211:
3665:
409:
322:
622:
647:
series of metallorganic clusters that comprise a long distance; the active site structures remain unchanged during the whole process. In -only hydrogenases, however, electrons are directly delivered to the active site via a short distance. Methenyl-H4MPT, a cofactor, directly accepts the hydride from H
646:
Unlike the other two types, -only hydrogenases are found only in some hydrogenotrophic methanogenic archaea. They also feature a fundamentally different enzymatic mechanism in terms of redox partners and how electrons are delivered to the active site. In and hydrogenases, electrons travel through a
685:
cleavage. The two approaches are complementary and can benefit one another. In fact, Cao and Hall combined both approaches in developing the model that describes how hydrogen molecules are oxidized or produced within the active site of hydrogenases. While more research and experimental data are
293:
Although originally believed to be "metal-free", the -only hydrogenases contain Fe at the active site and no iron-sulfur clusters. and hydrogenases have some common features in their structures: Each enzyme has an active site and a few Fe-S clusters that are buried in protein. The active site,
676:
in different steps of catalysis such as intramolecular transport of substrates. For instance, Cornish et al. conducted mutagenesis studies and found out that four amino acids located along the putative channel connecting the active site and protein surface are critical to enzymatic function of
659:
oxidation/production, which is the case for the other two types of hydrogenases. While the exact mechanism of the catalysis is still under study, recent finding suggests that molecular hydrogen is first heterolytically cleaved by Fe(II), followed by transfer of hydride to the carbocation of the
841:
Despite these findings, research is still under progress for engineering oxygen tolerance in hydrogenases. While researchers have found oxygen-tolerant hydrogenases, they are only efficient in hydrogen uptake and not production. Bingham et al.'s recent success in engineering hydrogenase from
382:
growth media. This finding increased hope that hydrogenases can be used in photosynthetic production of molecular hydrogen via splitting water. Another , called Huc or Hyd1 or cyanobacterial-type uptake hydrogenase, has been found to be oxygen insensitive while having a very high affinity for
474:
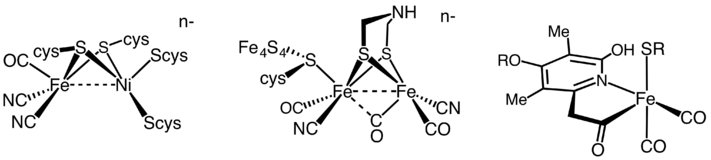
The active site of the diiron hydrogenase is known as the H-cluster. The H-cluster consists of a cubane-shaped structure, coupled to the low valent diiron co-factor by a cysteine derived thiol. The diiron co-factor includes two iron atoms, connected by a bridging aza-dithiolate ligand
824:
first converts into a reactive species at the active site of hydrogenases, and then damages its domain. Cohen et al. investigated how oxygen can reach the active site that is buried inside the protein body by molecular dynamics simulation approach; their results indicate that
709:
Recent studies have revealed other biological functions of hydrogenases. To begin with, bidirectional hydrogenases can also act as "valves" to control excess reducing equivalents, especially in photosynthetic microorganisms. Such a role makes hydrogenases play a vital role in
338:. On the basis of sequence similarity, however, the and hydrogenases should be considered a single superfamily. To date, periplasmic, cytoplasmic, and cytoplasmic membrane-bound hydrogenases have been found. The hydrogenases, when isolated, are found to catalyse both H
2551:
Cao Z, Hall MB (April 2001). "Modeling the active sites in metalloenzymes. 3. Density functional calculations on models for -hydrogenase: structures and vibrational frequencies of the observed redox forms and the reaction mechanism at the Diiron Active Center".
829:
diffuses through mainly two pathways that are formed by enlargement of and interconnection between cavities during dynamic motion. These works, in combination with other reports, suggest that inactivation is governed by two phenomena:
886:
catalyzed by hydrogenase allows for the capture and storage of renewable energy as fuel with use on demand. This can be demonstrated through the chemical storage of electricity obtained from a renewable source (e.g. solar, wind,
722:
uptake can help heavy metal contaminants to be recovered in intoxicated forms. These uptake hydrogenases have been recently discovered in pathogenic bacteria and parasites and are believed to be involved in their virulence.
655:-forming methylenetetrahydromethanopterin (methylene-H4MPT) dehydrogenase, because its function is the reversible reduction of methenyl-H4MPT to methylene-H4MPT. The hydrogenation of a methenyl-H4MPT+ occurs instead of H
594:). A closely related subclass from Group D has a similar location on the bacterial gene and share similar domain structure to a subclass from Group E but it lacks the PAS domain. Within Group D, the -hydrogenase from
714:. Moreover, hydrogenases may also be involved in membrane-linked energy conservation through the generation of a transmembrane protonmotive force.There is a possibility that hydrogenases have been responsible for
462:
In contrast to hydrogenases, hydrogenases are generally more active in production of molecular hydrogen. Turnover frequency (TOF) in the order of 10,000 s have been reported in literature for hydrogenases from
681:(CpI). On the other hand, one can also rely on computational analysis and simulations. Nilsson Lill and Siegbahn have recently taken this approach in investigating the mechanism by which hydrogenases catalyze H
333:
while the large subunit contains the active site, a nickel-iron centre which is connected to the solvent by a molecular tunnel. In some hydrogenases, one of the Ni-bound cysteine residues is replaced by
1457:
Burgdorf T, Lenz O, Buhrke T, van der Linden E, Jones AK, Albracht SP, et al. (2005). "-hydrogenases of
Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation".
686:
required to complete our understanding of the mechanism, these findings have allowed scientists to apply the knowledge in, e.g., building artificial catalysts mimicking active sites of hydrogenases.
668:
The molecular mechanism by which protons are converted into hydrogen molecules within hydrogenases is still under extensive study. One popular approach employs mutagenesis to elucidate roles of
207:
known as hydrogenases. Hydrogenases are sub-classified into three different types based on the active site metal content: iron-iron hydrogenase, nickel-iron hydrogenase, and iron hydrogenase.
2246:
Chongdar N, Birrell JA, Pawlak K, Sommer C, Reijerse EJ, Rüdiger O, et al. (January 2018). "Unique
Spectroscopic Properties of the H-Cluster in a Putative Sensory Hydrogenase".
1363:
Liebgott PP, Leroux F, Burlat B, Dementin S, Baffert C, Lautier T, et al. (January 2010). "Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase".
2847:
Hinnemann B, Moses PG, Bonde J, Jørgensen KP, Nielsen JH, Horch S, et al. (April 2005). "Biomimetic hydrogen evolution: MoS2 nanoparticles as catalyst for hydrogen evolution".
3202:
1505:"An analysis of the changes in soluble hydrogenase and global gene expression in Cupriavidus necator (Ralstonia eutropha) H16 grown in heterotrophic diauxic batch culture"
2330:
Shima S, Pilak O, Vogt S, Schick M, Stagni MS, Meyer-Klaucke W, et al. (July 2008). "The crystal structure of -hydrogenase reveals the geometry of the active site".
3323:
629:
706:
are not available. Based on these grounds, the primary role of hydrogenases are believed to be energy generation, and this can be sufficient to sustain an ecosystem.
2882:
Goris T, Wait AF, Saggu M, Fritsch J, Heidary N, Stein M, et al. (May 2011). "A unique iron-sulfur cluster is crucial for oxygen tolerance of a -hydrogenase".
765:
matrices. Understanding the catalytic mechanism of hydrogenase might help scientists design clean biological energy sources, such as algae, that produce hydrogen.
1822:
Madden C, Vaughn MD, Díez-Pérez I, Brown KA, King PW, Gust D, et al. (January 2012). "Catalytic turnover of -hydrogenase based on single-molecule imaging".
694:
Assuming that the Earth's atmosphere was initially rich in hydrogen, scientists hypothesize that hydrogenases were evolved to generate energy from/as molecular H
2430:
Shima S, Vogt S, Göbels A, Bill E (December 2010). "Iron-chromophore circular dichroism of -hydrogenase: the conformational change required for H2 activation".
781:. Different catalysts require unequal overpotential for this reduction reaction to take place. Hydrogenases are attractive since they require a relatively low
349:
acting as either electron donors or acceptors, depending on their oxidation state. Generally speaking, however, hydrogenases are more active in oxidizing H
2587:
Tard C, Liu X, Ibrahim SK, Bruschi M, De Gioia L, Davies SC, et al. (February 2005). "Synthesis of the H-cluster framework of iron-only hydrogenase".
789:
evolution reaction. Among three different types of hydrogenases, hydrogenases is considered as a strong candidate for an integral part of the solar H
1859:
1719:
2978:"Finding gas diffusion pathways in proteins: application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects"
572:
I). No representative examples of Group B has been characterized yet but it is phylogenetically distinct even when it shares similar amino acid
3195:
1940:
Land H, Senger M, Berggren G, Stripp ST (2020-05-28). "Current State of -Hydrogenase
Research: Biodiversity and Spectroscopic Investigations".
2191:"Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival"
2075:
Glick BR, Martin WG, Martin SM (October 1980). "Purification and properties of the periplasmic hydrogenase from
Desulfovibrio desulfuricans".
3174:
2516:
Lill SO, Siegbahn PE (February 2009). "An autocatalytic mechanism for NiFe-hydrogenase: reduction to Ni(I) followed by oxidative addition".
3328:
2110:
Nakos G, Mortenson L (March 1971). "Purification and properties of hydrogenase, an iron sulfur protein, from
Clostridium pasteurianum W5".
383:
hydrogen. Hydrogen is able to penetrate narrow channels in the enzyme that oxygen molecules cannot enter. This allows bacteria such as
1317:
Jugder BE, Welch J, Aguey-Zinsou KF, Marquis CP (2013-05-14). "Fundamentals and electrochemical applications of -uptake hydrogenases".
702:. Microbial communities driven by molecular hydrogen have, in fact, been found in deep-sea settings where other sources of energy from
698:. Accordingly, hydrogenases can either help microorganisms to proliferate under such conditions, or to set up ecosystems empowered by H
643:
contains neither nickel nor iron-sulfur clusters but an iron-containing cofactor that was recently characterized by X-ray diffraction.
2283:"Characterization of a putative sensory [FeFe]-hydrogenase provides new insight into the role of the active site architecture"
3685:
3384:
3188:
2734:
2717:
2808:"A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain"
3161:
3145:
3129:
3109:
3089:
1610:"Two uptake hydrogenases differentially interact with the aerobic respiratory chain during mycobacterial growth and persistence"
1561:"Production and purification of a soluble hydrogenase from Ralstonia eutropha H16 for potential hydrogen fuel cell applications"
1275:
Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y (October 2007). "Structure/function relationships of - and -hydrogenases".
3282:
3246:
3241:
1042:
329:
The hydrogenases are heterodimeric proteins consisting of small (S) and large (L) subunits. The small subunit contains three
2036:"Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii"
1400:"A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases"
816:
reaction. Past research efforts by various groups around the world have focused on understanding the mechanisms involved in O
590:
HydS) which shows only modest catalytic rates compared to Group A enzymes and an apparent high sensitivity toward hydrogen (H
3540:
777:
and H from incident sunlight. Likewise, numerous catalysts, either chemical or biological, can reduce the produced H into H
2638:
Vignais PM, Billoud B (October 2007). "Occurrence, classification, and biological function of hydrogenases: an overview".
762:
758:
3655:
2383:"The iron-site structure of [Fe]-hydrogenase and model systems: an X-ray absorption near edge spectroscopy study"
1985:"Complex Multimeric [FeFe] Hydrogenases: Biochemistry, Physiology and New Opportunities for the Hydrogen Economy"
294:
which is believed to be the place where catalysis takes place, is also a metallocluster, and each iron is coordinated by
820:-inactivation of hydrogenases. For instance, Stripp et al. relied on protein film electrochemistry and discovered that O
731:
Hydrogenases were first discovered in the 1930s, and they have since attracted interest from many researchers including
17:
533:
576:
around the H-cluster as Group A -hydrogenases. Group C has been classified as "sensory" based on the presence of a
552:
3525:
3641:
3628:
3615:
3602:
3589:
3576:
3563:
3343:
3318:
3310:
3292:
3274:
3264:
3256:
3228:
1087:
992:
524:
374:, and several other so-called Knallgas-bacteria, were found to be oxygen-tolerant. The soluble hydrogenase from
3535:
1148:
5,10-methenyltetrahydromethanopterin hydrogenase (hydrogen:5,10-methenyltetrahydromethanopterin oxidoreductase)
389:
to utilize the small amount of hydrogen in the atmosphere as a source of energy when other sources are lacking.
3489:
3432:
3219:
940:
928:
546:
522:. Group A comprises the best characterized and catalytically most active enzymes such as the -hydrogenase from
455:
3437:
385:
3351:
3300:
3236:
3017:
Bingham AS, Smith PR, Swartz JR (2012). "Evolution of an hydrogenase with decreased oxygen sensitivity".
1064:
949:
859:
417:
3180:
785:. In fact, its catalytic activity is more effective than platinum, which is the best known catalyst for H
3458:
3377:
2767:"Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture"
907:
This is one solution to the challenge in the development of technologies for the capture and storage of
454:
evolution. The ferredoxin functions as natural electron donor linking the enzyme to the photosynthetic
3530:
2930:
2596:
2339:
2202:
1778:
1731:
1672:
1411:
1326:
507:
491:
3494:
1857:
Smith PR, Bingham AS, Swartz JR (2012). "Generation of hydrogen from NADPH using an hydrogenase".
883:
732:
582:
515:
330:
467:. This has led to intense research focusing on use of hydrogenase for sustainable production of H
3427:
2917:
Stripp ST, Goldet G, Brandmayr C, Sanganas O, Vincent KA, Haumann M, et al. (October 2009).
2747:
2698:
2620:
2363:
1965:
1747:
1482:
741:
370:
125:
2381:
Salomone-Stagni M, Stellato F, Whaley CM, Vogt S, Morante S, Shima S, et al. (March 2010).
2281:
Land H, Sekretareva A, Huang P, Redman HJ, Németh B, Polidori N, et al. (September 2020).
502:
as a redox partner while bifurcating types perform the same reaction using both ferredoxin and
3695:
3117:
3097:
3066:
2999:
2958:
2899:
2864:
2829:
2788:
2739:
2690:
2655:
2612:
2569:
2533:
2498:
2447:
2412:
2355:
2312:
2263:
2228:
2171:
2127:
2092:
2057:
2016:
1957:
1919:
1839:
1804:
1700:
1641:
1590:
1536:
1474:
1439:
1380:
1342:
1292:
1257:
1219:
736:
511:
58:
1765:
Berggren G, Adamska A, Lambertz C, Simmons TR, Esselborn J, Atta M, et al. (July 2013).
3690:
3473:
3468:
3442:
3370:
3058:
3026:
2989:
2948:
2938:
2891:
2856:
2819:
2778:
2729:
2682:
2647:
2604:
2561:
2525:
2488:
2478:
2439:
2402:
2394:
2347:
2302:
2294:
2255:
2218:
2210:
2189:
Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. (March 2016).
2161:
2119:
2084:
2047:
2006:
1996:
1949:
1909:
1899:
1868:
1831:
1794:
1786:
1739:
1690:
1680:
1659:
Grinter R, Kropp A, Venugopal H, Senger M, Badley J, Cabotaje PR, et al. (March 2023).
1631:
1621:
1580:
1572:
1526:
1516:
1466:
1429:
1419:
1372:
1334:
1284:
1249:
1209:
573:
364:
Like hydrogenases, hydrogenases are known to be usually deactivated by molecular oxygen (O
2673:
Adams MW, Stiefel EI (December 1998). "Biological hydrogen production: not so elementary".
3520:
3504:
3417:
3333:
3165:
3149:
3133:
3113:
3093:
2467:"Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum"
1237:
863:
813:
495:
295:
2150:"The surprising diversity of clostridial hydrogenases: a comparative genomic perspective"
1608:
Cordero PR, Grinter R, Hards K, Cryle MJ, Warr CG, Cook GM, et al. (December 2019).
2934:
2600:
2343:
2206:
1782:
1735:
1695:
1676:
1660:
1415:
1330:
796:
Low overpotential and high catalytic activity of hydrogenases are accompanied by high O
3669:
3558:
3499:
3211:
2953:
2918:
2493:
2466:
2407:
2382:
2307:
2282:
2223:
2190:
2052:
2035:
2011:
1984:
1914:
1887:
1799:
1766:
1636:
1609:
1585:
1560:
1531:
1504:
1434:
1399:
1214:
1197:
715:
703:
519:
335:
141:
88:
1743:
793:
production system since they offer an additional advantage of high TOF (over 9000 s).
3679:
3463:
3422:
3158:
3142:
3126:
3106:
3086:
2367:
2123:
1969:
1751:
782:
487:
204:
2751:
2702:
919:
catalysts minus the catalyst poisoning, and thus is very efficient. In the case of H
210:
3412:
3030:
2624:
1872:
1486:
888:
506:
as electron donor or acceptor. In order to conserve energy, anaerobic bacteria use
2686:
1156:+ 5,10-methenyltetrahydromethanopterin ⇌ H + 5,10-methylenetetrahydromethanopterin
342:
evolution and uptake, with low-potential multihaem cytochromes such as cytochrome
3636:
3571:
3407:
2783:
2766:
754:
424:
cytoplasmic, soluble, monomeric hydrogenases, found in strict anaerobes such as
379:
3664:
2923:
Proceedings of the
National Academy of Sciences of the United States of America
1685:
1404:
Proceedings of the
National Academy of Sciences of the United States of America
408:
321:
2994:
2977:
2919:"How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms"
1576:
1521:
711:
669:
637:
577:
499:
167:
166:
fermentation). Both low-molecular weight compounds and proteins such as FNRs,
159:
3100:
Structure of the
Apoenzyme of the Iron-sulphur cluster-free hydrogenase from
2001:
1961:
1953:
1626:
1346:
214:
The structures of the active sites of the three types of hydrogenase enzymes.
3610:
3584:
2943:
2483:
2351:
1661:"Structural basis for bacterial energy extraction from atmospheric hydrogen"
1424:
908:
831:
621:
32:
3070:
3003:
2962:
2903:
2868:
2833:
2824:
2807:
2743:
2659:
2616:
2573:
2537:
2502:
2451:
2443:
2416:
2359:
2316:
2267:
2232:
2214:
2175:
2166:
2149:
2020:
1923:
1843:
1808:
1704:
1645:
1594:
1540:
1478:
1443:
1384:
1296:
1261:
1223:
1179:+ 2-(2,3-dihydropentaprenyloxy)phenazine ⇌ 2-dihydropentaprenyloxyphenazine
866:
at either both cathode and anode or at one electrode. In hydrogenase-based
2792:
2694:
2131:
2096:
2061:
483:
S-, adt), the iron atoms are coordinated by carbonyl and cyanide ligands.
420:
are called hydrogenases. Three families of hydrogenases are recognized:
3215:
2895:
2259:
1376:
916:
192:
163:
149:
40:
2735:
10.1002/1439-7633(20020301)3:2/3<153::AID-CBIC153>3.0.CO;2-B
2608:
1790:
1503:
Jugder BE, Chen Z, Ping DT, Lebhar H, Welch J, Marquis CP (March 2015).
1162:
1142:
1119:
1081:
633:
183:
can act as physiological electron donors or acceptors for hydrogenases.
3049:
Lubitz W, Ogata H, Rüdiger O, Reijerse E (April 2014). "Hydrogenases".
2298:
1904:
1338:
1058:
1036:
986:
966:
943:
911:
energy as fuel with use on demand. The generation of electricity from H
867:
846:
was also limited to retained activity (during exposure to oxygen) for H
640:
299:
137:
133:
3062:
2860:
2651:
2565:
2529:
1835:
1470:
1288:
1253:
494:-hydrogenases. In nature, prototypical -hydrogenases perform hydrogen
232:), the hydrogenases catalyze the reversible heterolytic cleavage of H
3623:
3393:
2398:
1720:"Soil bacteria enzyme generates electricity from hydrogen in the air"
838:
to the active site, and destructive modification of the active site.
673:
446:
soluble, monomeric hydrogenases, found in chloroplasts of green alga
162:(FNR), and serves to dispose excess electrons in cells (essential in
129:
28:
2088:
2465:
Cornish AJ, Gärtner K, Yang H, Peters JW, Hegg EL (November 2011).
1888:"Monitoring H-cluster assembly using a semi-synthetic HydF protein"
1767:"Biomimetic assembly and activation of [FeFe]-hydrogenases"
753:
oxidation and is relatively oxygen-tolerant. It can be produced on
3597:
620:
407:
320:
222:
uptake. The and hydrogenases are true redox catalysts, driving H
209:
36:
2976:
Cohen J, Kim K, King P, Seibert M, Schulten K (September 2005).
503:
3366:
3184:
1559:
Jugder BE, Lebhar H, Aguey-Zinsou KF, Marquis CP (2016-01-01).
1398:
Greening C, Berney M, Hards K, Cook GM, Conrad R (March 2014).
1240:, Ogata H, Rüdiger O, Reijerse E (April 2014). "Hydrogenases".
895:
during periods of low energy demands. When energy is desired, H
199:. Most of these species are microbes and their ability to use H
972:
hydrogen dehydrogenase (NADP) (hydrogen:NADPH oxidoreductase)
416:
The hydrogenases containing a di-iron center with a bridging
3362:
1125:
hydrogenase (acceptor) (hydrogen:acceptor oxidoreductase)
870:
cells, hydrogenase enzymes are present at the anode for H
158:) is coupled to the oxidation of electron donors such as
2148:
Calusinska M, Happe T, Joris B, Wilmotte A (June 2010).
191:
It has been estimated that 99% of all organisms utilize
2143:
2141:
718:
of chlorinated compounds. Hydrogenases proficient in H
1886:
Németh B, Esmieu C, Redman HJ, Berggren G (May 2019).
3653:
773:
Various systems are capable of splitting water into O
927:
fuel cells, where the product is water, there is no
651:
in the process. -only hydrogenase is also known as H
3549:
3513:
3482:
3451:
3400:
3342:
3309:
3291:
3273:
3255:
3227:
1983:Schuchmann K, Chowdhury NP, Müller V (2018-12-04).
1459:
Journal of
Molecular Microbiology and Biotechnology
486:-hydrogenases can be separated into four distinct
490:groups A−D. Group A consists of prototypical and
3324:5,10-Methenyltetrahydromethanopterin hydrogenase
2112:Biochimica et Biophysica Acta (BBA) - Enzymology
1554:
1552:
1550:
915:is comparable with the similar functionality of
630:5,10-methenyltetrahydromethanopterin hydrogenase
580:. One example of a Group C -hydrogenase is from
1075:+ oxidized ferredoxin ⇌ 2H + reduced ferredoxin
800:sensitivity. It is necessary to engineer them O
1358:
1356:
1198:"Classification and phylogeny of hydrogenases"
1196:Vignais PM, Billoud B, Meyer J (August 2001).
218:Hydrogenases catalyze, sometimes reversibly, H
203:as a metabolite arises from the expression of
3378:
3196:
2806:Florin L, Tsokoglou A, Happe T (March 2001).
1312:
1310:
1308:
1306:
439:periplasmic, heterodimeric hydrogenases from
226:oxidation and proton (H) reduction (equation
8:
2718:"Hydrogenases: hydrogen-activating enzymes"
1498:
1496:
3385:
3371:
3363:
3203:
3189:
3181:
749:-based biofuel application as it favours H
518:redox reactions are coupled to circumvent
3044:
3042:
3040:
2993:
2952:
2942:
2823:
2782:
2733:
2492:
2482:
2406:
2306:
2222:
2165:
2051:
2010:
2000:
1913:
1903:
1798:
1694:
1684:
1635:
1625:
1584:
1530:
1520:
1433:
1423:
1213:
745:H16 is a promising candidate enzyme for H
3019:International Journal of Hydrogen Energy
2849:Journal of the American Chemical Society
2554:Journal of the American Chemical Society
2248:Journal of the American Chemical Society
1860:International Journal of Hydrogen Energy
1824:Journal of the American Chemical Society
899:can be oxidized to produce electricity.
443:spp., which can be purified aerobically.
3660:
1188:
357:affinities have also been observed in H
152:. On the other hand, proton reduction (
3168:- PDB structure of -hydrogenase from
3152:- PDB structure of -hydrogenase from
3136:- PDB structure of -hydrogenase from
1935:
1933:
1067:(hydrogen:ferredoxin oxidoreductase)
7:
3329:Methanosarcina-phenazine hydrogenase
378:H16 can be conveniently produced on
268:
244:
83:
49:
2812:The Journal of Biological Chemistry
2471:The Journal of Biological Chemistry
1614:The Journal of Biological Chemistry
2053:10.1111/j.1432-1033.1993.tb17944.x
1215:10.1111/j.1574-6976.2001.tb00587.x
735:who have synthesized a variety of
14:
739:. The soluble hydrogenase from
625:Crystal structure of hydrogenase
412:Crystal structure of hydrogenase
325:Crystal structure of hydrogenase
124:) is coupled to the reduction of
3663:
2077:Canadian Journal of Microbiology
2040:European Journal of Biochemistry
862:involve the usage of enzymes as
3283:Hydrogen:quinone oxidoreductase
3120:structure of -hydrogenase from
2034:Happe T, Naber JD (June 1993).
1043:hydrogen:quinone oxidoreductase
854:Hydrogenase-based biofuel cells
3247:Hydrogenase (NAD+, ferredoxin)
3242:Hydrogen dehydrogenase (NADP+)
3102:Methanothermococcus jannaschii
3031:10.1016/j.ijhydene.2011.02.048
1873:10.1016/j.ijhydene.2011.03.172
952:(hydrogen:NAD oxidoreductase)
929:production of greenhouse gases
769:Biological hydrogen production
757:growth media and purified via
602:HydS) has been characterized.
1:
2687:10.1126/science.282.5395.1842
1744:10.1016/S0262-4079(23)00459-1
763:size exclusion chromatography
2765:Thauer RK (September 1998).
2124:10.1016/0005-2744(71)90008-8
804:-tolerant for use in solar H
596:Thermoanaerobacter mathranii
18:Hydrogenase (disambiguation)
3177:- Mechanism of -hydrogenase
3122:Desulfovibrio desulfuricans
2784:10.1099/00221287-144-9-2377
534:Desulfovibrio desulfuricans
238:
228:
154:
120:
3712:
3170:Desulfomicrobium baculatum
1686:10.1038/s41586-023-05781-7
1052:+ menaquinone ⇌ menaquinol
1002:(hydrogen:ferricytochrome-
935:Biochemical classification
553:Clostridium acetobutylicum
15:
3541:Michaelis–Menten kinetics
3319:Coenzyme F420 hydrogenase
3265:Cytochrome-c3 hydrogenase
2995:10.1016/j.str.2005.05.013
1989:Frontiers in Microbiology
1718:Wilkins A (Mar 8, 2023).
1577:10.1016/j.mex.2016.03.005
1522:10.1186/s12934-015-0226-4
1202:FEMS Microbiology Reviews
525:Chlamydomonas reinhardtii
361:-oxidizing hydrogenases.
187:Structural classification
3433:Diffusion-limited enzyme
3138:Clostridium pasteurianum
2002:10.3389/fmicb.2018.02911
1954:10.1021/acscatal.0c01614
1627:10.1074/jbc.RA119.011076
1509:Microbial Cell Factories
1171:-phenazine hydrogenase
844:Clostridium pasteurianum
679:Clostridium pasteurianum
547:Clostridium pasteurianum
465:Clostridium pasteurianum
456:electron transport chain
426:Clostridium pasteurianum
2944:10.1073/pnas.0905343106
2884:Nature Chemical Biology
2484:10.1074/jbc.M111.254664
2352:10.1126/science.1158978
1425:10.1073/pnas.1320586111
1365:Nature Chemical Biology
1024:⇌ 4H + ferrocytochrome
860:enzymatic biofuel cells
610:-only hydrogenase": -->
386:Mycobacterium smegmatis
3352:Hydrogenase (acceptor)
3301:Ferredoxin hydrogenase
3237:Hydrogen dehydrogenase
3154:Desulfovibrio vulgaris
2825:10.1074/jbc.M008470200
2444:10.1002/anie.201006255
2215:10.1038/ismej.2015.153
2167:10.1099/mic.0.032771-0
1065:ferredoxin hydrogenase
950:hydrogen dehydrogenase
626:
564:HydA1, referred to as
520:thermodynamic barriers
432:. They catalyse both H
413:
353:. A wide spectrum of H
326:
215:
3526:Eadie–Hofstee diagram
3459:Allosteric regulation
2716:Frey M (March 2002).
882:The bidirectional or
624:
436:evolution and uptake.
411:
324:
213:
3686:Iron–sulfur proteins
3536:Lineweaver–Burk plot
2896:10.1038/nchembio.555
2260:10.1021/jacs.7b11287
1377:10.1038/nchembio.276
1110:⇌ reduced coenzyme F
1094:(hydrogen:coenzyme F
712:anaerobic metabolism
508:electron bifurcation
448:Scenedesmus obliquus
430:Megasphaera elsdenii
368:). Hydrogenase from
331:iron-sulfur clusters
16:For other uses, see
2935:2009PNAS..10617331S
2929:(41): 17331–17336.
2777:(Pt 9): 2377–2406.
2681:(5395): 1842–1843.
2609:10.1038/nature03298
2601:2005Natur.433..610T
2477:(44): 38341–38347.
2387:Dalton Transactions
2344:2008Sci...321..572S
2293:(47): 12789–12801.
2207:2016ISMEJ..10..761G
2160:(Pt 6): 1575–1588.
1892:Dalton Transactions
1791:10.1038/nature12239
1783:2013Natur.499...66B
1736:2023NewSc.257...13W
1677:2023Natur.615..541G
1620:(50): 18980–18991.
1416:2014PNAS..111.4257G
1331:2013RSCAd...3.8142J
884:reversible reaction
850:consumption, only.
812:is a by-product of
690:Biological function
583:Thermotoga maritima
578:Per-Arnt-Sim domain
418:dithiolate cofactor
236:shown by reaction (
47:), as shown below:
3495:Enzyme superfamily
3428:Enzyme promiscuity
3164:2008-01-24 at the
3148:2008-01-24 at the
3132:2009-01-16 at the
3112:2009-01-16 at the
3092:2008-01-24 at the
2299:10.1039/D0SC03319G
1905:10.1039/C8DT04294B
1339:10.1039/c3ra22668a
1017:+ ferricytochrome
980:+ NADP ⇌ H + NADPH
808:production since O
742:Ralstonia eutropha
737:hydrogenase mimics
733:inorganic chemists
627:
414:
376:Ralstonia eutropha
371:Ralstonia eutropha
327:
216:
126:electron acceptors
3651:
3650:
3360:
3359:
3063:10.1021/cr4005814
2861:10.1021/ja0504690
2855:(15): 5308–5309.
2652:10.1021/cr050196r
2646:(10): 4206–4272.
2595:(7026): 610–613.
2566:10.1021/ja000116v
2560:(16): 3734–3742.
2530:10.1021/bi801218n
2438:(51): 9917–9921.
2432:Angewandte Chemie
2393:(12): 3057–3064.
2338:(5888): 572–575.
2083:(10): 1214–1223.
1948:(13): 7069–7086.
1898:(18): 5978–5986.
1836:10.1021/ja207461t
1671:(7952): 541–547.
1471:10.1159/000091564
1410:(11): 4257–4261.
1289:10.1021/cr050195z
1283:(10): 4273–4303.
1254:10.1021/cr4005814
677:hydrogenase from
606:-only hydrogenase
397:hydrogenase": -->
310:hydrogenase": -->
291:
290:
267:
266:
176:, and cytochrome
118:Hydrogen uptake (
116:
115:
82:
81:
3703:
3668:
3667:
3659:
3531:Hanes–Woolf plot
3474:Enzyme activator
3469:Enzyme inhibitor
3443:Enzyme catalysis
3387:
3380:
3373:
3364:
3205:
3198:
3191:
3182:
3075:
3074:
3057:(8): 4081–4148.
3051:Chemical Reviews
3046:
3035:
3034:
3025:(3): 2965–2976.
3014:
3008:
3007:
2997:
2988:(9): 1321–1329.
2973:
2967:
2966:
2956:
2946:
2914:
2908:
2907:
2879:
2873:
2872:
2844:
2838:
2837:
2827:
2818:(9): 6125–6132.
2803:
2797:
2796:
2786:
2762:
2756:
2755:
2737:
2728:(2–3): 153–160.
2713:
2707:
2706:
2670:
2664:
2663:
2640:Chemical Reviews
2635:
2629:
2628:
2584:
2578:
2577:
2548:
2542:
2541:
2524:(5): 1056–1066.
2513:
2507:
2506:
2496:
2486:
2462:
2456:
2455:
2427:
2421:
2420:
2410:
2399:10.1039/b922557a
2378:
2372:
2371:
2327:
2321:
2320:
2310:
2287:Chemical Science
2278:
2272:
2271:
2254:(3): 1057–1068.
2243:
2237:
2236:
2226:
2195:The ISME Journal
2186:
2180:
2179:
2169:
2145:
2136:
2135:
2107:
2101:
2100:
2072:
2066:
2065:
2055:
2031:
2025:
2024:
2014:
2004:
1980:
1974:
1973:
1937:
1928:
1927:
1917:
1907:
1883:
1877:
1876:
1867:(3): 2977–2983.
1854:
1848:
1847:
1830:(3): 1577–1582.
1819:
1813:
1812:
1802:
1762:
1756:
1755:
1715:
1709:
1708:
1698:
1688:
1656:
1650:
1649:
1639:
1629:
1605:
1599:
1598:
1588:
1556:
1545:
1544:
1534:
1524:
1500:
1491:
1490:
1465:(2–4): 181–196.
1454:
1448:
1447:
1437:
1427:
1395:
1389:
1388:
1360:
1351:
1350:
1314:
1301:
1300:
1277:Chemical Reviews
1272:
1266:
1265:
1248:(8): 4081–4148.
1242:Chemical Reviews
1234:
1228:
1227:
1217:
1193:
1098:oxidoreductase)
1009:oxidoreductase)
960:+ NAD ⇌ H + NADH
864:electrocatalysts
618:
617:
613:
598:(referred to as
405:
404:
400:
318:
317:
313:
285:
269:
261:
245:
110:
84:
76:
50:
3711:
3710:
3706:
3705:
3704:
3702:
3701:
3700:
3676:
3675:
3674:
3662:
3654:
3652:
3647:
3559:Oxidoreductases
3545:
3521:Enzyme kinetics
3509:
3505:List of enzymes
3478:
3447:
3418:Catalytic triad
3396:
3391:
3361:
3356:
3338:
3334:Sulfhydrogenase
3305:
3287:
3269:
3251:
3223:
3212:Oxidoreductases
3209:
3166:Wayback Machine
3150:Wayback Machine
3134:Wayback Machine
3114:Wayback Machine
3094:Wayback Machine
3083:
3078:
3048:
3047:
3038:
3016:
3015:
3011:
2975:
2974:
2970:
2916:
2915:
2911:
2881:
2880:
2876:
2846:
2845:
2841:
2805:
2804:
2800:
2764:
2763:
2759:
2715:
2714:
2710:
2672:
2671:
2667:
2637:
2636:
2632:
2586:
2585:
2581:
2550:
2549:
2545:
2515:
2514:
2510:
2464:
2463:
2459:
2429:
2428:
2424:
2380:
2379:
2375:
2329:
2328:
2324:
2280:
2279:
2275:
2245:
2244:
2240:
2188:
2187:
2183:
2147:
2146:
2139:
2109:
2108:
2104:
2089:10.1139/m80-203
2074:
2073:
2069:
2033:
2032:
2028:
1982:
1981:
1977:
1939:
1938:
1931:
1885:
1884:
1880:
1856:
1855:
1851:
1821:
1820:
1816:
1777:(7456): 66–69.
1764:
1763:
1759:
1717:
1716:
1712:
1658:
1657:
1653:
1607:
1606:
1602:
1558:
1557:
1548:
1502:
1501:
1494:
1456:
1455:
1451:
1397:
1396:
1392:
1362:
1361:
1354:
1316:
1315:
1304:
1274:
1273:
1269:
1236:
1235:
1231:
1195:
1194:
1190:
1186:
1178:
1155:
1136:
1132:
1113:
1109:
1105:
1097:
1091:
1074:
1051:
1030:
1023:
1016:
1008:
999:
979:
959:
937:
926:
922:
914:
905:
898:
894:
880:
873:
856:
849:
837:
828:
823:
819:
814:water splitting
811:
807:
803:
799:
792:
788:
780:
776:
771:
752:
748:
729:
721:
701:
697:
692:
684:
666:
658:
654:
650:
619:
615:
611:
609:
608:
593:
482:
478:
470:
453:
435:
406:
402:
398:
396:
395:
367:
360:
356:
352:
348:
341:
319:
315:
311:
309:
308:
296:carbon monoxide
283:
275:
259:
251:
235:
225:
221:
202:
198:
189:
182:
175:
147:
108:
101:
97:
93:
74:
67:
63:
56:
46:
35:the reversible
21:
12:
11:
5:
3709:
3707:
3699:
3698:
3693:
3688:
3678:
3677:
3673:
3672:
3649:
3648:
3646:
3645:
3632:
3619:
3606:
3593:
3580:
3567:
3553:
3551:
3547:
3546:
3544:
3543:
3538:
3533:
3528:
3523:
3517:
3515:
3511:
3510:
3508:
3507:
3502:
3497:
3492:
3486:
3484:
3483:Classification
3480:
3479:
3477:
3476:
3471:
3466:
3461:
3455:
3453:
3449:
3448:
3446:
3445:
3440:
3435:
3430:
3425:
3420:
3415:
3410:
3404:
3402:
3398:
3397:
3392:
3390:
3389:
3382:
3375:
3367:
3358:
3357:
3355:
3354:
3348:
3346:
3340:
3339:
3337:
3336:
3331:
3326:
3321:
3315:
3313:
3307:
3306:
3304:
3303:
3297:
3295:
3289:
3288:
3286:
3285:
3279:
3277:
3271:
3270:
3268:
3267:
3261:
3259:
3253:
3252:
3250:
3249:
3244:
3239:
3233:
3231:
3225:
3224:
3210:
3208:
3207:
3200:
3193:
3185:
3179:
3178:
3172:
3156:
3140:
3124:
3104:
3082:
3081:External links
3079:
3077:
3076:
3036:
3009:
2968:
2909:
2890:(5): 310–318.
2874:
2839:
2798:
2757:
2708:
2665:
2630:
2579:
2543:
2508:
2457:
2422:
2373:
2322:
2273:
2238:
2201:(3): 761–777.
2181:
2137:
2118:(3): 576–583.
2102:
2067:
2046:(2): 475–481.
2026:
1975:
1929:
1878:
1849:
1814:
1757:
1710:
1651:
1600:
1546:
1492:
1449:
1390:
1352:
1302:
1267:
1229:
1208:(4): 455–501.
1187:
1185:
1182:
1181:
1180:
1176:
1169:Methanosarcina
1166:
1165:
1158:
1157:
1153:
1146:
1145:
1138:
1137:
1134:
1130:
1123:
1122:
1115:
1114:
1111:
1107:
1103:
1095:
1089:
1085:
1084:
1077:
1076:
1072:
1062:
1061:
1054:
1053:
1049:
1040:
1039:
1032:
1031:
1028:
1021:
1014:
1006:
997:
990:
989:
982:
981:
977:
970:
969:
962:
961:
957:
947:
946:
936:
933:
924:
920:
912:
904:
901:
896:
892:
879:
876:
871:
855:
852:
847:
835:
826:
821:
817:
809:
805:
801:
797:
790:
786:
778:
774:
770:
767:
759:anion exchange
750:
746:
728:
725:
719:
716:bioremediation
704:photosynthesis
699:
695:
691:
688:
682:
665:
662:
656:
652:
648:
607:
604:
591:
480:
476:
468:
460:
459:
451:
444:
437:
433:
394:
391:
365:
358:
354:
350:
346:
339:
336:selenocysteine
307:
304:
302:(CN) ligands.
289:
288:
279:
277:
273:
265:
264:
255:
253:
249:
233:
223:
219:
205:metalloenzymes
200:
196:
188:
185:
180:
173:
145:
142:carbon dioxide
114:
113:
104:
102:
99:
95:
91:
80:
79:
70:
68:
65:
61:
54:
44:
13:
10:
9:
6:
4:
3:
2:
3708:
3697:
3694:
3692:
3689:
3687:
3684:
3683:
3681:
3671:
3666:
3661:
3657:
3643:
3639:
3638:
3633:
3630:
3626:
3625:
3620:
3617:
3613:
3612:
3607:
3604:
3600:
3599:
3594:
3591:
3587:
3586:
3581:
3578:
3574:
3573:
3568:
3565:
3561:
3560:
3555:
3554:
3552:
3548:
3542:
3539:
3537:
3534:
3532:
3529:
3527:
3524:
3522:
3519:
3518:
3516:
3512:
3506:
3503:
3501:
3500:Enzyme family
3498:
3496:
3493:
3491:
3488:
3487:
3485:
3481:
3475:
3472:
3470:
3467:
3465:
3464:Cooperativity
3462:
3460:
3457:
3456:
3454:
3450:
3444:
3441:
3439:
3436:
3434:
3431:
3429:
3426:
3424:
3423:Oxyanion hole
3421:
3419:
3416:
3414:
3411:
3409:
3406:
3405:
3403:
3399:
3395:
3388:
3383:
3381:
3376:
3374:
3369:
3368:
3365:
3353:
3350:
3349:
3347:
3345:
3341:
3335:
3332:
3330:
3327:
3325:
3322:
3320:
3317:
3316:
3314:
3312:
3308:
3302:
3299:
3298:
3296:
3294:
3290:
3284:
3281:
3280:
3278:
3276:
3272:
3266:
3263:
3262:
3260:
3258:
3254:
3248:
3245:
3243:
3240:
3238:
3235:
3234:
3232:
3230:
3226:
3221:
3217:
3213:
3206:
3201:
3199:
3194:
3192:
3187:
3186:
3183:
3176:
3173:
3171:
3167:
3163:
3160:
3157:
3155:
3151:
3147:
3144:
3141:
3139:
3135:
3131:
3128:
3125:
3123:
3119:
3115:
3111:
3108:
3105:
3103:
3099:
3095:
3091:
3088:
3085:
3084:
3080:
3072:
3068:
3064:
3060:
3056:
3052:
3045:
3043:
3041:
3037:
3032:
3028:
3024:
3020:
3013:
3010:
3005:
3001:
2996:
2991:
2987:
2983:
2979:
2972:
2969:
2964:
2960:
2955:
2950:
2945:
2940:
2936:
2932:
2928:
2924:
2920:
2913:
2910:
2905:
2901:
2897:
2893:
2889:
2885:
2878:
2875:
2870:
2866:
2862:
2858:
2854:
2850:
2843:
2840:
2835:
2831:
2826:
2821:
2817:
2813:
2809:
2802:
2799:
2794:
2790:
2785:
2780:
2776:
2772:
2768:
2761:
2758:
2753:
2749:
2745:
2741:
2736:
2731:
2727:
2723:
2719:
2712:
2709:
2704:
2700:
2696:
2692:
2688:
2684:
2680:
2676:
2669:
2666:
2661:
2657:
2653:
2649:
2645:
2641:
2634:
2631:
2626:
2622:
2618:
2614:
2610:
2606:
2602:
2598:
2594:
2590:
2583:
2580:
2575:
2571:
2567:
2563:
2559:
2555:
2547:
2544:
2539:
2535:
2531:
2527:
2523:
2519:
2512:
2509:
2504:
2500:
2495:
2490:
2485:
2480:
2476:
2472:
2468:
2461:
2458:
2453:
2449:
2445:
2441:
2437:
2433:
2426:
2423:
2418:
2414:
2409:
2404:
2400:
2396:
2392:
2388:
2384:
2377:
2374:
2369:
2365:
2361:
2357:
2353:
2349:
2345:
2341:
2337:
2333:
2326:
2323:
2318:
2314:
2309:
2304:
2300:
2296:
2292:
2288:
2284:
2277:
2274:
2269:
2265:
2261:
2257:
2253:
2249:
2242:
2239:
2234:
2230:
2225:
2220:
2216:
2212:
2208:
2204:
2200:
2196:
2192:
2185:
2182:
2177:
2173:
2168:
2163:
2159:
2155:
2151:
2144:
2142:
2138:
2133:
2129:
2125:
2121:
2117:
2113:
2106:
2103:
2098:
2094:
2090:
2086:
2082:
2078:
2071:
2068:
2063:
2059:
2054:
2049:
2045:
2041:
2037:
2030:
2027:
2022:
2018:
2013:
2008:
2003:
1998:
1994:
1990:
1986:
1979:
1976:
1971:
1967:
1963:
1959:
1955:
1951:
1947:
1943:
1942:ACS Catalysis
1936:
1934:
1930:
1925:
1921:
1916:
1911:
1906:
1901:
1897:
1893:
1889:
1882:
1879:
1874:
1870:
1866:
1862:
1861:
1853:
1850:
1845:
1841:
1837:
1833:
1829:
1825:
1818:
1815:
1810:
1806:
1801:
1796:
1792:
1788:
1784:
1780:
1776:
1772:
1768:
1761:
1758:
1753:
1749:
1745:
1741:
1737:
1733:
1729:
1725:
1724:New Scientist
1721:
1714:
1711:
1706:
1702:
1697:
1692:
1687:
1682:
1678:
1674:
1670:
1666:
1662:
1655:
1652:
1647:
1643:
1638:
1633:
1628:
1623:
1619:
1615:
1611:
1604:
1601:
1596:
1592:
1587:
1582:
1578:
1574:
1570:
1566:
1562:
1555:
1553:
1551:
1547:
1542:
1538:
1533:
1528:
1523:
1518:
1514:
1510:
1506:
1499:
1497:
1493:
1488:
1484:
1480:
1476:
1472:
1468:
1464:
1460:
1453:
1450:
1445:
1441:
1436:
1431:
1426:
1421:
1417:
1413:
1409:
1405:
1401:
1394:
1391:
1386:
1382:
1378:
1374:
1370:
1366:
1359:
1357:
1353:
1348:
1344:
1340:
1336:
1332:
1328:
1324:
1320:
1313:
1311:
1309:
1307:
1303:
1298:
1294:
1290:
1286:
1282:
1278:
1271:
1268:
1263:
1259:
1255:
1251:
1247:
1243:
1239:
1233:
1230:
1225:
1221:
1216:
1211:
1207:
1203:
1199:
1192:
1189:
1183:
1174:
1173:
1172:
1170:
1164:
1160:
1159:
1151:
1150:
1149:
1144:
1140:
1139:
1128:
1127:
1126:
1121:
1117:
1116:
1101:
1100:
1099:
1093:
1083:
1079:
1078:
1070:
1069:
1068:
1066:
1060:
1056:
1055:
1047:
1046:
1045:
1044:
1038:
1034:
1033:
1027:
1020:
1012:
1011:
1010:
1005:
1001:
996:
988:
984:
983:
975:
974:
973:
968:
964:
963:
955:
954:
953:
951:
945:
942:
939:
938:
934:
932:
930:
918:
910:
902:
900:
890:
885:
877:
875:
869:
865:
861:
853:
851:
845:
839:
833:
815:
794:
784:
783:overpotential
768:
766:
764:
760:
756:
755:heterotrophic
744:
743:
738:
734:
726:
724:
717:
713:
707:
705:
689:
687:
680:
675:
671:
663:
661:
644:
642:
639:
635:
631:
623:
614:
605:
603:
601:
597:
589:
585:
584:
579:
575:
571:
567:
563:
559:
555:
554:
549:
548:
543:
539:
535:
531:
527:
526:
521:
517:
513:
509:
505:
501:
497:
493:
489:
484:
472:
466:
457:
450:, catalyses H
449:
445:
442:
441:Desulfovibrio
438:
431:
427:
423:
422:
421:
419:
410:
401:
392:
390:
388:
387:
381:
380:heterotrophic
377:
373:
372:
362:
345:
337:
332:
323:
314:
305:
303:
301:
297:
287:
280:
278:
271:
270:
263:
256:
254:
247:
246:
243:
241:
240:
231:
230:
212:
208:
206:
194:
186:
184:
179:
172:
169:
165:
161:
157:
156:
151:
143:
139:
135:
131:
127:
123:
122:
112:
105:
103:
90:
86:
85:
78:
71:
69:
60:
52:
51:
48:
42:
39:of molecular
38:
34:
30:
26:
19:
3637:Translocases
3634:
3621:
3608:
3595:
3582:
3572:Transferases
3569:
3556:
3413:Binding site
3214:: Acting on
3169:
3153:
3137:
3121:
3101:
3054:
3050:
3022:
3018:
3012:
2985:
2981:
2971:
2926:
2922:
2912:
2887:
2883:
2877:
2852:
2848:
2842:
2815:
2811:
2801:
2774:
2771:Microbiology
2770:
2760:
2725:
2721:
2711:
2678:
2674:
2668:
2643:
2639:
2633:
2592:
2588:
2582:
2557:
2553:
2546:
2521:
2518:Biochemistry
2517:
2511:
2474:
2470:
2460:
2435:
2431:
2425:
2390:
2386:
2376:
2335:
2331:
2325:
2290:
2286:
2276:
2251:
2247:
2241:
2198:
2194:
2184:
2157:
2154:Microbiology
2153:
2115:
2111:
2105:
2080:
2076:
2070:
2043:
2039:
2029:
1992:
1988:
1978:
1945:
1941:
1895:
1891:
1881:
1864:
1858:
1852:
1827:
1823:
1817:
1774:
1770:
1760:
1730:(3430): 13.
1727:
1723:
1713:
1668:
1664:
1654:
1617:
1613:
1603:
1568:
1564:
1512:
1508:
1462:
1458:
1452:
1407:
1403:
1393:
1371:(1): 63–70.
1368:
1364:
1325:(22): 8142.
1322:
1319:RSC Advances
1318:
1280:
1276:
1270:
1245:
1241:
1232:
1205:
1201:
1191:
1168:
1167:
1147:
1124:
1106:+ coenzyme F
1086:
1063:
1041:
1025:
1018:
1003:
994:
991:
971:
948:
906:
889:hydrothermal
881:
857:
843:
840:
795:
772:
740:
730:
727:Applications
708:
693:
678:
667:
645:
638:methanogenic
628:
599:
595:
587:
581:
569:
565:
561:
557:
551:
545:
541:
537:
529:
523:
488:phylogenetic
485:
473:
464:
461:
447:
440:
429:
425:
415:
384:
375:
369:
363:
343:
328:
292:
281:
257:
237:
227:
217:
190:
177:
170:
153:
119:
117:
106:
72:
24:
22:
3408:Active site
2722:ChemBioChem
1571:: 242–250.
1092:hydrogenase
1000:hydrogenase
993:cytochrome-
874:oxidation.
670:amino acids
636:) found in
492:bifurcating
393:hydrogenase
306:hydrogenase
252:⇌ 2 H + 2 e
25:hydrogenase
3680:Categories
3611:Isomerases
3585:Hydrolases
3452:Regulation
3218:as donor (
1184:References
1088:coenzyme F
903:Advantages
660:acceptor.
560:HydA1 and
516:endergonic
500:ferredoxin
168:cytochrome
160:ferredoxin
3490:EC number
3175:Animation
2982:Structure
2368:206513302
1970:219749715
1962:2155-5435
1752:257625443
1515:(1): 42.
1347:2046-2069
1163:1.12.98.3
1143:1.12.98.2
1120:1.12.99.6
1082:1.12.98.1
909:renewable
878:Principle
832:diffusion
664:Mechanism
634:1.12.98.2
540:HydAB or
512:exergonic
298:(CO) and
37:oxidation
33:catalyses
3696:EC 1.2.1
3514:Kinetics
3438:Cofactor
3401:Activity
3216:hydrogen
3162:Archived
3146:Archived
3130:Archived
3110:Archived
3090:Archived
3071:24655035
3004:16154089
2963:19805068
2904:21390036
2869:15826154
2834:11096090
2752:36754174
2744:11921392
2703:38018712
2660:17927159
2617:15703741
2574:11457105
2538:19138102
2503:21900241
2452:21105038
2417:20221540
2360:18653896
2317:34094474
2268:29251926
2233:26405831
2176:20395274
2021:30564206
1995:: 2911.
1924:30632592
1844:21916466
1809:23803769
1705:36890228
1696:10017518
1646:31624148
1595:27077052
1565:MethodsX
1541:25880663
1479:16645314
1444:24591586
1385:19966788
1297:17850165
1262:24655035
1238:Lubitz W
1224:11524134
1133:+ A ⇌ AH
1059:1.12.7.2
1037:1.12.5.1
987:1.12.2.1
967:1.12.1.3
944:1.12.1.2
917:Platinum
858:Typical
544:H), and
532:HydA1),
496:turnover
193:hydrogen
164:pyruvate
150:fumarate
128:such as
64:→ 2H + A
41:hydrogen
3691:EC 1.12
3670:Biology
3624:Ligases
3394:Enzymes
3344:1.10.99
3311:1.10.98
2954:2765078
2931:Bibcode
2793:9782487
2695:9874636
2675:Science
2625:4430994
2597:Bibcode
2494:3207428
2408:3465567
2340:Bibcode
2332:Science
2308:8163306
2224:4817680
2203:Bibcode
2132:5569125
2097:7006765
2062:8513797
2012:6288185
1915:6509880
1800:3793303
1779:Bibcode
1732:Bibcode
1673:Bibcode
1637:6916507
1586:4816682
1532:4377017
1487:8030367
1435:3964045
1412:Bibcode
1327:Bibcode
868:biofuel
674:ligands
672:and/or
641:Archaea
300:cyanide
276:⇌ H + H
148:), and
138:sulfate
134:nitrate
3656:Portal
3598:Lyases
3293:1.12.7
3275:1.12.5
3257:1.12.2
3229:1.12.1
3069:
3002:
2961:
2951:
2902:
2867:
2832:
2791:
2750:
2742:
2701:
2693:
2658:
2623:
2615:
2589:Nature
2572:
2536:
2501:
2491:
2450:
2415:
2405:
2366:
2358:
2315:
2305:
2266:
2231:
2221:
2174:
2130:
2095:
2060:
2019:
2009:
1968:
1960:
1922:
1912:
1842:
1807:
1797:
1771:Nature
1750:
1703:
1693:
1665:Nature
1644:
1634:
1593:
1583:
1539:
1529:
1485:
1477:
1442:
1432:
1383:
1345:
1295:
1260:
1222:
891:) as H
574:motifs
568:I and
510:where
504:NAD(H)
498:using
479:-NH-CH
130:oxygen
29:enzyme
27:is an
3550:Types
3222:1.12)
2748:S2CID
2699:S2CID
2621:S2CID
2364:S2CID
1966:S2CID
1748:S2CID
1483:S2CID
475:(-SCH
87:2H +
31:that
3642:list
3635:EC7
3629:list
3622:EC6
3616:list
3609:EC5
3603:list
3596:EC4
3590:list
3583:EC3
3577:list
3570:EC2
3564:list
3557:EC1
3159:1CC1
3143:1UBR
3127:1C4A
3107:1HFE
3087:2B0J
3067:PMID
3000:PMID
2959:PMID
2900:PMID
2865:PMID
2830:PMID
2789:PMID
2740:PMID
2691:PMID
2656:PMID
2613:PMID
2570:PMID
2534:PMID
2499:PMID
2448:PMID
2413:PMID
2356:PMID
2313:PMID
2264:PMID
2229:PMID
2172:PMID
2128:PMID
2093:PMID
2058:PMID
2017:PMID
1958:ISSN
1920:PMID
1840:PMID
1805:PMID
1701:PMID
1642:PMID
1591:PMID
1537:PMID
1475:PMID
1440:PMID
1381:PMID
1343:ISSN
1293:PMID
1258:PMID
1220:PMID
834:of O
761:and
632:(EC
612:edit
550:and
514:and
428:and
399:edit
312:edit
3118:PDB
3098:PDB
3059:doi
3055:114
3027:doi
2990:doi
2949:PMC
2939:doi
2927:106
2892:doi
2857:doi
2853:127
2820:doi
2816:276
2779:doi
2775:144
2730:doi
2683:doi
2679:282
2648:doi
2644:107
2605:doi
2593:433
2562:doi
2558:123
2526:doi
2489:PMC
2479:doi
2475:286
2440:doi
2403:PMC
2395:doi
2348:doi
2336:321
2303:PMC
2295:doi
2256:doi
2252:140
2219:PMC
2211:doi
2162:doi
2158:156
2120:doi
2116:227
2085:doi
2048:doi
2044:214
2007:PMC
1997:doi
1950:doi
1910:PMC
1900:doi
1869:doi
1832:doi
1828:134
1795:PMC
1787:doi
1775:499
1740:doi
1728:257
1691:PMC
1681:doi
1669:615
1632:PMC
1622:doi
1618:294
1581:PMC
1573:doi
1527:PMC
1517:doi
1467:doi
1430:PMC
1420:doi
1408:111
1373:doi
1335:doi
1285:doi
1281:107
1250:doi
1246:114
1210:doi
1161:EC
1141:EC
1118:EC
1112:420
1108:420
1096:420
1090:420
1080:EC
1057:EC
1035:EC
985:EC
965:EC
600:Tam
242:).
195:, H
144:(CO
98:+ D
94:→ H
92:red
66:red
3682::
3220:EC
3116:-
3096:-
3065:.
3053:.
3039:^
3023:37
3021:.
2998:.
2986:13
2984:.
2980:.
2957:.
2947:.
2937:.
2925:.
2921:.
2898:.
2886:.
2863:.
2851:.
2828:.
2814:.
2810:.
2787:.
2773:.
2769:.
2746:.
2738:.
2724:.
2720:.
2697:.
2689:.
2677:.
2654:.
2642:.
2619:.
2611:.
2603:.
2591:.
2568:.
2556:.
2532:.
2522:48
2520:.
2497:.
2487:.
2473:.
2469:.
2446:.
2436:49
2434:.
2411:.
2401:.
2391:39
2389:.
2385:.
2362:.
2354:.
2346:.
2334:.
2311:.
2301:.
2291:11
2289:.
2285:.
2262:.
2250:.
2227:.
2217:.
2209:.
2199:10
2197:.
2193:.
2170:.
2156:.
2152:.
2140:^
2126:.
2114:.
2091:.
2081:26
2079:.
2056:.
2042:.
2038:.
2015:.
2005:.
1991:.
1987:.
1964:.
1956:.
1946:10
1944:.
1932:^
1918:.
1908:.
1896:48
1894:.
1890:.
1865:37
1863:.
1838:.
1826:.
1803:.
1793:.
1785:.
1773:.
1769:.
1746:.
1738:.
1726:.
1722:.
1699:.
1689:.
1679:.
1667:.
1663:.
1640:.
1630:.
1616:.
1612:.
1589:.
1579:.
1567:.
1563:.
1549:^
1535:.
1525:.
1513:14
1511:.
1507:.
1495:^
1481:.
1473:.
1463:10
1461:.
1438:.
1428:.
1418:.
1406:.
1402:.
1379:.
1367:.
1355:^
1341:.
1333:.
1321:.
1305:^
1291:.
1279:.
1256:.
1244:.
1218:.
1206:25
1204:.
1200:.
1013:2H
941:EC
931:.
923:/O
588:Tm
570:Ca
566:Cp
562:Ca
558:Cp
542:Dd
538:Dd
530:Cr
471:.
140:,
136:,
132:,
100:ox
62:ox
57:+
43:(H
23:A
3658::
3644:)
3640:(
3631:)
3627:(
3618:)
3614:(
3605:)
3601:(
3592:)
3588:(
3579:)
3575:(
3566:)
3562:(
3386:e
3379:t
3372:v
3204:e
3197:t
3190:v
3073:.
3061::
3033:.
3029::
3006:.
2992::
2965:.
2941::
2933::
2906:.
2894::
2888:7
2871:.
2859::
2836:.
2822::
2795:.
2781::
2754:.
2732::
2726:3
2705:.
2685::
2662:.
2650::
2627:.
2607::
2599::
2576:.
2564::
2540:.
2528::
2505:.
2481::
2454:.
2442::
2419:.
2397::
2370:.
2350::
2342::
2319:.
2297::
2270:.
2258::
2235:.
2213::
2205::
2178:.
2164::
2134:.
2122::
2099:.
2087::
2064:.
2050::
2023:.
1999::
1993:9
1972:.
1952::
1926:.
1902::
1875:.
1871::
1846:.
1834::
1811:.
1789::
1781::
1754:.
1742::
1734::
1707:.
1683::
1675::
1648:.
1624::
1597:.
1575::
1569:3
1543:.
1519::
1489:.
1469::
1446:.
1422::
1414::
1387:.
1375::
1369:6
1349:.
1337::
1329::
1323:3
1299:.
1287::
1264:.
1252::
1226:.
1212::
1177:2
1175:H
1154:2
1152:H
1135:2
1131:2
1129:H
1104:2
1102:H
1073:2
1071:H
1050:2
1048:H
1029:3
1026:c
1022:3
1019:c
1015:2
1007:3
1004:c
998:3
995:c
978:2
976:H
958:2
956:H
925:2
921:2
913:2
897:2
893:2
872:2
848:2
836:2
827:2
825:O
822:2
818:2
810:2
806:2
802:2
798:2
791:2
787:2
779:2
775:2
751:2
747:2
720:2
700:2
696:2
683:2
657:2
653:2
649:2
616:]
592:2
586:(
556:(
536:(
528:(
481:2
477:2
469:2
458:.
452:2
434:2
403:]
366:2
359:2
355:2
351:2
347:3
344:c
340:2
316:]
286:)
284:4
282:(
274:2
272:H
262:)
260:3
258:(
250:2
248:H
239:4
234:2
229:3
224:2
220:2
201:2
197:2
181:6
178:c
174:3
171:c
155:2
146:2
121:1
111:)
109:2
107:(
96:2
89:D
77:)
75:1
73:(
59:A
55:2
53:H
45:2
20:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.