926:
that overlap the variation. Then it uses a rule-based approach to predict the effects that each allele of the variation may have on the transcript. The set of consequence terms, defined by the
Sequence Ontology (SO) can be currently assigned to each combination of an allele and a transcript. Each allele of each variation may have a different effect in different transcripts. A variety of different tools are used to predict human mutations in the Ensembl database, one of the most widely used is SIFT, that predicts whether an amino acid substitution is likely to affect protein function based on sequence homology and the physic-chemical similarity between the alternate amino acids. The data provided for each amino acid substitution is a score and a qualitative prediction (either 'tolerated' or 'deleterious'). The score is the normalized probability that the amino acid change is tolerated so scores near 0 are more likely to be deleterious. The qualitative prediction is derived from this score such that substitutions with a score < 0.05 are called 'deleterious' and all others are called 'tolerated'. SIFT can be applied to naturally occurring nonsynonymous polymorphisms and laboratory-induced missense mutations, that will lead to build relationships in phenotype characteristics,
121:. Many genetic diseases are developed from before birth. Genetic disorders account for a significant number of the health care problems in our society. Advances in the understanding of this diseases have increased both the life span and quality of life for many of those affected by genetic disorders. Recent developments in bioinformatics and laboratory genetics have made possible the better delineation of certain malformation and mental retardation syndromes, so that their mode of inheritance can be understood. This information enables the genetic counselor to predict the risk for occurrence of a large number of genetic disorders. Most genetic counseling is done, however, only after the birth of at least one affected individual has alerted the family to their predilection for having children with a genetic disorder. The association of a single gene to a disease is rare and a genetic disease may or may not be a transmissible disorder. Some genetic diseases are inherited from the parent's genes, but others are caused by new
1822:
approaches to respond to new techniques, such as next-generation sequencing technologies. For instance, the availability of large numbers of individual human genomes will promote the development of computational analyses of rare variants, including the statistical mining of their relations to lifestyles, drug interactions and other factors. Biomedical research will also be driven by our ability to efficiently mine the large body of existing and continuously generated biomedical data. Text-mining techniques, in particular, when combined with other molecular data, can provide information about gene mutations and interactions and will become crucial to stay ahead of the exponential growth of data generated in biomedical research. Another field that is benefiting from the advances in mining and integration of molecular, clinical and drug analysis is pharmacogenomics.
696:(RGD) began as a collaborative effort between leading research institutions involved in rat genetic and genomic research. The rat continues to be extensively used by researchers as a model organism for investigating the biology and pathophysiology of disease. In the past several years, there has been a rapid increase in rat genetic and genomic data. This explosion of information highlighted the need for a centralized database to efficiently and effectively collect, manage, and distribute a rat-centric view of this data to researchers around the world. The Rat Genome Database was created to serve as a repository of rat genetic and genomic data, as well as mapping, strain, and physiological information. It also facilitates investigators research efforts by providing tools to search, mine, and predict this data.
1141:
that leads to disease) or is a biomarker for a disease. #Genetic
Variation Association: Used when a sequence variation (a mutation, a SNP) is associated to the disease phenotype, but there is still no evidence to say that the variation causes the disease. In some cases the presence of the variants increase the susceptibility to the disease. In general, the NCBI SNP identifiers are provided. #Altered Expression Association: Alterations in the function of the protein by means of altered expression of the gene are associated with the disease phenotype. #Post-translational Modification Association: Alterations in the function of the protein by means of post-translational modifications (methylation or phosphorylation of the protein) are associated with the disease phenotype.
210:, helps to understand about the effects of environmental compounds on human health by integrating data from curated scientific literature to describe biochemical interactions with genes and proteins, and links between diseases and chemicals, and diseases and genes or proteins. CTD contains curated data defining cross-species chemical–gene/protein interactions and chemical– and gene–disease associations to illuminate molecular mechanisms underlying variable susceptibility and environmentally influenced diseases. These data deliver insights into complex chemical–gene and protein interaction networks. One of the main sources in this Database is curated information from OMIM.
1124:
assess the concept of modularity of human diseases, this database performs a systematic study of the emergent properties of human gene-disease networks by means of network topology and functional annotation analysis. The results indicate a highly shared genetic origin of human diseases and show that for most diseases, including
Mendelian, complex and environmental diseases, functional modules exist. Moreover, a core set of biological pathways is found to be associated with most human diseases. Obtaining similar results when studying
1810:
multiple diseases. The next step is to produce a complete picture of the mechanistic aspects of the diseases and the design of drugs against them. For that, a combination of two approaches will be needed: a systematic search and in-depth study of each gene. The future of the field will be defined by new techniques to integrate large bodies of data from different sources and to incorporate functional information into the analysis of large-scale data generated by bioinformatics studies.
836:
1800:
809:
181:
the
Comparative Toxicogenomics Database (CTD), Online Mendelian Inheritance in Man (OMIM), the genetic Association Database (GAD) or the Disease genetic Association Database (DisGeNET). Each of these databases focuses on different aspects of the phenotype-genotype relationship, and due to the nature of the database curation process, they are not complete, but in a way they are fully complementary between each other.
559:), and to other related genes or genomes, which the same as prediction over time is not necessarily. When information is transferred across time, often to specific points in time, the process is known as forecasting. Three of the main examples of databases that can be considered in this category include: The Mouse genome Database (MGD), The Rat genome Database (RGD), OMIM and the SIFT Tool from Ensembl.
1839:
1133:
1818:. Often, such identification is made with the aim of better understanding the genetic basis of disease, unique adaptations, desirable properties, or differences between populations. In a less formal way, bioinformatics also tries to understand the organisational principles within nucleic acid and protein sequences.
198:
activity The implication is that the resulting database is of high quality. The contrast is with data which may have been gathered through some automated process or using particularly low or inexpert unsupported data quality and possibly untrustworthy. Some of the most common examples include: CTD and UNIPROT.
154:
1821:
The response of bioinformatics to new experimental techniques brings a new perspective into the analysis of the experimental data, as demonstrated by the advances in the analysis of information from gene disease databases and other technologies. It is expected that this trend will continue with novel
1140:
The description of each association type in this ontology is: #Therapeutic
Association: The gene/protein has a therapeutic role in the amelioration of the disease. #Biomarker Association: The gene/protein either plays a role in the etiology of the disease (e.g. participates in the molecular mechanism
925:
is one of several well-known genome browsers for the retrieval of genomic-disease information. Ensembl imports variation data from a variety of different sources, Ensembl predicts the effects of variants. For each variation that is mapped to the reference genome, each
Ensembl transcript is identified
699:
Data at RGD that is useful for researchers investigating disease genes include disease annotations for rat, mouse and human genes. Annotations are manually curated from the literature, or downloaded via automated pipelines from other disease-related databases. Downloaded annotations are mapped to the
1813:
Bioinformatics is both a term for the body of biological gene disease studies that use computer programming as part of their methodology, as well as a reference to specific analysis pipelines that are repeatedly used, particularly in the fields of genetics and genomics. Common uses of bioinformatics
978:
This sort of databases include
Mendelian, compound and environmental diseases in an integrated gene-disease association archive and show that the concept of modularity applies for all of them They provide a functional analysis of diseases in case of important new biological insights, which might not
960:
The literature-derived human gene-disease network (LHGDN) is a text mining derived database with focus on extracting and classifying gene-disease associations with respect to several biomolecular conditions. It uses a machine learning based algorithm to extract semantic gene-disease relations from a
180:
However, increasingly evidences point out that most human diseases cannot be attributed to a single gene but arise due to complex interactions among multiple genetic variants and environmental risk factors. Several databases have been developed storing associations between genes and diseases such as
804:
Supported by the NCBI, The Online
Mendelian Inheritance in Man (OMIM) is a database that catalogues all the known diseases with a genetic component, and predicts their relationship to relevant genes in the human genome and provides references for further research and tools for genomic analysis of a
554:
A predictive database is one based on statistical inference. One particular approach to such inference is known as predictive inference, but the prediction can be undertaken within any of the several approaches to statistical inference. Indeed, one description of biostatistics is that it provides a
197:
The term curated data refers to information, that may comprise the most sophisticated computational formats for structured data, scientific updates, and curated knowledge, that has been composed and prepared under the regulation of one or more experts considered to be qualified to engage in such an
1826:
studies of the relationships between human variations and their effect on diseases will be key to the development of personalized medicine. In summary, Gene
Disease Databases have already transformed the search for disease genes and has the potential to become a crucial component of other areas of
1809:
The completion of the human genome has changed the way the search for disease genes is performed. In the past, the approach was to focus on one or a few genes at a time. Now, projects like the DisGeNET exemplify the efforts to systematically analyze all the gene alterations involved in a single or
1128:
of diseases, the findings in this database suggest that related diseases might arise due to dysfunction of common biological processes in the cell. The network analysis of this integrated database points out that data integration is needed to obtain a comprehensive view of the genetic landscape of
172:
to investigate further experimentally and which to leave out because of limited resources. Computational methods that integrate complex, heterogeneous data sets, such as expression data, sequence information, functional annotation and the biomedical literature, allow prioritizing genes for future
1123:
is a comprehensive gene-disease association database that integrates associations from several sources that covers different biomedical aspects of diseases. In particular, it is focused on the current knowledge of human genetic diseases including
Mendelian, complex and environmental diseases. To
567:
The Mouse genome Database (MGD) is the international community resource for integrated genetic, genomic and biological data about the laboratory mouse. MGD provides full annotation of phenotypes and human disease associations for mouse models (genotypes) using terms from the Mammalian Phenotype
110:
have been trying to comprehend the molecular mechanisms of diseases to design preventive and therapeutic strategies for a long time. For some illnesses, it has become apparent that it is the right amount of animosity is made for not enough to obtain an index of the disease-related genes but to
947:
The Genetic Association Database is an archive of human genetic association studies of complex diseases. GAD is primarily focused on archiving information on common complex human disease rather than rare Mendelian disorders as found in the OMIM. It includes curated summary data extracted from
140:
There are more than six thousand known single-gene disorders (monogenic), which occur in about 1 out of every 200 births. As their term suggests, these diseases are caused by a mutation in one gene. By contrast, polygenic disorders are caused by several genes, regularly in combination with
160:: Typical lists come from linkage regions, chromosomal aberrations, association study loci, deferentially expressed gene lists or genes identified by sequencing variants. Alternatively, the complete genome can be prioritized, but substantially more false positives would then be expected.
1654:
Gu, Ying; Liu, Guang-Hui; Plongthongkum, Nongluk; Benner, Christopher; Yi, Fei; Qu, Jing; Suzuki, Keiichiro; Yang, Jiping; Zhang, Weiqi; Li, Mo; Montserrat, Nuria; Crespo, Isaac; Del Sol, Antonio; Esteban, Concepcion Rodriguez; Zhang, Kun; Izpisua Belmonte, Juan Carlos (2014).
979:
be discovered when considering each of the gene-disease associations independently. Hence, they present a suitable framework for the study of how genetic and environmental factors, such as drugs, contribute to diseases. The best example for this sort of database is DisGeNET.
920:
This one of the largest resources available for all genomic and genetic studies, it provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other vertebrates and model disease organisms.
538:
969:
Extracts gene-disease associations from MEDLINE abstract using the BeFree system. BeFree is composed of a biomedical Named Entity Recognition (BioNER) module to detect diseases and genes and a relation extraction module based on morphosyntactic information.
805:
catalogued gene. OMIM is a comprehensive, authoritative compendium of human genes and genetic phenotypes that is freely available and updated daily. The database has been used as a resource for predicting relevant information to inherited conditions.
815:
Showing the concept that diseases have large association with a variety of genes, a mean pathway homogeneity values of single diseases and random controls are plotted for four networks binned by the number of associated gene products per disease.
239:) is an inclusive resource for protein sequence and annotation data. It is a comprehensive, first-class and freely accessible database of protein sequence and functional information, that has many entries being derived from
85:
is a systematized collection of data, typically structured to model aspects of reality, in a way to comprehend the underlying mechanisms of complex diseases, by understanding multiple composite interactions between
111:
uncover how disruptions of molecular grids in the cell give rise to disease phenotypes. Moreover, even with the unprecedented wealth of information available, obtaining such catalogues is extremely difficult.
243:
sequencing projects. It contains a large amount of information about the biological function of proteins derived from the study literature, which can hint to a direct connection between gene-protein-disease.
1188:
Kaikkonen, Minna U.; Niskanen, Henri; Romanoski, Casey E.; Kansanen, Emilia; Kivelä, Annukka M.; Laitalainen, Jarkko; Heinz, Sven; Benner, Christopher; Glass, Christopher K.; Ylä-Herttuala, Seppo (2014).
938:
This sort of databases summarize books, articles, book reviews, dissertations, and annotations about gene-disease databases. Some of the following are examples of this type: GAD, LGHDN and BeFree Data.
1472:
Lee, In-Hee; Lee, Kyungjoon; Hsing, Michael; Choe, Yongjoon; Park, Jin-Ho; Kim, Shu Hee; Bohn, Justin M.; Neu, Matthew B.; Hwang, Kyu-Baek; Green, Robert C.; Kohane, Isaac S.; Kong, Sek Won (2014).
173:
study in a more informed way. Such methods can substantially increase the yield of downstream studies and are becoming invaluable to researchers. So one of the main concerns in biological and
700:
same disease vocabulary used for manual annotations to provide consistency across the dataset. RGD also maintains disease-related quantitative phenotype data for the rat (PhenoMiner).
818:
This graph shows how difficult is to correlate a bigger number of diseases vs concordance in 4 different databases, hence Gene Disease Databases test these relationships
1611:
Koczor, Christopher A.; Lee, Eva K.; Torres, Rebecca A.; Boyd, Amy; Vega, J. David; Uppal, Karan; Yuan, Fan; Fields, Earl J.; Samarel, Allen M.; Lewis, William (2013).
1913:
177:
research is to recognise the underlying mechanisms behind this intricate genetic phenotypes. Great effort has been spent on finding the genes related to diseases
1232:
Grosdidier, Solène; Ferrer, Antoni; Faner, Rosa; Piñero, Janet; Roca, Josep; Cosío, Borja; Agustí, Alvar; Gea, Joaquim; Sanz, Ferran; Furlong, Laura I. (2014).
755:
1312:
Gallagher, Suzanne Renick; Dombrower, Micah; Goldberg, Debra S. (2014). "Using 2-node hypergraph clustering coefficients to analyze disease-gene networks".
1933:
2798:
145:, breast cancer, leukemia, Down syndrome, heart defects, and deafness; therefore, cataloguing to sort out all the diseases related to genes is needed.
2016:
Botstein, D (2003). "Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease".
94:
relationships and gene-disease mechanisms. Gene Disease Databases integrate human gene-disease associations from various expert curated databases and
2218:"Text Mining Effectively Scores and Ranks the Literature for Improving Chemical-Gene-Disease Curation at the Comparative Toxicogenomics Database"
137:, may stem from an inbred condition in some people, from new changes in other people, and from non-genetic causes in still other individuals.
1435:
Santiago, Jose A.; Potashkin, Judith A. (2014). "System-based approaches to decode the molecular links in Parkinson's disease and diabetes".
1613:"Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis"
207:
189:
Essentially, there are four types of databases: curated databases, predictive databases, literature databases and integrative databases
213:
CTD is a unique resource where bioinformatics specialists read the scientific literature and manually curate four types of core data:
2814:"Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research"
1277:
Cristiano, Francesca; Veltri, Pierangelo (2014). "An R-based tool for miRNA data analysis and correlation with clinical ontologies".
1888:
1329:
1294:
876:
315:
311:
1700:"Integrated analysis of transcript-level regulation of metabolism reveals disease-relevant nodes of the human metabolic network"
1923:
871:
354:
949:
545:
The curated data may comprise a process from practical experience and literature review to web publication of the database
631:
1928:
621:
319:
61:
17:
521:
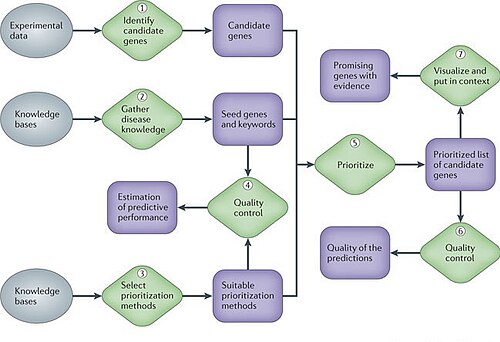
Yes – manual and automatic. Rules for automatic annotation generated by database curators and computational algorithms.
2970:
1883:
431:
2772:
2100:
1908:
1741:
Tieri, Paolo; Termanini, Alberto; Bellavista, Elena; Salvioli, Stefano; Capri, Miriam; Franceschi, Claudio (2012).
1948:
1873:
1392:"Molecularly and clinically related drugs and diseases are enriched in phenotypically similar drug-disease pairs"
1314:
Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics - BCB '14
1279:
Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics - BCB '14
2652:
A. Homosh, "Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders,"
1858:
1853:
1698:
Galhardo, Mafalda; Sinkkonen, Lasse; Berninger, Philipp; Lin, Jake; Sauter, Thomas; Heinäniemi, Merja (2014).
1474:"Prioritizing Disease-Linked Variants, Genes, and Pathways with an Interactive Whole-Genome Analysis Pipeline"
2401:"Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical–gene–disease networks"
2558:
S. Hunter and P. Jones, "InterPro in 2011: new developments in the family and domain prediction database,"
1878:
684:
350:
142:
1349:"Organ system heterogeneity DB: A database for the visualization of phenotypes at the organ system level"
1868:
468:
2325:"Systematic analysis of gene properties influencing organ system phenotypes in mammalian perturbations"
1150:
Some of the most interesting cases using Gene-Disease Databases can be found in the following papers:
835:
2587:
M. Dwinell, E. Worthey and S. M, "The Rat genome Database 2009: variation, ontologies and pathways,"
2229:
1903:
1754:
1528:
1893:
693:
498:
24:
1799:
1129:
human diseases and that the genetic origin of complex diseases is much more common than expected.
948:
published papers in peer reviewed journals on candidate gene and genome Wide Association Studies (
2149:
2041:
1898:
1460:
1335:
1300:
2868:"DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes"
2284:"DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene–disease networks"
1568:
Zhao, Yilei; Wang, Chen; Wu, Jianwei; Wang, Yan; Zhu, Wenliang; Zhang, Yong; Du, Zhimin (2013).
961:
textual source of interest. It is part of the Linked Life Data, of the LMU in Munchen, Germany.
555:
means of transferring knowledge about a sample of a genetic population to the whole population (
2946:
2897:
2845:
2792:
2754:
2697:
2632:
2528:
2479:
2430:
2356:
2305:
2257:
2198:
2141:
2082:
2033:
1782:
1729:
1686:
1642:
1599:
1556:
1503:
1452:
1423:
1378:
1325:
1290:
1265:
1220:
1176:
1064:
765:
649:
165:
2936:
2928:
2887:
2879:
2835:
2825:
2744:
2687:
2679:
2622:
2614:
2603:"The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease"
2518:
2510:
2469:
2461:
2420:
2412:
2346:
2336:
2295:
2247:
2237:
2188:
2180:
2131:
2072:
2025:
1772:
1762:
1719:
1711:
1676:
1668:
1632:
1624:
1589:
1581:
1546:
1536:
1493:
1485:
1444:
1413:
1403:
1368:
1360:
1317:
1282:
1255:
1245:
1210:
1202:
1168:
1156:
490:
485:
153:
2571:
C. Bult and J. Eppig, "The Mouse genome Database (MGD): mouse biology and model systems,"
1918:
1125:
1035:
863:
750:
616:
462:
306:
1657:"Global DNA methylation and transcriptional analyses of human ESC-derived cardiomyocytes"
2233:
1758:
1532:
1191:"Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization"
2941:
2916:
2892:
2867:
2840:
2813:
2627:
2602:
2523:
2498:
2474:
2449:
2425:
2400:
2351:
2324:
2252:
2217:
2193:
2168:
1844:
1777:
1742:
1724:
1699:
1681:
1656:
1637:
1628:
1612:
1594:
1569:
1551:
1516:
1498:
1473:
1418:
1391:
1373:
1348:
1260:
1233:
1215:
1190:
107:
78:
41:
2964:
2692:
2667:
2136:
2119:
1943:
1159:(2014). "A network approach to clinical intervention in neurodegenerative diseases".
2341:
2300:
2283:
2153:
2077:
2060:
2045:
1464:
1339:
1304:
537:
808:
2242:
1767:
1541:
1517:"A Computational Framework to Infer Human Disease-Associated Long Noncoding RNAs"
1172:
1863:
424:
340:
118:
95:
1838:
2883:
2830:
1995:
A. Bauer-Mehren, "Gene-Disease network Analysis Reveals Functional Modules in
1834:
1672:
1570:"Choline Protects Against Cardiac Hypertrophy Induced by Increased After-load"
1448:
1408:
1250:
927:
174:
98:
derived associations including Mendelian, complex and environmental diseases.
168:
need to choose, even after careful statistical data analysis, which genes or
2932:
1996:
1938:
1321:
1286:
952:). The GAD was frozen as of 09/01/2014 but is still available for download.
439:
130:
122:
87:
51:
2950:
2901:
2849:
2758:
2701:
2636:
2532:
2483:
2434:
2360:
2309:
2261:
2202:
2145:
2086:
2037:
1786:
1733:
1690:
1646:
1603:
1560:
1515:
Liu, Ming-Xi; Chen, Xing; Chen, Geng; Cui, Qing-Hua; Yan, Gui-Ying (2014).
1507:
1456:
1427:
1382:
1269:
1224:
1180:
1068:
769:
653:
2683:
2618:
2514:
1206:
2465:
2416:
2184:
2000:
1715:
1364:
1120:
1016:
731:
598:
556:
291:
273:
117:
Broadly speaking, genetic diseases are caused by aberrations in genes or
114:
91:
2776:
2104:
1347:
Mannil, Deepthi; Vogt, Ingo; Prinz, Jeanette; Campillos, Monica (2015).
2749:
2732:
1489:
922:
472:
269:
236:
169:
1585:
129:. In other occurrences, the same disease, for instance, some forms of
240:
134:
1132:
785:
2499:"Ongoing and future developments at the Universal Protein Resource"
2029:
400:
1798:
1131:
807:
536:
346:
330:
Ongoing and future developments at the Universal Protein Resource
1815:
705:
449:
444:
1814:
include the identification of candidate genes and nucleotides,
380:
434:
358:
141:
environmental factors. Examples of genetic phenotypes include
126:
2917:"Predicting disease genes using protein-protein interactions"
272:
data created by combining the Swiss-Prot, TrEMBL and PIR-PSD
1040:
Research Programme on Biomedical Informatics (GRIB) IMIM-UPF
1084:
726:
OMIM is a compendium of human genes and genetic phenotypes.
2103:. American Medical Informatics Association. Archived from
907:
2545:
K. Brown, "Online Predicted human Interaction Database,"
2101:"American Medical Informatics Association Strategic Plan"
412:
370:
2601:
Shimoyama M, De Pons J, Hayman GT, et al. (2015).
1390:
Vogt, Ingo; Prinz, Jeanette; Campillos, Mónica (2014).
2169:"The Comparative Toxicogenomics Database: update 2011"
669:
1056:
Ferran Sanz and Laura I. Furlong (Pinero et al, 2015)
956:
Literature-derived human gene-disease network (LHGDN)
1234:"Network medicine analysis of COPD multimorbidities"
1110:
1100:
1095:
1079:
1074:
1060:
1052:
1044:
1034:
1029:
1015:
1005:
997:
992:
902:
897:
885:
862:
857:
847:
842:
780:
775:
761:
749:
744:
730:
722:
717:
680:
664:
659:
645:
637:
627:
615:
610:
597:
586:
581:
525:
517:
507:
497:
484:
479:
461:
456:
423:
395:
365:
339:
334:
326:
305:
300:
290:
280:
257:
252:
67:
57:
47:
37:
1795:Remarks about the future in Gene Disease Databases
531:Yes – both individual protein entries and searches
164:At different stages of any gene disease project,
542:The process of database compilation and curation
1914:International Society for Computational Biology
983:The Gene Disease Associations Database DisGeNET
158:Gene prioritization workflow of human diseases
202:The Comparative Toxicogenomics Database (CTD)
8:
2061:"Databases, data tombs and dust in the wind"
1991:
1989:
1987:
1985:
1743:"Charting the NF-κB Pathway Interactome Map"
1574:International Journal of Biological Sciences
987:
828:
712:
641:Mary E. Shimoyama, PhD; Howard J. Jacob, PhD
576:
247:
32:
2277:
2275:
2273:
2271:
2011:
2009:
1983:
1981:
1979:
1977:
1975:
1973:
1971:
1969:
1967:
1965:
1934:List of open-source bioinformatics software
1137:DisGeNET gene-disease association ontology
2583:
2581:
2450:"The Universal Protein Resource (UniProt)"
2372:
2370:
1001:Integrates human gene-disease associations
986:
827:
711:
575:
246:
106:Experts in different areas of biology and
2940:
2891:
2861:
2859:
2839:
2829:
2748:
2717:P. Flicek and M. Ridwan, "Ensembl 2012,"
2713:
2711:
2691:
2648:
2646:
2626:
2522:
2473:
2424:
2394:
2392:
2377:Buneman, P. (2008). "Curated Databases".
2350:
2340:
2299:
2251:
2241:
2192:
2135:
2076:
1776:
1766:
1723:
1680:
1636:
1593:
1550:
1540:
1497:
1417:
1407:
1372:
1259:
1249:
1214:
704:The Online Mendelian Inheritance in Man (
410:& for downloading complete data sets
2120:"The modular nature of genetic diseases"
1048:Integrative Biomedical Informatics Group
231:The Universal Protein Resource (UNIPROT)
152:
23:For broader coverage of this topic, see
2666:Hubbard T, et al. (January 2002).
2282:Bauer-Mehren, A.; Rautscha, M. (2010).
1961:
713:The Online Mendelian Inheritance in Man
2797:: CS1 maint: archived copy as title (
2790:
2549:, vol. 21, no. 9, pp. 2076-2082, 2005.
568:Ontology and disease names from OMIM.
475:O, bulk retrieval/download, ID mapping
31:
2668:"The Ensembl genome database project"
813:Pathway Hogeneity vs Associated Genes
7:
829:The Ensembl genome database project.
2656:, vol. 33, no. 1, pp. 514-517, 2005
2591:, vol. 37, no. 1, pp. 744-749, 2009
2575:, vol. 36, no. 1, pp. 724-728, 2007
632:Human Molecular and Genetics Center
208:Comparative Toxicogenomics Database
2733:"The genetic Association Database"
1629:10.1152/physiolgenomics.00013.2013
943:Genetic Association Database (GAD)
268:resource, a central repository of
16:For bioinformatics databases, see
14:
2721:, vol. 40, no. 1, pp. 84-90, 2012
2562:, vol. 10, no. 1, pp. 12-22, 2011
1889:European Bioinformatics Institute
877:European Bioinformatics Institute
2137:10.1111/j.1399-0004.2006.00708.x
1837:
834:
235:The Universal Protein Resource (
2731:Becker, K.; Barnes, K. (2004).
2216:Davis, A.; Wiegers, T. (2013).
1924:List of bioinformatics journals
872:Wellcome Trust Sanger Institute
563:The Mouse genome Database (MGD)
226:Chemical-phenotype associations
2812:Bravo, A; et al. (2014).
2399:Murphy, C.; Davis, A. (2009).
1804:Relationships in Gene Diseases
1:
2509:(Database issue): D214–D219.
2342:10.1093/bioinformatics/btu487
2301:10.1093/bioinformatics/btq538
2078:10.1093/bioinformatics/btn464
1359:(Database issue): D900–D906.
572:The Rat Genome Database (RGD)
220:Chemical-disease associations
2866:Piñero; et al. (2015).
2448:Uniprot, Consortium (2008).
2243:10.1371/journal.pone.0058201
2167:Davis, A.; King, B. (2011).
1999:, Complex and Environmental
1929:List of biological databases
1768:10.1371/journal.pone.0032678
1542:10.1371/journal.pone.0084408
1173:10.1016/j.molmed.2014.10.002
1161:Trends in Molecular Medicine
622:Medical College of Wisconsin
18:List of biological databases
2613:(Database issue): D743–50.
2059:Wren JD, Bateman A (2008).
2003:," PLOS One, pp. 1-3, 2011.
1884:Disease gene identification
2987:
1909:Integrative bioinformatics
217:Chemical-gene interactions
22:
15:
2831:10.1186/s12859-015-0472-9
1949:Structural bioinformatics
1874:Computational biomodeling
1673:10.1007/s13238-013-0016-x
1449:10.1016/j.nbd.2014.03.019
1409:10.1186/s13073-014-0052-z
1251:10.1186/s12931-014-0111-4
833:
223:Gene-disease associations
1859:Bioinformatics companies
1854:Biodiversity informatics
149:Challenges with creation
2933:10.1136/jmg.2006.041376
2884:10.1093/database/bav028
1437:Neurobiology of Disease
1322:10.1145/2649387.2660817
1287:10.1145/2649387.2660847
2719:Nucleic Acids Research
2672:Nucleic Acids Research
2654:Nucleic Acids Research
2607:Nucleic Acids Research
2589:Nucleic Acids Research
2573:Nucleic Acids Research
2560:Nucleic Acids Research
2503:Nucleic Acids Research
2454:Nucleic Acids Research
1879:Computational genomics
1806:
1704:Nucleic Acids Research
1617:Physiological Genomics
1353:Nucleic Acids Research
1195:Nucleic Acids Research
1142:
820:
546:
161:
1869:Computational biology
1802:
1135:
1011:Associations Database
974:Integrative databases
811:
540:
156:
83:Gene Disease Database
33:Gene Disease Database
2497:Uniprot, C. (2010).
1904:Human Genome Project
1316:. pp. 647–648.
1281:. pp. 768–773.
1238:Respiratory Research
1157:Potashkin, Judith A.
934:Literature databases
550:Predictive databases
493:Attribution-NoDerivs
166:molecular biologists
68:Subtype of Databases
2779:on 24 February 2021
2684:10.1093/nar/30.1.38
2619:10.1093/nar/gku1026
2515:10.1093/nar/gkq1020
2234:2013PLoSO...858201D
2107:on 26 October 2009.
1894:Functional genomics
1759:2012PLoSO...732678T
1533:2014PLoSO...984408L
1207:10.1093/nar/gku1036
1201:(20): 12570–12584.
1155:Santiago, Jose A.;
989:
830:
714:
694:Rat Genome Database
592:Rat Genome Database
578:
249:
143:Alzheimer's disease
34:
25:Biological database
2971:Genetics databases
2818:BMC Bioinformatics
2750:10.1038/ng0504-431
2466:10.1093/nar/gkm895
2417:10.1093/nar/gkn580
2185:10.1093/nar/gkq813
1899:Health informatics
1827:medical research.
1807:
1716:10.1093/nar/gkt989
1661:Protein & Cell
1490:10.1002/humu.22520
1365:10.1093/nar/gku948
1143:
821:
547:
345:Custom flat file,
286:Protein annotation
185:Types of databases
162:
125:or changes to the
2335:(21): 3093–3100.
2323:Vogt, I. (2014).
2294:(22): 2924–2926.
2173:Nucleic Acids Res
2124:Clinical Genetics
1586:10.7150/ijbs.5976
1118:
1117:
918:
917:
824:Ensembl SIFT tool
802:
801:
690:
689:
604:Rattus norvegicus
535:
534:
467:Advanced search,
266:universal protein
193:Curated databases
75:
74:
58:Type of Databases
48:Subclassification
2978:
2955:
2954:
2944:
2912:
2906:
2905:
2895:
2863:
2854:
2853:
2843:
2833:
2809:
2803:
2802:
2796:
2788:
2786:
2784:
2775:. Archived from
2769:
2763:
2762:
2752:
2728:
2722:
2715:
2706:
2705:
2695:
2663:
2657:
2650:
2641:
2640:
2630:
2598:
2592:
2585:
2576:
2569:
2563:
2556:
2550:
2543:
2537:
2536:
2526:
2494:
2488:
2487:
2477:
2445:
2439:
2438:
2428:
2396:
2387:
2386:
2374:
2365:
2364:
2354:
2344:
2320:
2314:
2313:
2303:
2279:
2266:
2265:
2255:
2245:
2213:
2207:
2206:
2196:
2179:(1): 1067–1072.
2164:
2158:
2157:
2139:
2115:
2109:
2108:
2097:
2091:
2090:
2080:
2056:
2050:
2049:
2013:
2004:
1993:
1847:
1842:
1841:
1790:
1780:
1770:
1737:
1727:
1710:(3): 1474–1496.
1694:
1684:
1650:
1640:
1607:
1597:
1564:
1554:
1544:
1511:
1501:
1468:
1431:
1421:
1411:
1386:
1376:
1343:
1308:
1273:
1263:
1253:
1228:
1218:
1184:
1091:
1088:
1086:
1061:Primary citation
990:
914:
911:
909:
886:Primary citation
838:
831:
798:
795:
793:
791:
789:
787:
762:Primary citation
715:
685:RGD Data Release
676:
673:
671:
646:Primary citation
579:
491:Creative Commons
452:
442:
419:
416:
414:
409:
406:
404:
402:
391:
388:
386:
384:
382:
377:
374:
372:
327:Primary citation
250:
35:
2986:
2985:
2981:
2980:
2979:
2977:
2976:
2975:
2961:
2960:
2959:
2958:
2915:Oti, M (2006).
2914:
2913:
2909:
2865:
2864:
2857:
2811:
2810:
2806:
2789:
2782:
2780:
2773:"Archived copy"
2771:
2770:
2766:
2737:Nature Genetics
2730:
2729:
2725:
2716:
2709:
2665:
2664:
2660:
2651:
2644:
2600:
2599:
2595:
2586:
2579:
2570:
2566:
2557:
2553:
2544:
2540:
2496:
2495:
2491:
2447:
2446:
2442:
2398:
2397:
2390:
2376:
2375:
2368:
2322:
2321:
2317:
2281:
2280:
2269:
2215:
2214:
2210:
2166:
2165:
2161:
2118:Oti, M (2007).
2117:
2116:
2112:
2099:
2098:
2094:
2058:
2057:
2053:
2018:Nature Genetics
2015:
2014:
2007:
1994:
1963:
1958:
1953:
1919:Jumping library
1843:
1836:
1833:
1797:
1740:
1697:
1653:
1623:(14): 597–605.
1610:
1567:
1514:
1471:
1434:
1396:Genome Medicine
1389:
1346:
1332:
1311:
1297:
1276:
1231:
1187:
1167:(12): 694–703.
1154:
1148:
1139:
1102:
1083:
1036:Research center
1007:
985:
976:
967:
958:
945:
936:
906:
881:
864:Research center
826:
784:
751:Research center
710:
668:
617:Research center
574:
565:
552:
544:
527:
518:Curation policy
509:
448:
438:
411:
399:
379:
378:
369:
318:, Switzerland;
307:Research center
282:
233:
204:
195:
187:
151:
104:
28:
21:
12:
11:
5:
2984:
2982:
2974:
2973:
2963:
2962:
2957:
2956:
2927:(8): 691–698.
2907:
2855:
2804:
2764:
2743:(5): 431–432.
2723:
2707:
2658:
2642:
2593:
2577:
2564:
2551:
2547:Bioinformatics
2538:
2489:
2460:(1): 190–195.
2440:
2411:(1): 786–792.
2405:Bioinformatics
2388:
2366:
2329:Bioinformatics
2315:
2288:Bioinformatics
2267:
2208:
2159:
2110:
2092:
2071:(19): 2127–8.
2065:Bioinformatics
2051:
2030:10.1038/ng1090
2024:(1): 228–237.
2005:
1960:
1959:
1957:
1954:
1952:
1951:
1946:
1941:
1936:
1931:
1926:
1921:
1916:
1911:
1906:
1901:
1896:
1891:
1886:
1881:
1876:
1871:
1866:
1861:
1856:
1850:
1849:
1848:
1845:Biology portal
1832:
1829:
1796:
1793:
1792:
1791:
1738:
1695:
1651:
1608:
1580:(3): 295–302.
1565:
1512:
1484:(5): 537–547.
1478:Human Mutation
1469:
1432:
1387:
1344:
1330:
1309:
1295:
1274:
1229:
1185:
1147:
1146:Some use cases
1144:
1116:
1115:
1112:
1108:
1107:
1104:
1098:
1097:
1093:
1092:
1081:
1077:
1076:
1072:
1071:
1062:
1058:
1057:
1054:
1050:
1049:
1046:
1042:
1041:
1038:
1032:
1031:
1027:
1026:
1019:
1013:
1012:
1009:
1003:
1002:
999:
995:
994:
984:
981:
975:
972:
966:
963:
957:
954:
944:
941:
935:
932:
930:and genomics.
916:
915:
904:
900:
899:
895:
894:
887:
883:
882:
880:
879:
874:
868:
866:
860:
859:
855:
854:
849:
845:
844:
840:
839:
825:
822:
800:
799:
782:
778:
777:
773:
772:
763:
759:
758:
753:
747:
746:
742:
741:
734:
728:
727:
724:
720:
719:
709:
702:
688:
687:
682:
678:
677:
666:
662:
661:
657:
656:
647:
643:
642:
639:
635:
634:
629:
625:
624:
619:
613:
612:
608:
607:
601:
595:
594:
588:
584:
583:
573:
570:
564:
561:
551:
548:
533:
532:
529:
523:
522:
519:
515:
514:
511:
505:
504:
501:
495:
494:
488:
482:
481:
477:
476:
465:
459:
458:
454:
453:
428:
421:
420:
397:
393:
392:
367:
363:
362:
343:
337:
336:
332:
331:
328:
324:
323:
309:
303:
302:
298:
297:
294:
288:
287:
284:
278:
277:
259:
255:
254:
232:
229:
228:
227:
224:
221:
218:
203:
200:
194:
191:
186:
183:
150:
147:
108:bioinformatics
103:
100:
79:bioinformatics
73:
72:
69:
65:
64:
59:
55:
54:
49:
45:
44:
42:Bioinformatics
39:
38:Classification
13:
10:
9:
6:
4:
3:
2:
2983:
2972:
2969:
2968:
2966:
2952:
2948:
2943:
2938:
2934:
2930:
2926:
2922:
2921:J. Med. Genet
2918:
2911:
2908:
2903:
2899:
2894:
2889:
2885:
2881:
2877:
2873:
2869:
2862:
2860:
2856:
2851:
2847:
2842:
2837:
2832:
2827:
2823:
2819:
2815:
2808:
2805:
2800:
2794:
2778:
2774:
2768:
2765:
2760:
2756:
2751:
2746:
2742:
2738:
2734:
2727:
2724:
2720:
2714:
2712:
2708:
2703:
2699:
2694:
2689:
2685:
2681:
2677:
2673:
2669:
2662:
2659:
2655:
2649:
2647:
2643:
2638:
2634:
2629:
2624:
2620:
2616:
2612:
2608:
2604:
2597:
2594:
2590:
2584:
2582:
2578:
2574:
2568:
2565:
2561:
2555:
2552:
2548:
2542:
2539:
2534:
2530:
2525:
2520:
2516:
2512:
2508:
2504:
2500:
2493:
2490:
2485:
2481:
2476:
2471:
2467:
2463:
2459:
2455:
2451:
2444:
2441:
2436:
2432:
2427:
2422:
2418:
2414:
2410:
2406:
2402:
2395:
2393:
2389:
2385:(1): 152–162.
2384:
2380:
2379:Bibliometrics
2373:
2371:
2367:
2362:
2358:
2353:
2348:
2343:
2338:
2334:
2330:
2326:
2319:
2316:
2311:
2307:
2302:
2297:
2293:
2289:
2285:
2278:
2276:
2274:
2272:
2268:
2263:
2259:
2254:
2249:
2244:
2239:
2235:
2231:
2227:
2223:
2219:
2212:
2209:
2204:
2200:
2195:
2190:
2186:
2182:
2178:
2174:
2170:
2163:
2160:
2155:
2151:
2147:
2143:
2138:
2133:
2129:
2125:
2121:
2114:
2111:
2106:
2102:
2096:
2093:
2088:
2084:
2079:
2074:
2070:
2066:
2062:
2055:
2052:
2047:
2043:
2039:
2035:
2031:
2027:
2023:
2019:
2012:
2010:
2006:
2002:
1998:
1992:
1990:
1988:
1986:
1984:
1982:
1980:
1978:
1976:
1974:
1972:
1970:
1968:
1966:
1962:
1955:
1950:
1947:
1945:
1944:Phylogenetics
1942:
1940:
1937:
1935:
1932:
1930:
1927:
1925:
1922:
1920:
1917:
1915:
1912:
1910:
1907:
1905:
1902:
1900:
1897:
1895:
1892:
1890:
1887:
1885:
1882:
1880:
1877:
1875:
1872:
1870:
1867:
1865:
1862:
1860:
1857:
1855:
1852:
1851:
1846:
1840:
1835:
1830:
1828:
1825:
1819:
1817:
1811:
1805:
1801:
1794:
1788:
1784:
1779:
1774:
1769:
1764:
1760:
1756:
1753:(3): e32678.
1752:
1748:
1744:
1739:
1735:
1731:
1726:
1721:
1717:
1713:
1709:
1705:
1701:
1696:
1692:
1688:
1683:
1678:
1674:
1670:
1666:
1662:
1658:
1652:
1648:
1644:
1639:
1634:
1630:
1626:
1622:
1618:
1614:
1609:
1605:
1601:
1596:
1591:
1587:
1583:
1579:
1575:
1571:
1566:
1562:
1558:
1553:
1548:
1543:
1538:
1534:
1530:
1527:(1): e84408.
1526:
1522:
1518:
1513:
1509:
1505:
1500:
1495:
1491:
1487:
1483:
1479:
1475:
1470:
1466:
1462:
1458:
1454:
1450:
1446:
1442:
1438:
1433:
1429:
1425:
1420:
1415:
1410:
1405:
1401:
1397:
1393:
1388:
1384:
1380:
1375:
1370:
1366:
1362:
1358:
1354:
1350:
1345:
1341:
1337:
1333:
1331:9781450328944
1327:
1323:
1319:
1315:
1310:
1306:
1302:
1298:
1296:9781450328944
1292:
1288:
1284:
1280:
1275:
1271:
1267:
1262:
1257:
1252:
1247:
1243:
1239:
1235:
1230:
1226:
1222:
1217:
1212:
1208:
1204:
1200:
1196:
1192:
1186:
1182:
1178:
1174:
1170:
1166:
1162:
1158:
1153:
1152:
1151:
1145:
1138:
1134:
1130:
1127:
1122:
1113:
1109:
1105:
1099:
1096:Miscellaneous
1094:
1090:
1082:
1078:
1073:
1070:
1066:
1063:
1059:
1055:
1051:
1047:
1043:
1039:
1037:
1033:
1028:
1024:
1020:
1018:
1014:
1010:
1004:
1000:
996:
991:
982:
980:
973:
971:
964:
962:
955:
953:
951:
942:
940:
933:
931:
929:
924:
913:
905:
901:
896:
892:
888:
884:
878:
875:
873:
870:
869:
867:
865:
861:
856:
853:
850:
846:
841:
837:
832:
823:
819:
814:
810:
806:
797:
783:
779:
774:
771:
767:
764:
760:
757:
754:
752:
748:
743:
739:
735:
733:
729:
725:
721:
716:
707:
703:
701:
697:
695:
686:
683:
679:
675:
667:
663:
658:
655:
651:
648:
644:
640:
636:
633:
630:
626:
623:
620:
618:
614:
609:
605:
602:
600:
596:
593:
589:
585:
580:
571:
569:
562:
560:
558:
549:
543:
539:
530:
524:
520:
516:
512:
506:
502:
500:
496:
492:
489:
487:
483:
480:Miscellaneous
478:
474:
470:
466:
464:
460:
455:
451:
446:
441:
436:
433:
429:
426:
422:
418:
408:
398:
394:
390:
376:
368:
364:
360:
356:
352:
348:
344:
342:
338:
333:
329:
325:
321:
317:
313:
310:
308:
304:
299:
295:
293:
289:
285:
279:
275:
271:
267:
263:
260:
256:
251:
245:
242:
238:
230:
225:
222:
219:
216:
215:
214:
211:
209:
201:
199:
192:
190:
184:
182:
178:
176:
171:
167:
159:
155:
148:
146:
144:
138:
136:
132:
128:
124:
120:
116:
112:
109:
101:
99:
97:
93:
89:
84:
80:
70:
66:
63:
60:
56:
53:
50:
46:
43:
40:
36:
30:
26:
19:
2924:
2920:
2910:
2875:
2871:
2821:
2817:
2807:
2781:. Retrieved
2777:the original
2767:
2740:
2736:
2726:
2718:
2678:(1): 38–41.
2675:
2671:
2661:
2653:
2610:
2606:
2596:
2588:
2572:
2567:
2559:
2554:
2546:
2541:
2506:
2502:
2492:
2457:
2453:
2443:
2408:
2404:
2382:
2378:
2332:
2328:
2318:
2291:
2287:
2225:
2221:
2211:
2176:
2172:
2162:
2127:
2123:
2113:
2105:the original
2095:
2068:
2064:
2054:
2021:
2017:
1823:
1820:
1812:
1808:
1803:
1750:
1746:
1707:
1703:
1667:(1): 59–68.
1664:
1660:
1620:
1616:
1577:
1573:
1524:
1520:
1481:
1477:
1440:
1436:
1399:
1395:
1356:
1352:
1313:
1278:
1241:
1237:
1198:
1194:
1164:
1160:
1149:
1136:
1119:
1101:Data release
1022:
977:
968:
959:
946:
937:
919:
890:
851:
817:
812:
803:
737:
698:
691:
681:Download URL
603:
591:
566:
553:
541:
526:Bookmarkable
508:Data release
396:Download URL
265:
261:
234:
212:
205:
196:
188:
179:
163:
157:
139:
113:
105:
102:Introduction
82:
76:
71:Gene-Disease
29:
2783:18 November
2228:(4): 1–29.
2130:(1): 1–11.
1864:Biomedicine
998:Description
965:BeFree Data
848:Description
723:Description
587:Description
425:Web service
341:Data format
258:Description
119:chromosomes
96:text mining
2878:: bav028.
1956:References
1244:(1): 111.
1045:Laboratory
1023:H. Sapiens
1006:Data types
928:proteomics
738:H. Sapiens
628:Laboratory
499:Versioning
407:/downloads
281:Data types
175:biomedical
62:Biological
2824:(1): 55.
1997:Mendelian
1939:Pathology
1824:In silico
1443:: 84–91.
1402:(7): 52.
1103:frequency
1087:.disgenet
1017:Organisms
889:Hubbard,
732:Organisms
599:Organisms
510:frequency
447:see info
437:see info
292:Organisms
274:databases
131:carcinoma
123:mutations
88:phenotype
52:Databases
2965:Category
2951:16611749
2902:25877637
2872:Database
2850:25886734
2793:cite web
2759:15118671
2702:11752248
2637:25355511
2533:21051339
2484:18045787
2435:18782832
2361:25061072
2310:20861032
2262:23613709
2222:PLOS ONE
2203:20864448
2154:24615025
2146:17204041
2087:18819940
2046:10599219
2038:12610532
2001:diseases
1831:See also
1787:22403694
1747:PLOS ONE
1734:24198249
1691:24474197
1647:23695888
1604:23493786
1561:24392133
1521:PLOS ONE
1508:24478219
1465:41944859
1457:24718034
1428:25276232
1383:25313158
1340:30593231
1305:17123912
1270:25248857
1225:25352550
1181:25455073
1126:clusters
1121:DisGeNET
1069:25877637
1008:captured
988:DisGeNET
910:.ensembl
770:25398906
654:25355511
557:genomics
528:entities
415:.uniprot
403:.uniprot
383:.uniprot
373:.uniprot
312:EMBL-EBI
283:captured
170:proteins
135:melanoma
92:genotype
2942:2564594
2893:4397996
2841:4466840
2628:4383884
2524:3013648
2475:1669721
2426:2686584
2352:4609011
2253:3629079
2230:Bibcode
2194:3013756
1778:3293857
1755:Bibcode
1725:3919568
1682:3938846
1638:3727018
1595:3596715
1552:3879311
1529:Bibcode
1499:4130156
1419:4165361
1374:4384019
1261:4177421
1216:4227755
1111:Version
1080:Website
1053:Authors
1030:Contact
1021:Human (
993:Content
923:Ensembl
903:Website
858:Contact
852:Ensembl
843:Content
781:Website
745:Contact
736:Human (
718:Content
665:Website
638:Authors
611:Contact
582:Content
513:4 weeks
486:License
473:Clustal
366:Website
301:Contact
270:protein
264:is the
262:UniProt
253:Content
248:UniProt
237:UniProt
115:Genetic
2949:
2939:
2900:
2890:
2848:
2838:
2757:
2700:
2690:
2635:
2625:
2531:
2521:
2482:
2472:
2433:
2423:
2359:
2349:
2308:
2260:
2250:
2201:
2191:
2152:
2144:
2085:
2044:
2036:
1785:
1775:
1732:
1722:
1689:
1679:
1645:
1635:
1602:
1592:
1559:
1549:
1506:
1496:
1463:
1455:
1426:
1416:
1381:
1371:
1338:
1328:
1303:
1293:
1268:
1258:
1223:
1213:
1179:
1106:annual
1075:Access
1067:
898:Access
893:(2002)
891:et al.
776:Access
768:
660:Access
652:
443:&
430:Yes –
335:Access
314:, UK;
241:genome
2693:99161
2150:S2CID
2042:S2CID
1461:S2CID
1336:S2CID
1301:S2CID
796:/omim
788:.ncbi
606:(rat)
469:BLAST
457:Tools
387:/news
347:FASTA
322:, US.
2947:PMID
2898:PMID
2876:2015
2846:PMID
2799:link
2785:2016
2755:PMID
2698:PMID
2633:PMID
2529:PMID
2480:PMID
2431:PMID
2357:PMID
2306:PMID
2258:PMID
2199:PMID
2142:PMID
2083:PMID
2034:PMID
1816:SNPs
1783:PMID
1730:PMID
1687:PMID
1643:PMID
1600:PMID
1557:PMID
1504:PMID
1453:PMID
1424:PMID
1379:PMID
1326:ISBN
1291:ISBN
1266:PMID
1221:PMID
1177:PMID
1089:.org
1065:PMID
950:GWAS
912:.org
794:.gov
792:.nih
790:.nlm
766:PMID
756:NCBI
706:OMIM
692:The
674:.edu
672:.mcw
650:PMID
590:The
450:here
445:REST
440:here
432:JAVA
417:.org
405:.org
385:.org
375:.org
206:The
81:, a
2937:PMC
2929:doi
2888:PMC
2880:doi
2836:PMC
2826:doi
2745:doi
2688:PMC
2680:doi
2623:PMC
2615:doi
2519:PMC
2511:doi
2470:PMC
2462:doi
2421:PMC
2413:doi
2383:978
2347:PMC
2337:doi
2296:doi
2248:PMC
2238:doi
2189:PMC
2181:doi
2132:doi
2073:doi
2026:doi
1773:PMC
1763:doi
1720:PMC
1712:doi
1677:PMC
1669:doi
1633:PMC
1625:doi
1590:PMC
1582:doi
1547:PMC
1537:doi
1494:PMC
1486:doi
1445:doi
1414:PMC
1404:doi
1369:PMC
1361:doi
1318:doi
1283:doi
1256:PMC
1246:doi
1211:PMC
1203:doi
1169:doi
1085:www
908:www
786:www
670:rgd
577:RGD
503:Yes
463:Web
435:API
427:URL
413:ftp
401:www
381:www
371:www
359:XML
355:RDF
351:GFF
320:PIR
316:SIB
296:All
133:or
127:DNA
77:In
2967::
2945:.
2935:.
2925:43
2923:.
2919:.
2896:.
2886:.
2874:.
2870:.
2858:^
2844:.
2834:.
2822:16
2820:.
2816:.
2795:}}
2791:{{
2753:.
2741:36
2739:.
2735:.
2710:^
2696:.
2686:.
2676:30
2674:.
2670:.
2645:^
2631:.
2621:.
2611:43
2609:.
2605:.
2580:^
2527:.
2517:.
2507:39
2505:.
2501:.
2478:.
2468:.
2458:36
2456:.
2452:.
2429:.
2419:.
2409:37
2407:.
2403:.
2391:^
2381:.
2369:^
2355:.
2345:.
2333:30
2331:.
2327:.
2304:.
2292:26
2290:.
2286:.
2270:^
2256:.
2246:.
2236:.
2224:.
2220:.
2197:.
2187:.
2177:39
2175:.
2171:.
2148:.
2140:.
2128:71
2126:.
2122:.
2081:.
2069:24
2067:.
2063:.
2040:.
2032:.
2022:33
2020:.
2008:^
1964:^
1781:.
1771:.
1761:.
1749:.
1745:.
1728:.
1718:.
1708:42
1706:.
1702:.
1685:.
1675:.
1663:.
1659:.
1641:.
1631:.
1621:45
1619:.
1615:.
1598:.
1588:.
1576:.
1572:.
1555:.
1545:.
1535:.
1523:.
1519:.
1502:.
1492:.
1482:35
1480:.
1476:.
1459:.
1451:.
1441:72
1439:.
1422:.
1412:.
1398:.
1394:.
1377:.
1367:.
1357:43
1355:.
1351:.
1334:.
1324:.
1299:.
1289:.
1264:.
1254:.
1242:15
1240:.
1236:.
1219:.
1209:.
1199:42
1197:.
1193:.
1175:.
1165:20
1163:.
471:,
357:,
353:,
349:,
2953:.
2931::
2904:.
2882::
2852:.
2828::
2801:)
2787:.
2761:.
2747::
2704:.
2682::
2639:.
2617::
2535:.
2513::
2486:.
2464::
2437:.
2415::
2363:.
2339::
2312:.
2298::
2264:.
2240::
2232::
2226:8
2205:.
2183::
2156:.
2134::
2089:.
2075::
2048:.
2028::
1789:.
1765::
1757::
1751:7
1736:.
1714::
1693:.
1671::
1665:5
1649:.
1627::
1606:.
1584::
1578:9
1563:.
1539::
1531::
1525:9
1510:.
1488::
1467:.
1447::
1430:.
1406::
1400:6
1385:.
1363::
1342:.
1320::
1307:.
1285::
1272:.
1248::
1227:.
1205::
1183:.
1171::
1114:3
1025:)
740:)
708:)
389:/
361:.
276:.
90:-
27:.
20:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.