischer EH, Krebs EG, Neurath H, Stadtman ER, eds. (1974).
1564:
The interactive pathway map can be edited at WikiPathways:
4024:
812:
Three different groups of FBPases have been identified in
1904:
Donahue JL, Bownas JL, Niehaus WG, Larson TJ (Oct 2000).
882:
assisted catalysis mechanism proposed for other FBPases.
848:
for FBP and are suggested to be the true FBPase in these
701:
being preferred) and the enzyme is potently inhibited by
2047:
Nishimasu H, Fushinobu S, Shoun H, Wakagi T (Jun 2004).
820:(FBPase I-III). None of these groups have been found in
2824:
This article incorporates text from the public domain
2810:
This article incorporates text from the public domain
2796:
This article incorporates text from the public domain
844:. The characterised members of this group show strict
4315:
2764:(5th ed.). New York: W. H. Freeman and Company.
1583:
Fructose 1,6-bisphosphatase also plays a key role in
4211:
4175:
4144:
4113:
4062:
3981:
3869:
3821:
3807:
3785:
3767:
3753:
3733:
3672:
3576:
3415:
3383:
3233:
3144:
3115:
3091:
3067:
3042:
3035:
2890:
1812:
1777:
605:
585:
567:
562:
538:
526:
514:
506:
501:
484:
464:
444:
426:
421:
405:
392:
380:
372:
367:
362:
342:
322:
304:
299:
275:
263:
251:
238:
226:
218:
213:
196:
178:
168:
163:
142:
130:
125:
113:
101:
89:
77:
65:
57:
49:
44:
23:
2757:
2669:
2667:
2665:
2663:
885:The fructose 1,6-bisphosphatases found within the
828:activity has recently been identified in Archaea.
3512:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
3152:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
2312:
2310:
2308:
2306:
2220:
2218:
878:was found to be consistent with the three-metal
496:crystal structure of fructose-1,6-bisphosphatase
2487:
2485:
2483:
2481:
1452:
990:
2270:
2268:
2266:
2264:
2182:
2180:
2178:
2176:
2174:
2042:
2040:
1998:
1996:
831:A new group of FBPases (FBPase V) is found in
4040:
3202:
2853:
2622:: CS1 maint: DOI inactive as of April 2024 (
2594:. Cambridge: The Royal Society of Chemistry.
632:-fructose-1,6-bisphosphate 1-phosphohydrolase
8:
2637:
2635:
2633:
2584:
2582:
2580:
2578:
2576:
1845:York JD, Ponder JW, Majerus PW (May 1995).
4047:
4033:
4025:
3818:
3764:
3750:
3209:
3195:
3187:
3039:
2894:
2860:
2846:
2838:
1628:, and therefore FBPase, while stimulating
1624:in the tissues. Anoxic conditions inhibit
559:
490:
418:
296:
202:
160:
2788:at the U.S. National Library of Medicine
2556:
2546:
2461:
2412:
2227:Respiratory Physiology & Neurobiology
2189:Comparative Biochemistry and Physiology A
2154:
2105:
2064:
2020:
1933:
1880:
1870:
856:study suggests that FBPase V has a novel
2994:Glyceraldehyde 3-phosphate dehydrogenase
2517:Dawson NJ, Biggar KK, Storey KB (2013).
2436:Underwood AH, Newsholme EA (July 1967).
4322:
3402:Ubiquitin carboxy-terminal hydrolase L1
2720:European Journal of Medicinal Chemistry
2644:Diabetes - Perspectives in Drug Therapy
2319:Archives of Biochemistry and Biophysics
1742:Archives of Biochemistry and Biophysics
1732:
1557:
935:
769:is also found in the distantly-related
2615:
1587:, which requires strict regulation of
895:bacteria) do not show any significant
481:
359:
193:
20:
3982:either deoxy- or ribo-
2395:Taketa K, Pogell BM (February 1965).
1535:|alt=Glycolysis and Gluconeogenesis
793:conserved family involved in diverse
363:Firmicute fructose-1,6-bisphosphatase
7:
3563:Protein serine/threonine phosphatase
717:of fructose-1,6-bisphosphatase from
3664:Cyclic nucleotide phosphodiesterase
3658:Clostridium perfringens alpha toxin
3459:Tartrate-resistant acid phosphatase
2401:The Journal of Biological Chemistry
2277:Journal of Comparative Physiology B
2094:The Journal of Biological Chemistry
2009:The Journal of Biological Chemistry
785:. It has been suggested that these
765:and participate in catalysis. This
3507:Pyruvate dehydrogenase phosphatase
1707:Fructose bisphosphatase deficiency
835:archaea and the hyperthermophilic
14:
3407:4-hydroxybenzoyl-CoA thioesterase
3059:Phosphoenolpyruvate carboxykinase
1567:"GlycolysisGluconeogenesis_WP534"
917:, though this can be overcome by
669:-fructose 6-phosphate + phosphate
4325:
2496:. Berlin, Heidelberg: Springer.
2147:10.1128/JB.180.16.4309-4313.1998
1926:10.1128/jb.182.19.5624-5627.2000
1613:, ADP, inorganic phosphate, and
1548:Glycolysis and Gluconeogenesis
952:
938:
29:
3725:N-acetylglucosamine-6-sulfatase
3613:Sphingomyelin phosphodiesterase
2088:Fujita Y, Freese E (Jun 1979).
1579:Hibernation and cold adaptation
3534:Inositol-phosphate phosphatase
3397:Palmitoyl protein thioesterase
2963:Fructose-bisphosphate aldolase
921:, and is dependent on Mn(2+).
757:/Ser) which has been shown to
661:-fructose 1,6-bisphosphate + H
634:) catalyses the conversion of
627:(EC 3.1.3.11; systematic name
1:
3897:RNA-induced silencing complex
2414:10.1016/S0021-9258(17)45224-0
2331:10.1016/S0003-9861(02)00579-9
2201:10.1016/S0300-9629(97)00238-7
2107:10.1016/S0021-9258(18)50601-3
563:Available protein structures:
422:Available protein structures:
300:Available protein structures:
24:fructose-1,6-bisphosphatase 1
4001:Serratia marcescens nuclease
3568:Dual-specificity phosphatase
3558:Protein tyrosine phosphatase
2732:10.1016/j.ejmech.2017.09.029
2548:10.1371/journal.pone.0068830
2366:10.1016/0014-5793(84)80127-1
1789:10.1016/0006-291X(86)90005-7
1754:10.1016/0003-9861(90)90475-E
4348:Genes on human chromosome 9
3478:Fructose 1,6-bisphosphatase
2949:Fructose 1,6-bisphosphatase
2600:10.1039/9781849735322-00306
485:Fructose-1,6-bisphosphatase
197:Fructose-1-6-bisphosphatase
4374:
3176:Bisphosphoglycerate mutase
2823:
2809:
2795:
2786:Fructose-1,6-Biphosphatase
2239:10.1016/j.resp.2004.03.014
4203:Michaelis–Menten kinetics
3715:Galactosamine-6 sulfatase
3271:6-phosphogluconolactonase
2989:
2981:Triosephosphate isomerase
2958:
2897:
2521:Trachemys scripta elegans
2066:10.1016/j.str.2004.03.026
1959:Nature Structural Biology
1615:fructose-2,6-bisphosphate
946:Fructose 1,6-bisphosphate
678:fructose 2,6-bisphosphate
636:fructose-1,6-bisphosphate
558:
489:
417:
295:
201:
159:
28:
4095:Diffusion-limited enzyme
3463:Purple acid phosphatases
2790:Medical Subject Headings
826:inositol monophosphatase
689:. FBPase requires metal
3004:Phosphoglycerate mutase
2999:Phosphoglycerate kinase
2873:carbohydrate metabolism
2602:(inactive 2024-04-11).
2442:The Biochemical Journal
2135:Journal of Bacteriology
1914:Journal of Bacteriology
1872:10.1073/pnas.92.11.5149
967:Interactive pathway map
913:enzyme is inhibited by
625:fructose bisphosphatase
3888:Microprocessor complex
3527:Beta-propeller phytase
3132:Glycerol dehydrogenase
2022:10.1074/jbc.M202868200
1824:10.1006/bbrc.1993.1159
1677:
1529:
1067:
4188:Eadie–Hofstee diagram
4121:Allosteric regulation
3823:Endodeoxyribonuclease
3720:Iduronate-2-sulfatase
3473:Glucose 6-phosphatase
3259:Butyrylcholinesterase
3083:Lactate dehydrogenase
2933:Phosphofructokinase 1
2922:Glucose 6-phosphatase
1675:
1528:
1066:
846:substrate specificity
4198:Lineweaver–Burk plot
4006:Micrococcal nuclease
3841:Deoxyribonuclease IV
3836:Deoxyribonuclease II
3769:Exodeoxyribonuclease
3429:Alkaline phosphatase
3254:Acetylcholinesterase
3107:Alanine transaminase
3054:Pyruvate carboxylase
3036:Gluconeogenesis only
960:Fructose 6-phosphate
808:Species distribution
640:fructose 6-phosphate
3861:UvrABC endonuclease
3831:Deoxyribonuclease I
3554:Protein phosphatase
3490:Protein phosphatase
3288:Bile salt-dependent
3276:PAF acetylhydrolase
2539:2013PLoSO...868830D
1863:1995PNAS...92.5149Y
1684:. In this disease,
1609:inhibitors such as
919:phosphoenolpyruvate
674:Phosphofructokinase
4157:Enzyme superfamily
4090:Enzyme promiscuity
3994:Mung bean nuclease
3853:Restriction enzyme
3846:Restriction enzyme
2289:10.1007/BF00257932
1678:
1530:
1068:
899:similarity to the
789:define an ancient
685:and found in most
683:metabolic pathways
4313:
4312:
4022:
4021:
4018:
4017:
4014:
4013:
3803:
3802:
3795:Oligonucleotidase
3740:deoxyribonuclease
3708:Steroid sulfatase
3583:Phosphodiesterase
3312:Hormone-sensitive
3184:
3183:
3140:
3139:
3031:
3030:
2928:Glucose isomerase
2689:10.2337/db05-1443
2653:978-3-642-17214-4
2609:978-1-84973-414-1
2503:978-3-642-80817-3
2454:10.1042/bj1040300
910:Bacillus subtilis
668:
660:
652:anabolic pathways
650:, which are both
631:
621:
620:
617:
616:
612:structure summary
480:
479:
476:
475:
471:structure summary
358:
357:
354:
353:
349:structure summary
192:
191:
188:
187:
4365:
4358:Protein families
4330:
4329:
4321:
4193:Hanes–Woolf plot
4136:Enzyme activator
4131:Enzyme inhibitor
4105:Enzyme catalysis
4049:
4042:
4035:
4026:
3871:Endoribonuclease
3857:
3851:
3819:
3765:
3751:
3451:Acid phosphatase
3332:Monoacylglycerol
3242:ester hydrolases
3211:
3204:
3197:
3188:
3040:
2895:
2862:
2855:
2848:
2839:
2775:
2763:
2744:
2743:
2715:
2709:
2708:
2671:
2658:
2657:
2639:
2628:
2627:
2621:
2613:
2586:
2571:
2570:
2560:
2550:
2514:
2508:
2507:
2489:
2476:
2475:
2465:
2433:
2427:
2426:
2416:
2392:
2386:
2385:
2349:
2343:
2342:
2314:
2301:
2300:
2272:
2259:
2258:
2222:
2213:
2212:
2184:
2169:
2168:
2158:
2126:
2120:
2119:
2109:
2085:
2079:
2078:
2068:
2044:
2035:
2034:
2024:
2015:(34): 30649–55.
2000:
1991:
1990:
1954:
1948:
1947:
1937:
1908:Escherichia coli
1901:
1895:
1894:
1884:
1874:
1842:
1836:
1835:
1807:
1801:
1800:
1772:
1766:
1765:
1737:
1571:
1570:
1562:
1444:
1439:
1434:
1429:
1424:
1419:
1414:
1409:
1404:
1399:
1394:
1389:
1384:
1379:
1374:
1369:
1364:
1359:
1354:
1349:
1344:
1339:
1334:
1329:
1324:
1319:
1314:
1309:
1304:
1299:
1294:
1289:
1284:
1279:
1274:
1269:
1264:
1259:
1254:
1249:
1244:
1239:
1234:
1229:
1224:
1219:
1214:
1209:
1204:
1199:
1194:
1189:
1184:
1179:
1174:
1169:
1164:
1159:
1154:
1149:
1144:
1139:
1134:
1129:
1124:
1119:
1114:
1109:
1104:
1099:
1094:
1089:
1084:
1079:
1074:
956:
942:
841:Aquifex aeolicus
666:
658:
629:
560:
494:
482:
419:
360:
297:
206:
194:
161:
33:
21:
16:Class of enzymes
4373:
4372:
4368:
4367:
4366:
4364:
4363:
4362:
4338:
4337:
4336:
4324:
4316:
4314:
4309:
4221:Oxidoreductases
4207:
4183:Enzyme kinetics
4171:
4167:List of enzymes
4140:
4109:
4080:Catalytic triad
4058:
4053:
4023:
4010:
3977:
3865:
3855:
3849:
3812:
3799:
3787:Exoribonuclease
3781:
3758:
3742:
3738:
3729:
3703:Arylsulfatase L
3698:Arylsulfatase B
3693:Arylsulfatase A
3668:
3581:
3572:
3411:
3379:
3241:
3229:
3215:
3185:
3180:
3136:
3127:Glycerol kinase
3111:
3087:
3063:
3027:
3014:Pyruvate kinase
2985:
2954:
2886:
2881:gluconeogenesis
2866:
2836:
2822:
2808:
2782:
2772:
2755:
2752:
2750:Further reading
2747:
2717:
2716:
2712:
2673:
2672:
2661:
2654:
2641:
2640:
2631:
2614:
2610:
2588:
2587:
2574:
2516:
2515:
2511:
2504:
2491:
2490:
2479:
2435:
2434:
2430:
2394:
2393:
2389:
2351:
2350:
2346:
2316:
2315:
2304:
2274:
2273:
2262:
2224:
2223:
2216:
2186:
2185:
2172:
2141:(16): 4309–13.
2128:
2127:
2123:
2087:
2086:
2082:
2046:
2045:
2038:
2002:
2001:
1994:
1965:(11): 1046–50.
1956:
1955:
1951:
1903:
1902:
1898:
1857:(11): 5149–53.
1844:
1843:
1839:
1809:
1808:
1804:
1774:
1773:
1769:
1739:
1738:
1734:
1730:
1717:Gluconeogenesis
1703:
1682:type 2 diabetes
1670:
1634:pyruvate kinase
1626:gluconeogenesis
1603:
1581:
1576:
1575:
1574:
1565:
1563:
1559:
1554:
1553:
1552:
1547:
1542:
1541:
1540:
1534:
1533:
1532:
1531:
1527:
1446:
1445:
1442:
1440:
1437:
1435:
1432:
1430:
1427:
1425:
1422:
1420:
1417:
1415:
1412:
1410:
1407:
1405:
1402:
1400:
1397:
1395:
1392:
1390:
1387:
1385:
1382:
1380:
1377:
1375:
1372:
1370:
1367:
1365:
1362:
1360:
1357:
1355:
1352:
1350:
1347:
1345:
1342:
1340:
1337:
1335:
1332:
1330:
1327:
1325:
1322:
1320:
1317:
1315:
1312:
1310:
1307:
1305:
1302:
1300:
1297:
1295:
1292:
1290:
1287:
1285:
1282:
1280:
1277:
1275:
1272:
1270:
1267:
1265:
1262:
1260:
1257:
1255:
1252:
1250:
1247:
1245:
1242:
1240:
1237:
1235:
1232:
1230:
1227:
1225:
1222:
1220:
1217:
1215:
1212:
1210:
1207:
1205:
1202:
1200:
1197:
1195:
1192:
1190:
1187:
1185:
1182:
1180:
1177:
1175:
1172:
1170:
1167:
1165:
1162:
1160:
1157:
1155:
1152:
1150:
1147:
1145:
1142:
1140:
1137:
1135:
1132:
1130:
1127:
1125:
1122:
1120:
1117:
1115:
1112:
1110:
1107:
1105:
1102:
1100:
1097:
1095:
1092:
1090:
1087:
1085:
1082:
1080:
1077:
1075:
1072:
1069:
1065:
969:
962:
957:
948:
943:
810:
711:
693:for catalysis (
664:
644:gluconeogenesis
497:
209:
40:
17:
12:
11:
5:
4371:
4369:
4361:
4360:
4355:
4350:
4340:
4339:
4335:
4334:
4311:
4310:
4308:
4307:
4294:
4281:
4268:
4255:
4242:
4229:
4215:
4213:
4209:
4208:
4206:
4205:
4200:
4195:
4190:
4185:
4179:
4177:
4173:
4172:
4170:
4169:
4164:
4159:
4154:
4148:
4146:
4145:Classification
4142:
4141:
4139:
4138:
4133:
4128:
4123:
4117:
4115:
4111:
4110:
4108:
4107:
4102:
4097:
4092:
4087:
4082:
4077:
4072:
4066:
4064:
4060:
4059:
4054:
4052:
4051:
4044:
4037:
4029:
4020:
4019:
4016:
4015:
4012:
4011:
4009:
4008:
4003:
3998:
3997:
3996:
3985:
3983:
3979:
3978:
3976:
3975:
3970:
3969:
3968:
3963:
3958:
3953:
3943:
3938:
3933:
3928:
3927:
3926:
3921:
3916:
3911:
3901:
3900:
3899:
3890:
3875:
3873:
3867:
3866:
3864:
3863:
3858:
3843:
3838:
3833:
3827:
3825:
3816:
3805:
3804:
3801:
3800:
3798:
3797:
3791:
3789:
3783:
3782:
3780:
3779:
3773:
3771:
3762:
3748:
3731:
3730:
3728:
3727:
3722:
3717:
3712:
3711:
3710:
3705:
3700:
3695:
3682:
3680:
3670:
3669:
3667:
3666:
3661:
3651:
3646:
3637:
3632:
3627:
3622:
3621:
3620:
3610:
3609:
3608:
3603:
3593:
3587:
3585:
3574:
3573:
3571:
3570:
3565:
3560:
3551:
3550:
3549:
3531:
3530:
3529:
3519:
3514:
3509:
3504:
3499:
3498:
3497:
3487:
3486:
3485:
3475:
3470:
3465:
3448:
3447:
3446:
3441:
3436:
3425:
3423:
3413:
3412:
3410:
3409:
3404:
3399:
3393:
3391:
3381:
3380:
3378:
3377:
3372:
3366:
3365:
3364:
3363:
3358:
3353:
3342:
3341:
3340:
3339:
3337:Diacylglycerol
3334:
3329:
3324:
3319:
3314:
3309:
3304:
3299:
3290:
3279:
3278:
3273:
3268:
3266:Pectinesterase
3263:
3262:
3261:
3256:
3249:Cholinesterase
3245:
3243:
3231:
3230:
3216:
3214:
3213:
3206:
3199:
3191:
3182:
3181:
3179:
3178:
3173:
3172:
3171:
3148:
3146:
3142:
3141:
3138:
3137:
3135:
3134:
3129:
3123:
3121:
3113:
3112:
3110:
3109:
3103:
3101:
3089:
3088:
3086:
3085:
3079:
3077:
3065:
3064:
3062:
3061:
3056:
3050:
3048:
3037:
3033:
3032:
3029:
3028:
3026:
3025:
3011:
3006:
3001:
2996:
2990:
2987:
2986:
2984:
2983:
2978:
2959:
2956:
2955:
2953:
2952:
2930:
2925:
2898:
2892:
2888:
2887:
2867:
2865:
2864:
2857:
2850:
2842:
2794:
2793:
2781:
2780:External links
2778:
2777:
2776:
2770:
2751:
2748:
2746:
2745:
2710:
2683:(6): 1747–54.
2659:
2652:
2629:
2608:
2572:
2509:
2502:
2477:
2428:
2387:
2344:
2302:
2260:
2214:
2195:(4): 1115–24.
2170:
2121:
2100:(12): 5340–9.
2080:
2036:
1992:
1949:
1920:(19): 5624–7.
1896:
1837:
1802:
1767:
1731:
1729:
1726:
1725:
1724:
1719:
1714:
1709:
1702:
1699:
1669:
1666:
1601:
1580:
1577:
1573:
1572:
1556:
1555:
1544:
1543:
1447:
1441:
1436:
1431:
1426:
1421:
1416:
1411:
1406:
1401:
1396:
1391:
1386:
1381:
1376:
1371:
1366:
1361:
1356:
1351:
1346:
1341:
1336:
1331:
1326:
1321:
1316:
1311:
1306:
1301:
1296:
1291:
1286:
1281:
1276:
1271:
1266:
1261:
1256:
1251:
1246:
1241:
1236:
1231:
1226:
1221:
1216:
1211:
1206:
1201:
1196:
1191:
1186:
1181:
1176:
1171:
1166:
1161:
1156:
1151:
1146:
1141:
1136:
1131:
1126:
1121:
1116:
1111:
1106:
1101:
1096:
1091:
1086:
1081:
1076:
1071:
1070:
985:
984:
983:
982:
980:
979:
978:
977:
976:
975:
968:
965:
964:
963:
958:
951:
949:
944:
937:
809:
806:
727:sequence motif
710:
707:
671:
670:
662:
619:
618:
615:
614:
609:
603:
602:
589:
583:
582:
572:
565:
564:
556:
555:
542:
536:
535:
530:
524:
523:
518:
512:
511:
508:
504:
503:
499:
498:
495:
487:
486:
478:
477:
474:
473:
468:
462:
461:
448:
442:
441:
431:
424:
423:
415:
414:
409:
403:
402:
397:
390:
389:
384:
378:
377:
374:
370:
369:
365:
364:
356:
355:
352:
351:
346:
340:
339:
326:
320:
319:
309:
302:
301:
293:
292:
279:
273:
272:
267:
261:
260:
255:
249:
248:
243:
236:
235:
230:
224:
223:
220:
216:
215:
211:
210:
207:
199:
198:
190:
189:
186:
185:
180:
176:
175:
170:
166:
165:
157:
156:
146:
140:
139:
134:
128:
127:
123:
122:
117:
111:
110:
105:
99:
98:
93:
87:
86:
81:
75:
74:
69:
63:
62:
59:
55:
54:
51:
47:
46:
42:
41:
34:
26:
25:
15:
13:
10:
9:
6:
4:
3:
2:
4370:
4359:
4356:
4354:
4351:
4349:
4346:
4345:
4343:
4333:
4328:
4323:
4319:
4305:
4301:
4300:
4295:
4292:
4288:
4287:
4282:
4279:
4275:
4274:
4269:
4266:
4262:
4261:
4256:
4253:
4249:
4248:
4243:
4240:
4236:
4235:
4230:
4227:
4223:
4222:
4217:
4216:
4214:
4210:
4204:
4201:
4199:
4196:
4194:
4191:
4189:
4186:
4184:
4181:
4180:
4178:
4174:
4168:
4165:
4163:
4162:Enzyme family
4160:
4158:
4155:
4153:
4150:
4149:
4147:
4143:
4137:
4134:
4132:
4129:
4127:
4126:Cooperativity
4124:
4122:
4119:
4118:
4116:
4112:
4106:
4103:
4101:
4098:
4096:
4093:
4091:
4088:
4086:
4085:Oxyanion hole
4083:
4081:
4078:
4076:
4073:
4071:
4068:
4067:
4065:
4061:
4057:
4050:
4045:
4043:
4038:
4036:
4031:
4030:
4027:
4007:
4004:
4002:
3999:
3995:
3992:
3991:
3990:
3987:
3986:
3984:
3980:
3974:
3971:
3967:
3964:
3962:
3959:
3957:
3954:
3952:
3949:
3948:
3947:
3944:
3942:
3939:
3937:
3934:
3932:
3929:
3925:
3922:
3920:
3917:
3915:
3912:
3910:
3907:
3906:
3905:
3902:
3898:
3894:
3891:
3889:
3885:
3882:
3881:
3880:
3877:
3876:
3874:
3872:
3868:
3862:
3859:
3854:
3847:
3844:
3842:
3839:
3837:
3834:
3832:
3829:
3828:
3826:
3824:
3820:
3817:
3815:
3810:
3806:
3796:
3793:
3792:
3790:
3788:
3784:
3778:
3775:
3774:
3772:
3770:
3766:
3763:
3761:
3756:
3752:
3749:
3746:
3741:
3736:
3732:
3726:
3723:
3721:
3718:
3716:
3713:
3709:
3706:
3704:
3701:
3699:
3696:
3694:
3691:
3690:
3689:
3688:
3687:arylsulfatase
3684:
3683:
3681:
3679:
3675:
3671:
3665:
3662:
3659:
3655:
3652:
3650:
3647:
3645:
3641:
3638:
3636:
3633:
3631:
3628:
3626:
3623:
3619:
3616:
3615:
3614:
3611:
3607:
3604:
3602:
3599:
3598:
3597:
3596:Phospholipase
3594:
3592:
3589:
3588:
3586:
3584:
3579:
3575:
3569:
3566:
3564:
3561:
3559:
3555:
3552:
3548:
3544:
3540:
3537:
3536:
3535:
3532:
3528:
3525:
3524:
3523:
3520:
3518:
3515:
3513:
3510:
3508:
3505:
3503:
3500:
3496:
3493:
3492:
3491:
3488:
3484:
3481:
3480:
3479:
3476:
3474:
3471:
3469:
3466:
3464:
3460:
3456:
3452:
3449:
3445:
3442:
3440:
3437:
3435:
3432:
3431:
3430:
3427:
3426:
3424:
3422:
3418:
3414:
3408:
3405:
3403:
3400:
3398:
3395:
3394:
3392:
3390:
3386:
3382:
3376:
3373:
3371:
3368:
3367:
3362:
3359:
3357:
3354:
3352:
3349:
3348:
3347:
3346:Phospholipase
3344:
3343:
3338:
3335:
3333:
3330:
3328:
3325:
3323:
3320:
3318:
3315:
3313:
3310:
3308:
3305:
3303:
3300:
3298:
3294:
3291:
3289:
3286:
3285:
3284:
3281:
3280:
3277:
3274:
3272:
3269:
3267:
3264:
3260:
3257:
3255:
3252:
3251:
3250:
3247:
3246:
3244:
3240:
3236:
3232:
3227:
3223:
3219:
3212:
3207:
3205:
3200:
3198:
3193:
3192:
3189:
3177:
3174:
3170:
3166:
3162:
3158:
3155:
3154:
3153:
3150:
3149:
3147:
3143:
3133:
3130:
3128:
3125:
3124:
3122:
3119:
3114:
3108:
3105:
3104:
3102:
3099:
3098:Alanine cycle
3095:
3090:
3084:
3081:
3080:
3078:
3075:
3071:
3066:
3060:
3057:
3055:
3052:
3051:
3049:
3046:
3041:
3038:
3034:
3023:
3019:
3015:
3012:
3010:
3007:
3005:
3002:
3000:
2997:
2995:
2992:
2991:
2988:
2982:
2979:
2976:
2972:
2968:
2964:
2961:
2960:
2957:
2950:
2946:
2942:
2938:
2934:
2931:
2929:
2926:
2923:
2919:
2915:
2911:
2907:
2903:
2900:
2899:
2896:
2893:
2889:
2885:
2882:
2878:
2874:
2870:
2863:
2858:
2856:
2851:
2849:
2844:
2843:
2840:
2835:
2831:
2827:
2821:
2817:
2813:
2807:
2803:
2799:
2791:
2787:
2784:
2783:
2779:
2773:
2771:0-7167-3051-0
2767:
2762:
2761:
2754:
2753:
2749:
2741:
2737:
2733:
2729:
2725:
2721:
2714:
2711:
2706:
2702:
2698:
2694:
2690:
2686:
2682:
2678:
2670:
2668:
2666:
2664:
2660:
2655:
2649:
2645:
2638:
2636:
2634:
2630:
2625:
2619:
2611:
2605:
2601:
2597:
2593:
2585:
2583:
2581:
2579:
2577:
2573:
2568:
2564:
2559:
2554:
2549:
2544:
2540:
2536:
2533:(7): e68830.
2532:
2528:
2524:
2522:
2513:
2510:
2505:
2499:
2495:
2488:
2486:
2484:
2482:
2478:
2473:
2469:
2464:
2459:
2455:
2451:
2447:
2443:
2439:
2432:
2429:
2424:
2420:
2415:
2410:
2407:(2): 651–62.
2406:
2402:
2398:
2391:
2388:
2383:
2379:
2375:
2371:
2367:
2363:
2359:
2355:
2348:
2345:
2340:
2336:
2332:
2328:
2325:(2): 279–85.
2324:
2320:
2313:
2311:
2309:
2307:
2303:
2298:
2294:
2290:
2286:
2282:
2278:
2271:
2269:
2267:
2265:
2261:
2256:
2252:
2248:
2244:
2240:
2236:
2233:(3): 317–29.
2232:
2228:
2221:
2219:
2215:
2210:
2206:
2202:
2198:
2194:
2190:
2183:
2181:
2179:
2177:
2175:
2171:
2166:
2162:
2157:
2152:
2148:
2144:
2140:
2136:
2132:
2125:
2122:
2117:
2113:
2108:
2103:
2099:
2095:
2091:
2084:
2081:
2076:
2072:
2067:
2062:
2059:(6): 949–59.
2058:
2054:
2050:
2043:
2041:
2037:
2032:
2028:
2023:
2018:
2014:
2010:
2006:
1999:
1997:
1993:
1988:
1984:
1980:
1976:
1972:
1971:10.1038/80968
1968:
1964:
1960:
1953:
1950:
1945:
1941:
1936:
1931:
1927:
1923:
1919:
1915:
1911:
1909:
1900:
1897:
1892:
1888:
1883:
1878:
1873:
1868:
1864:
1860:
1856:
1852:
1848:
1841:
1838:
1833:
1829:
1825:
1821:
1818:(3): 1080–3.
1817:
1813:
1806:
1803:
1798:
1794:
1790:
1786:
1783:(2): 374–81.
1782:
1778:
1771:
1768:
1763:
1759:
1755:
1751:
1747:
1743:
1736:
1733:
1727:
1723:
1720:
1718:
1715:
1713:
1710:
1708:
1705:
1704:
1700:
1698:
1694:
1692:
1687:
1686:hyperglycemia
1683:
1674:
1667:
1665:
1662:
1658:
1652:
1650:
1646:
1643:
1637:
1635:
1631:
1627:
1623:
1618:
1616:
1612:
1608:
1598:
1594:
1590:
1586:
1578:
1568:
1561:
1558:
1551:
1546:
1538:
1451:
989:
974:
973:
966:
961:
955:
950:
947:
941:
936:
934:
932:
928:
924:
920:
916:
912:
911:
906:
902:
898:
894:
893:Gram-positive
890:
889:
883:
881:
877:
873:
870:
866:
863:
859:
855:
851:
847:
843:
842:
838:
834:
829:
827:
823:
819:
815:
807:
805:
803:
800:
796:
792:
788:
784:
780:
776:
772:
768:
764:
760:
756:
752:
748:
744:
740:
736:
732:
728:
724:
720:
716:
708:
706:
704:
700:
696:
692:
688:
684:
679:
675:
657:
656:
655:
653:
649:
645:
641:
637:
633:
626:
613:
610:
608:
604:
601:
597:
593:
590:
588:
584:
580:
576:
573:
570:
566:
561:
557:
554:
550:
546:
543:
541:
537:
534:
531:
529:
525:
522:
519:
517:
513:
509:
505:
500:
493:
488:
483:
472:
469:
467:
463:
460:
456:
452:
449:
447:
443:
439:
435:
432:
429:
425:
420:
416:
413:
410:
408:
404:
401:
398:
395:
391:
388:
385:
383:
379:
375:
371:
366:
361:
350:
347:
345:
341:
338:
334:
330:
327:
325:
321:
317:
313:
310:
307:
303:
298:
294:
291:
287:
283:
280:
278:
274:
271:
268:
266:
262:
259:
256:
254:
250:
247:
244:
241:
237:
234:
231:
229:
225:
221:
217:
212:
205:
200:
195:
184:
181:
177:
174:
171:
167:
162:
158:
155:
154:
150:
147:
145:
141:
138:
135:
133:
129:
124:
121:
118:
116:
112:
109:
106:
104:
100:
97:
94:
92:
88:
85:
82:
80:
76:
73:
70:
68:
64:
60:
56:
52:
48:
43:
38:
32:
27:
22:
19:
4299:Translocases
4296:
4283:
4270:
4257:
4244:
4234:Transferases
4231:
4218:
4075:Binding site
3856:}}
3850:{{
3814:Endonuclease
3745:ribonuclease
3685:
3477:
3468:Nucleotidase
3389:Thioesterase
3045:oxaloacetate
2948:
2760:Biochemistry
2759:
2723:
2719:
2713:
2680:
2676:
2643:
2591:
2530:
2526:
2520:
2512:
2493:
2448:(1): 300–5.
2445:
2441:
2431:
2404:
2400:
2390:
2360:(2): 203–9.
2357:
2354:FEBS Letters
2353:
2347:
2322:
2318:
2283:(1): 23–28.
2280:
2276:
2230:
2226:
2192:
2188:
2138:
2134:
2124:
2097:
2093:
2083:
2056:
2052:
2012:
2008:
1962:
1958:
1952:
1917:
1913:
1907:
1899:
1854:
1850:
1840:
1815:
1811:
1805:
1780:
1776:
1770:
1748:(1): 151–7.
1745:
1741:
1735:
1695:
1691:hypoglycemia
1679:
1653:
1638:
1619:
1582:
1560:
971:
970:
908:
886:
884:
839:
833:thermophilic
830:
811:
791:structurally
712:
672:
648:Calvin cycle
628:
624:
622:
151:
58:Alt. symbols
18:
4070:Active site
3989:Nuclease S1
3760:Exonuclease
3654:Lecithinase
3483:Calcineurin
3421:Phosphatase
3327:Lipoprotein
3317:Endothelial
2918:Glucokinase
2726:: 473–505.
1649:proteolytic
1585:hibernation
903:from other
872:side chains
865:phosphatase
623:The enzyme
502:Identifiers
368:Identifiers
214:Identifiers
173:Swiss-model
45:Identifiers
4342:Categories
4273:Isomerases
4247:Hydrolases
4114:Regulation
3302:Pancreatic
3239:Carboxylic
3145:Regulatory
3074:Cori cycle
2965:(Aldolase
2902:Hexokinase
2891:Glycolysis
2877:glycolysis
2869:Metabolism
1728:References
1722:Metabolism
1630:glycolysis
1607:allosteric
874:and metal
854:structural
814:eukaryotes
802:metabolism
783:homologues
575:structures
434:structures
312:structures
169:Structures
164:Search for
126:Other data
4152:EC number
3879:RNase III
3737:(includes
3678:Sulfatase
3591:Autotaxin
3455:Prostatic
3307:Lysosomal
3222:esterases
3218:Hydrolase
2834:IPR002803
2820:IPR009164
2806:IPR000146
2618:cite book
2053:Structure
1657:substrate
1645:proteases
1642:lysosomal
1597:euthermic
1589:metabolic
905:organisms
888:Bacillota
869:catalytic
850:organisms
837:bacterium
795:metabolic
775:bacterial
709:Structure
687:organisms
533:IPR002803
412:IPR009164
270:PDOC00114
258:IPR000146
132:EC number
108:NM_000507
67:NCBI gene
4353:EC 3.1.3
4176:Kinetics
4100:Cofactor
4063:Activity
3973:RNase T1
3735:Nuclease
3370:Cutinase
3118:glycerol
2945:Platelet
2830:InterPro
2816:InterPro
2802:InterPro
2740:29055870
2705:19832318
2697:16731838
2677:Diabetes
2567:23874782
2527:PLOS ONE
2423:14275118
2382:22515761
2339:12464282
2255:32940046
2247:15288602
2075:15274916
2031:12065581
1979:11062561
1944:10986273
1712:Fructose
1701:See also
1668:Diabetes
1661:Aldolase
1593:dormancy
931:glycerol
897:sequence
891:(low GC
818:bacteria
787:proteins
723:Inositol
646:and the
592:RCSB PDB
528:InterPro
510:FBPase_3
451:RCSB PDB
407:InterPro
376:FBPase_2
329:RCSB PDB
253:InterPro
183:InterPro
137:3.1.3.11
4332:Biology
4286:Ligases
4056:Enzymes
3946:RNase E
3941:RNase Z
3936:RNase A
3931:RNase P
3904:RNase H
3522:Phytase
3322:Hepatic
3297:Lingual
3293:Gastric
3094:alanine
3070:lactate
3009:Enolase
2884:enzymes
2558:3715522
2535:Bibcode
2472:4292000
2463:1270577
2374:6321241
2297:1881399
2209:9505421
2165:9696785
1987:7617099
1891:7761465
1859:Bibcode
1832:8382485
1797:3008716
1762:2159755
981:[[File:
923:Mutants
901:enzymes
876:ligands
822:Archaea
799:quinone
781:IMPase
521:PF01950
387:PF06874
265:PROSITE
233:PF00316
179:Domains
115:UniProt
4318:Portal
4260:Lyases
3884:Drosha
3809:3.1.21
3777:RecBCD
3755:3.1.11
3375:PETase
3283:Lipase
3169:PFKFB4
3165:PFKFB3
3161:PFKFB2
3157:PFKFB1
2941:Muscle
2792:(MeSH)
2768:
2738:
2703:
2695:
2650:
2606:
2565:
2555:
2500:
2470:
2460:
2421:
2380:
2372:
2337:
2295:
2253:
2245:
2207:
2163:
2156:107433
2153:
2116:221467
2114:
2073:
2029:
1985:
1977:
1942:
1935:111013
1932:
1889:
1879:
1830:
1795:
1760:
1622:anoxia
927:malate
907:. The
860:for a
771:fungal
761:metal
607:PDBsum
581:
571:
553:SUPFAM
507:Symbol
466:PDBsum
440:
430:
400:CL0163
373:Symbol
344:PDBsum
318:
308:
290:SUPFAM
246:CL0171
222:FBPase
219:Symbol
149:Chr. 9
120:P09467
103:RefSeq
96:229700
50:Symbol
4212:Types
3893:Dicer
3848:;see
3674:3.1.6
3644:PDE4B
3640:PDE4A
3578:3.1.4
3547:IMPA3
3543:IMPA2
3539:IMPA1
3417:3.1.3
3385:3.1.2
3235:3.1.1
3116:from
3092:from
3068:from
2937:Liver
2701:S2CID
2378:S2CID
2293:S2CID
2251:S2CID
1983:S2CID
1882:41866
862:sugar
779:yeast
767:motif
745:-Asp-
549:SCOPe
540:SCOP2
286:SCOPe
277:SCOP2
153:q22.3
144:Locus
4304:list
4297:EC7
4291:list
4284:EC6
4278:list
4271:EC5
4265:list
4258:EC4
4252:list
4245:EC3
4239:list
4232:EC2
4226:list
4219:EC1
3811:-31:
3757:-16:
3743:and
3649:PDE5
3635:PDE3
3630:PDE2
3625:PDE1
3517:PTEN
3502:OCRL
3495:PP2A
3444:ALPP
3439:ALPL
3434:ALPI
3228:3.1)
3022:PKM2
3018:PKLR
2828:and
2826:Pfam
2814:and
2812:Pfam
2800:and
2798:Pfam
2766:ISBN
2736:PMID
2693:PMID
2648:ISBN
2624:link
2604:ISBN
2563:PMID
2498:ISBN
2468:PMID
2419:PMID
2370:PMID
2335:PMID
2243:PMID
2205:PMID
2161:PMID
2112:PMID
2071:PMID
2027:PMID
1975:PMID
1940:PMID
1887:PMID
1828:PMID
1793:PMID
1758:PMID
1550:edit
1537:edit
929:and
858:fold
852:. A
816:and
777:and
763:ions
759:bind
719:pigs
715:fold
713:The
697:and
691:ions
665:O =
642:in
600:PDBj
596:PDBe
579:ECOD
569:Pfam
545:1umg
516:Pfam
459:PDBj
455:PDBe
438:ECOD
428:Pfam
396:clan
394:Pfam
382:Pfam
337:PDBj
333:PDBe
316:ECOD
306:Pfam
282:1frp
242:clan
240:Pfam
228:Pfam
91:OMIM
84:3606
79:HGNC
72:2203
53:FBP1
37:3FBP
3966:4/5
3043:to
2947:)→/
2920:)→/
2914:HK3
2910:HK2
2906:HK1
2728:doi
2724:141
2685:doi
2596:doi
2553:PMC
2543:doi
2458:PMC
2450:doi
2446:104
2409:doi
2405:240
2362:doi
2358:167
2327:doi
2323:408
2285:doi
2281:162
2235:doi
2231:141
2197:doi
2193:118
2151:PMC
2143:doi
2139:180
2102:doi
2098:254
2061:doi
2017:doi
2013:277
1967:doi
1930:PMC
1922:doi
1918:182
1877:PMC
1867:doi
1820:doi
1816:190
1785:doi
1781:135
1750:doi
1746:279
1676:AMP
1611:AMP
915:AMP
880:ion
755:Thr
751:Ser
747:Gly
743:Leu
739:Ile
735:Pro
731:Asp
638:to
587:PDB
446:PDB
324:PDB
61:FBP
4344::
3924:2C
3919:2B
3914:2A
3895::
3886::
3676::
3556::
3545:,
3541:,
3457:)/
3419::
3387::
3356:A2
3351:A1
3237::
3226:EC
3220::
3167:,
3163:,
3159:,
3100:):
3076:):
3020:,
2973:,
2969:,
2943:,
2939:,
2916:,
2912:,
2908:,
2875::
2871::
2832::
2818::
2804::
2734:.
2722:.
2699:.
2691:.
2681:55
2679:.
2662:^
2632:^
2620:}}
2616:{{
2575:^
2561:.
2551:.
2541:.
2529:.
2525:.
2480:^
2466:.
2456:.
2444:.
2440:.
2417:.
2403:.
2399:.
2376:.
2368:.
2356:.
2333:.
2321:.
2305:^
2291:.
2279:.
2263:^
2249:.
2241:.
2229:.
2217:^
2203:.
2191:.
2173:^
2159:.
2149:.
2137:.
2133:.
2110:.
2096:.
2092:.
2069:.
2057:12
2055:.
2051:.
2039:^
2025:.
2011:.
2007:.
1995:^
1981:.
1973:.
1961:.
1938:.
1928:.
1916:.
1912:.
1885:.
1875:.
1865:.
1855:92
1853:.
1849:.
1826:.
1814:.
1791:.
1779:.
1756:.
1744:.
1539:]]
933:.
804:.
773:,
705:.
703:Li
699:Mn
695:Mg
654::
598:;
594:;
577:/
551:/
547:/
457:;
453:;
436:/
335:;
331:;
314:/
288:/
284:/
4320::
4306:)
4302:(
4293:)
4289:(
4280:)
4276:(
4267:)
4263:(
4254:)
4250:(
4241:)
4237:(
4228:)
4224:(
4048:e
4041:t
4034:v
3961:3
3956:2
3951:1
3909:1
3747:)
3660:)
3656:(
3642:/
3618:1
3606:D
3601:C
3580::
3461:/
3453:(
3361:B
3295:/
3224:(
3210:e
3203:t
3196:v
3120::
3096:(
3072:(
3047::
3024:)
3016:(
2977:)
2975:C
2971:B
2967:A
2951:←
2935:(
2924:←
2904:(
2879:/
2861:e
2854:t
2847:v
2774:.
2742:.
2730::
2707:.
2687::
2656:.
2626:)
2612:.
2598::
2569:.
2545::
2537::
2531:8
2506:.
2474:.
2452::
2425:.
2411::
2384:.
2364::
2341:.
2329::
2299:.
2287::
2257:.
2237::
2211:.
2199::
2167:.
2145::
2118:.
2104::
2077:.
2063::
2033:.
2019::
1989:.
1969::
1963:7
1946:.
1924::
1910:"
1893:.
1869::
1861::
1834:.
1822::
1799:.
1787::
1764:.
1752::
1602:m
1600:K
1569:.
1443:]
1438:]
1433:]
1428:]
1423:]
1418:]
1413:]
1408:]
1403:]
1398:]
1393:]
1388:]
1383:]
1378:]
1373:]
1368:]
1363:]
1358:]
1353:]
1348:]
1343:]
1338:]
1333:]
1328:]
1323:]
1318:]
1313:]
1308:]
1303:]
1298:]
1293:]
1288:]
1283:]
1278:]
1273:]
1268:]
1263:]
1258:]
1253:]
1248:]
1243:]
1238:]
1233:]
1228:]
1223:]
1218:]
1213:]
1208:]
1203:]
1198:]
1193:]
1188:]
1183:]
1178:]
1173:]
1168:]
1163:]
1158:]
1153:]
1148:]
1143:]
1138:]
1133:]
1128:]
1123:]
1118:]
1113:]
1108:]
1103:]
1098:]
1093:]
1088:]
1083:]
1078:]
1073:]
753:-
749:/
741:/
737:-
733:-
729:(
667:D
663:2
659:D
630:D
39:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.