246:
heating time also means that the procedure is not compatible for certain procedures such as the one tube, single buffer reverse transcription-PCR method which requires lower temperature to undergo the reverse transcription step. In chemically modified hot start PCR, the amplification process of DNA can be negatively affected firstly due to a significant increase in the reactivation time required for the polymerase to activate and secondly if the length of the target DNA template is too long. In antibody based procedures, each enzyme requires a different antibody and therefore the cost to perform the procedure is higher. There is also evidence that many commercial hot start enzymes actually have some level of activity prior to denaturation, and few suppliers provide any information about testing for this residual activity. This means that the benefits attributed to hot start enzymes may not be realized, or at best will vary between batches or manufacturer.
233:
temperature and significantly decreases the formation of primer-dimers by preventing primers from binding to one another before the PCR process has begun as well as limiting non-specific priming. Similarly, hot start PCR inhibits the binding of primers to the template sequences which have a low homology which leads to mispriming. It can also improve specificity and sensitivity, due to the stringent conditions, as well as increase the product yield of the targeted fragment. In antibody based hot start PCR, the polymerase is activated after the initial denaturation step during the cycling process, therefore decreasing the time required. This also leads to a high
191:
protecting group will be removed during the heat activation step. The hot start dNTP, dA, dT, dC and dG replace the natural nucleotides. Using all four of the modified nucleotides is recommended, however, previous research shows that by replacing either one or two of the natural nucleotides with the modified dNTPs would be enough to ensure that non-specific amplification does not occur. Another chemical modification of nucleic acid is through the heat-reversible covalent modification which acts to impede the hybridisation of the primers to the template of interest. The guanosine amino group interact with glyoxal to form dG.
87:
mixture, are the cause behind the synthesis of by-products such as primer dimer and mis-priming. Mis-priming greatly impedes and reduces the efficiency of PCR amplification through actively competing with the target sequences for amplification. Similarly, primer dimers form complexes which decreases the amount of copy number amplifications obtained. This can be controlled by implementing hot start PCR which allows primer extensions to be blocked until the optimal temperatures are met.
54:
135:
Taq DNA polymerase into the reaction and allowing the amplification process to start. Platinum Taq DNA polymerase and AccuStart Taq DNA polymerase ( both developed by Ayoub
Rashtchian at Life technologies and Quanta BioSciences, respectively) are examples of commercially available antibody based hot start Taq DNA polymerases. These Taq DNA polymerase are precomplexed with a mixture of monoclonal antibodies specific to Taq DNA polymerase.
1520:
94:) are prevented from reacting in the PCR mixture until the optimal temperatures are met through physical separation or chemical modifications. Hot start PCR can also occur when the Taq polymerase is inhibited/inactivated or its addition is delayed until optimal annealing temperatures, through deoxyribonucleotide triphosphate modifications or by modifying the primers through caging and
974:
203:
form a linear structure instead, which enables the primer to attach to the target segment and begin PCR. Actually, there is a more stable configuration to the hairpin primers termed ‘double-bubble’ primers, that form a head to tail homodimer configuration that can be utilized both for reverse- transcription and for hot- start PCR.
115:
45:
inactive or are inhibited until the optimal annealing temperature is reached. Inhibiting formation of non-specific PCR products, especially in early cycles, results in a substantial increase in sensitivity of amplification by PCR. This is of utmost importance in diagnostic applications of PCR or RT-PCR.
245:
Along with its advantages, hot start PCR also has limitations which must be considered before implementing the method. Hot start PCR requires the addition of heat for longer periods of time as opposed to conventional PCR, therefore, the template DNA is more susceptible to being damaged. The increased
171:
Freezing acts as a form of physical separation much like the wax beads. The reaction mixture containing primers, the template strand, water and deoxyribonucleotide triphosphate (dNTP) is frozen before Taq polymerase and the remaining PCR components are added on top of the frozen mixture. This acts to
141:
A physical barrier is created between Taq DNA polymerase and the remainder of the PCR components by the wax beads which are temperature dependent. Once the temperature rises over 70 °C, during the denaturation step in the first cycle, the wax bead melts, allowing the Taq DNA polymerase to escape
134:
The enzyme linked antibodies inactivate the Taq DNA polymerase. The antibodies link and bind to the polymerase, preventing early DNA amplification which could occur at lower temperatures. Once the optimal annealing temperature is met, the antibodies will begin to degrade and dissociate, releasing the
164:
The PCR machine is heated in advance whilst the components are mixed over ice and then immediately placed into the PCR machine once it reaches optimum temperature. This would eliminate the warm-up process required, reduce non-specific annealing of the primers and ensures that any miss paired primers
122:
Hot start PCR is a method which prevents DNA polymerase extension at lower temperature to prevent non-specific binding to minimise yield loss. Hot start PCR reduces the amount of non-specific binding through limiting reagents until the heating steps of PCR – limit the reaction early by limiting Taq
68:
technique used to amplify specific DNA segments by several orders of magnitude. The specific segments of DNA is amplified over three processes, denaturation, annealing and extension – where the DNA strands are separated by raising the temperature to the optimal from room temperature before primers
27:
due to non-specific DNA amplification at room (or colder) temperatures. Many variations and modifications of the PCR procedure have been developed in order to achieve higher yields; hot start PCR is one of them. Hot start PCR follows the same principles as the conventional PCR - in that it uses DNA
232:
Hot start PCR is advantageous in that it requires less handling and reduces the risk of contamination. Hot start PCR can either be chemically modified or antibody based which provide different advantages to the procedure. In chemically modified hot start PCR, the procedure can be taken under room
202:
Certain secondary structure may impede the functions of the primers. For example, oligonucleotides with a hairpin structure cannot act efficiently as a primer. However, after heating the reaction mix to the annealing temperature the primer will undergo a conformation change allowing the primer to
148:
Oligonucleotides are short polymers of nucleic acid which easily bind. Highly specific oligonucleotides, such as aptamers, bind to Taq DNA polymerase at lower temperatures making it inactive in the mixture. Only at higher temperatures will the oligonucleotides separate from the Taq allowing it to
190:
Hot start dNTP can be chemically modified to include a heat sensitive protecting group at the 3 prime terminus. This modification will prevent the nucleotides from interacting with the Taq polymerase to bind to the template strand until after the optimal temperatures are reached therefore, the
44:
Through these additional methods, hot start PCR is able to decrease the amount of non-specific amplifications which naturally occur during lower temperatures – which remains a problem for conventional PCR. These modifications work overall to ensure that specific enzymes in solution will remain
86:
In conventional PCR, lower temperatures below the optimal annealing temperature (50-65 °C) results in off target modifications such as non-specific amplifications where primers will bind non-specifically to the nucleic acid. These non-specific primer complexes, which are in excess in the
210:
A caging group which is a protecting group that is photochemically removable, such as caged thymidine phosphoramidites, is incorporated into a oligonucleotide primer. This allows the function of the primer to be activated and deactivated through the use of UV irradiation (365 nm).
224:, providing a hot start for the reaction as there is no magnesium for the DNA polymerase until during the thermal cycling stage. During thermal cycling, the magnesium will dissolve back into solution and become available for the polymerase to use allowing it to function normally.
152:
These are the most effective methods for hot start PCR, the enzyme linked antibodies and highly specific oligonucleotides methods in particular are most suited during procedures which require a shorter inactivation time. However, other methods are known to be implemented such as:
178:
The components of PCR in the reaction mix are prepared and heated without the addition of Taq. Taq is only later introduced into the mixture once the optimal temperature is reached. However, this method is the least reliable and may lead to a contamination of the components.
82:
are kept separate until the mixture is heated to the specific annealing temperature. This reduces annealing time, which in turn reduces the likelihood of non-specific DNA extension and the influence of non-specific primer binding prior to denaturation.
801:
530:
988:
Ailenberg, M and
Silverman, M, 2000-11-1, Controlled hot start and improved specificity in carrying out PCR utilizing touch-up and loop incorporated primers (TULIPS),Biotechniques, 29(5):1018- 1022,doi: 10.2144/00295st03,PMID: 11084864
77:
or anneal to DNA non-specifically. During the PCR procedure, DNA polymerase will extend any piece of DNA with bound primers, generating target products but also nonspecific products which lower the yield. In hot start PCR, some of the
32:
and late addition of Taq polymerase, to increase product yield as well as provide a higher specificity and sensitivity. Non-specific binding and priming or formation of primer dimers are minimized by completing the reaction mix after
101:
Hot start PCR is often a better approach opposed to traditional PCR in circumstances where there is a low concentration of DNA in the reaction mix, the DNA template is highly complex, or if there are several pairs of
142:
past the barrier and be released into the reaction – starting the amplification process. The wax layer then moves to the top of the reaction mixture during the amplification stage to later act as a vapour barrier.
219:
Magnesium is required in PCR and acts as a co-factor because Taq polymerase is magnesium dependent. Increasing the concentration of magnesium and phosphate to the standard buffer reagents creates a magnesium
972:, Bonner, Alex, "Reversible chemical modification of nucleic acids and improved method for nucleic acid hybridization", published 2003-08-28, assigned to BioLink Partners Inc.
1055:
Ailenberg, M, Kapus, A and
Rotstein, OD, 2021-09-14, Improved SARS-CoV-2 PCR detection and genotyping with double-bubble primers, Biotechniques,71 (6):587-597, DOI: 10.2144/btn-2021-0063
28:
polymerase to synthesise DNA from a single stranded template. However, it utilizes additional heating and separation methods, such as inactivating or inhibiting the binding of
1269:
123:
DNA polymerase in a reaction. Non-specific binding often leads to primer dimers and mis-primed/false primed targets. These can be rectified through modified methods such as:
73:, which is slightly active at low temperatures. In conventional PCR, the reaction mix is completed at room temperature, and due to DNA polymerase activity, primers may form
268:
Sharkey DJ, Scalice ER, Christy KG, Atwood SM, Daiss JL (May 1994). "Antibodies as thermolabile switches: high temperature triggering for the polymerase chain reaction".
613:
Lebedev, Alexandre V.; Paul, Natasha; Yee, Joyclyn; Timoshchuk, Victor A.; Shum, Jonathan; Miyagi, Kei; Kellum, Jack; Hogrefe, Richard I.; Zon, Gerald (November 2008).
41:
activity in low temperatures, use of modified deoxyribonucleotide triphosphates (dNTPs), and the physical addition of one of the essential reagents after denaturation.
1448:
1408:
1436:
408:
1466:
1262:
182:
Another method is through deoxyribonucleotide triphosphate mediated hot start PCR which modifies the nucleotide bases through a protecting group.
1389:
1477:
1442:
1426:
1540:
1255:
1460:
1414:
584:
514:
337:
1420:
1510:
862:"3′-Protected 2′-Deoxynucleoside 5′-Triphosphates as a Tool for Heat-Triggered Activation of Polymerase Chain Reaction"
1299:
34:
118:
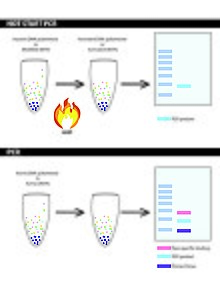
PCR vs. hot start PCR: contrasting PCR to hot start PCR, by showing their methods and resulting PCR product on a gel.
234:
1278:
497:
van Pelt-Verkuil E, van Belkum A, Hays JP, eds. (2008). "Variants and
Adaptations of the Standard PCR Protocol".
221:
61:
20:
1309:
91:
1065:
Young, Douglas D.; Edwards, Wesleigh F.; Lusic, Hrvoje; Lively, Mark O.; Deiters, Alexander (2008-01-10).
969:
1379:
920:
Koukhareva, I.; Haoqiang, H.; Yee, J.; Shum, J.; Paul, N.; Hogrefe, R. I.; Lebedev, A. V. (2008-09-01).
1340:
452:
1175:
Barnes, Wayne M; Rowlyk, Katherine R (June 2002). "Magnesium precipitate hot start method for PCR".
1364:
1304:
95:
1497:
1369:
1359:
1354:
476:
293:
131:
Enzyme linked antibodies/Taq DNA polymerase complexed with Anti Taq DNA polymerase antibodies:
53:
1344:
1233:
1192:
1157:
1139:
1100:
1082:
1038:
1020:
1001:"PCR hot start using primers with the structure of molecular beacons (hairpin-like structure)"
951:
943:
899:
881:
769:
751:
709:
691:
652:
634:
590:
580:
510:
468:
389:
381:
343:
333:
285:
65:
364:
Green, Michael R.; Sambrook, Joseph (May 2018). "Hot Start
Polymerase Chain Reaction (PCR)".
37:. Some ways to complete reaction mixes at high temperatures involve modifications that block
1402:
1223:
1184:
1147:
1131:
1090:
1074:
1028:
1012:
933:
889:
873:
759:
743:
699:
683:
642:
626:
615:"Hot Start PCR with heat-activatable primers: a novel approach for improved PCR performance"
502:
460:
373:
325:
277:
1394:
1335:
832:
409:"Polymerase Chain Reaction (PCR)- Principle, Procedure, Types, Applications and Animation"
103:
1212:"Many commercial hot-start polymerases demonstrate activity prior to thermal activation"
456:
1524:
1374:
1349:
1247:
1152:
1119:
1095:
1066:
894:
861:
764:
731:
730:
Schoenbrunner, Nancy J; Gupta, Amar P; Young, Karen K Y; Will, Stephen G (2017-01-01).
647:
614:
70:
38:
29:
1033:
1000:
704:
671:
1534:
1492:
732:"Covalent modification of primers improves PCR amplification specificity and yield"
480:
297:
74:
24:
999:
Kaboev, O. K.; Luchkina, L. A.; Tret’iakov, A. N.; Bahrmand, A. R. (2000-11-01).
922:"Heat Activatable 3'-modified dNTPs: Synthesis and Application for Hot Start PCR"
506:
329:
1487:
747:
551:
211:
Therefore, primers can be activated after the annealing temperature is reached.
1519:
1327:
324:. Methods in Molecular Biology. Vol. 630. Humana Press. pp. 301–18.
1143:
1086:
1024:
947:
885:
755:
695:
638:
594:
385:
1016:
1237:
1196:
1188:
1161:
1104:
1042:
955:
903:
773:
713:
656:
393:
377:
347:
90:
In hot start PCR, important reagents (such as DNA polymerase and magnesium
938:
921:
672:"Cold‐sensitive mutants of Taq DNA polymerase provide a hot start for PCR"
472:
289:
1454:
687:
630:
281:
670:
Kermekchiev, Milko B.; Tzekov, Anatoly; Barnes, Wayne M. (2003-11-01).
79:
69:
bind and polymerase aligns nucleotides to the template strand. It uses
1228:
1211:
1135:
877:
1078:
464:
113:
114:
1251:
1120:"Multiplex polymerase chain reaction: A practical approach"
802:"How is Hot-Start Technology Beneficial For Your PCR - AU"
23:(PCR) that reduces the presence of undesired products and
57:
Procedure of traditional polymerase chain reaction (PCR)
1455:
Co-amplification at lower denaturation temperature PCR
1508:
531:"How is Hot-Start Technology Beneficial For Your PCR"
499:
Principles and
Technical Aspects of PCR Amplification
186:
Deoxyribonucleotide triphosphate (dNTP) modifications
1476:
1388:
1326:
1286:
1118:Markoulatos, P.; Siafakas, N.; Moncany, M. (2002).
860:Koukhareva, Inna; Lebedev, Alexandre (2009-06-15).
443:Birch DE (May 1996). "Simplified hot start PCR".
1449:Multiplex ligation-dependent probe amplification
1409:Reverse transcription polymerase chain reaction
552:"DNTP - The School of Biomedical Sciences Wiki"
320:Paul N, Shum J, Le T (2010). "Hot start PCR".
1263:
127:Inactivation/inhibition of Taq DNA polymerase
8:
786:Westfall et.al. (1997), Focus 19.3, page 46.
1437:Overlap extension polymerase chain reaction
1067:"Light-triggered polymerase chain reaction"
1270:
1256:
1248:
501:. Springer Netherlands. pp. 231–276.
1227:
1151:
1094:
1032:
937:
893:
763:
703:
646:
52:
1515:
1467:Random amplification of polymorphic DNA
1124:Journal of Clinical Laboratory Analysis
255:
1403:Quantitative polymerase chain reaction
915:
913:
855:
853:
827:
825:
823:
821:
796:
794:
792:
725:
723:
608:
606:
604:
263:
261:
259:
7:
1210:Stevens AJ, et al. (Aug 2016).
492:
490:
438:
436:
434:
432:
430:
428:
359:
357:
315:
313:
311:
309:
307:
1443:Multiplex polymerase chain reaction
1427:Touchdown polymerase chain reaction
157:Late addition of Taq DNA polymerase
19:is a modified form of conventional
145:Highly specific oligonucleotides:
14:
1461:Digital polymerase chain reaction
1415:Inverse polymerase chain reaction
207:Photochemically removable cages:
1518:
1421:Nested polymerase chain reaction
215:Controlled addition of magnesium
970:US application 20030162199
926:Nucleic Acids Symposium Series
577:Diagnostic molecular pathology
172:prevent non-specific binding.
165:in the mixture are separated.
1:
1177:Molecular and Cellular Probes
736:Biology Methods and Protocols
579:. : Elsevier Academic Press.
507:10.1007/978-1-4020-6241-4_12
366:Cold Spring Harbor Protocols
330:10.1007/978-1-60761-629-0_19
407:Aryal, Sagar (2015-04-23).
1557:
833:"What is a hot start PCR?"
1541:Polymerase chain reaction
1279:Polymerase chain reaction
748:10.1093/biomethods/bpx011
62:Polymerase chain reaction
21:polymerase chain reaction
1071:Chemical Communications
175:Later addition of Taq:
1189:10.1006/mcpr.2002.0407
1005:Nucleic Acids Research
676:Nucleic Acids Research
619:Nucleic Acids Research
378:10.1101/pdb.prot095125
119:
58:
1017:10.1093/nar/28.21.e94
413:Microbiology Info.com
372:(5): pdb.prot095125.
199:Secondary structure:
117:
56:
866:Analytical Chemistry
806:www.thermofisher.com
106:primers in the PCR.
939:10.1093/nass/nrn131
575:Coleman WB (2016).
457:1996Natur.381..445B
282:10.1038/nbt0594-506
96:secondary structure
979:, since abandoned.
688:10.1093/nar/gkg813
631:10.1093/nar/gkn575
556:teaching.ncl.ac.uk
120:
59:
1506:
1505:
1229:10.2144/000114481
1136:10.1002/jcla.2058
878:10.1021/ac8026977
872:(12): 4955–4962.
837:Genetic Education
682:(21): 6139–6147.
66:molecular biology
1548:
1523:
1522:
1514:
1316:Final elongation
1272:
1265:
1258:
1249:
1242:
1241:
1231:
1207:
1201:
1200:
1172:
1166:
1165:
1155:
1115:
1109:
1108:
1098:
1079:10.1039/B715152G
1062:
1056:
1053:
1047:
1046:
1036:
996:
990:
986:
980:
978:
977:
973:
966:
960:
959:
941:
917:
908:
907:
897:
857:
848:
847:
845:
844:
829:
816:
815:
813:
812:
798:
787:
784:
778:
777:
767:
727:
718:
717:
707:
667:
661:
660:
650:
610:
599:
598:
572:
566:
565:
563:
562:
548:
542:
541:
539:
538:
527:
521:
520:
494:
485:
484:
465:10.1038/381445a0
440:
423:
422:
420:
419:
404:
398:
397:
361:
352:
351:
322:RT-PCR Protocols
317:
302:
301:
265:
195:Modified primers
1556:
1555:
1551:
1550:
1549:
1547:
1546:
1545:
1531:
1530:
1529:
1517:
1509:
1507:
1502:
1480:
1472:
1445:(multiplex PCR)
1392:
1384:
1322:
1282:
1276:
1246:
1245:
1209:
1208:
1204:
1174:
1173:
1169:
1117:
1116:
1112:
1064:
1063:
1059:
1054:
1050:
998:
997:
993:
987:
983:
975:
968:
967:
963:
919:
918:
911:
859:
858:
851:
842:
840:
831:
830:
819:
810:
808:
800:
799:
790:
785:
781:
729:
728:
721:
669:
668:
664:
612:
611:
602:
587:
574:
573:
569:
560:
558:
550:
549:
545:
536:
534:
529:
528:
524:
517:
496:
495:
488:
451:(6581): 445–6.
442:
441:
426:
417:
415:
406:
405:
401:
363:
362:
355:
340:
319:
318:
305:
267:
266:
257:
252:
243:
230:
217:
206:
197:
188:
159:
129:
112:
104:oligonucleotide
51:
12:
11:
5:
1554:
1552:
1544:
1543:
1533:
1532:
1528:
1527:
1504:
1503:
1501:
1500:
1495:
1490:
1484:
1482:
1474:
1473:
1471:
1470:
1464:
1458:
1452:
1446:
1440:
1434:
1429:
1424:
1418:
1412:
1406:
1399:
1397:
1386:
1385:
1383:
1382:
1377:
1372:
1367:
1362:
1357:
1352:
1347:
1338:
1332:
1330:
1324:
1323:
1321:
1320:
1317:
1314:
1313:
1312:
1307:
1302:
1294:
1293:Initialization
1290:
1288:
1284:
1283:
1277:
1275:
1274:
1267:
1260:
1252:
1244:
1243:
1222:(6): 293–296.
1202:
1183:(3): 167–171.
1167:
1110:
1073:(4): 462–464.
1057:
1048:
991:
981:
961:
932:(1): 259–260.
909:
849:
817:
788:
779:
719:
662:
600:
585:
567:
543:
533:. Thermofisher
522:
515:
486:
424:
399:
353:
338:
303:
270:Bio/Technology
254:
253:
251:
248:
242:
239:
229:
226:
216:
213:
196:
193:
187:
184:
158:
155:
128:
125:
111:
108:
98:manipulation.
71:DNA polymerase
50:
47:
39:DNA polymerase
30:Taq polymerase
13:
10:
9:
6:
4:
3:
2:
1553:
1542:
1539:
1538:
1536:
1526:
1521:
1516:
1512:
1499:
1496:
1494:
1491:
1489:
1486:
1485:
1483:
1479:
1475:
1468:
1465:
1462:
1459:
1456:
1453:
1450:
1447:
1444:
1441:
1438:
1435:
1433:
1432:Hot start PCR
1430:
1428:
1425:
1422:
1419:
1417:(inverse PCR)
1416:
1413:
1410:
1407:
1404:
1401:
1400:
1398:
1396:
1391:
1387:
1381:
1378:
1376:
1373:
1371:
1368:
1366:
1363:
1361:
1358:
1356:
1353:
1351:
1348:
1346:
1342:
1339:
1337:
1334:
1333:
1331:
1329:
1325:
1318:
1315:
1311:
1308:
1306:
1303:
1301:
1298:
1297:
1295:
1292:
1291:
1289:
1285:
1280:
1273:
1268:
1266:
1261:
1259:
1254:
1253:
1250:
1239:
1235:
1230:
1225:
1221:
1217:
1216:BioTechniques
1213:
1206:
1203:
1198:
1194:
1190:
1186:
1182:
1178:
1171:
1168:
1163:
1159:
1154:
1149:
1145:
1141:
1137:
1133:
1129:
1125:
1121:
1114:
1111:
1106:
1102:
1097:
1092:
1088:
1084:
1080:
1076:
1072:
1068:
1061:
1058:
1052:
1049:
1044:
1040:
1035:
1030:
1026:
1022:
1018:
1014:
1010:
1006:
1002:
995:
992:
985:
982:
971:
965:
962:
957:
953:
949:
945:
940:
935:
931:
927:
923:
916:
914:
910:
905:
901:
896:
891:
887:
883:
879:
875:
871:
867:
863:
856:
854:
850:
838:
834:
828:
826:
824:
822:
818:
807:
803:
797:
795:
793:
789:
783:
780:
775:
771:
766:
761:
757:
753:
749:
745:
742:(1): bpx011.
741:
737:
733:
726:
724:
720:
715:
711:
706:
701:
697:
693:
689:
685:
681:
677:
673:
666:
663:
658:
654:
649:
644:
640:
636:
632:
628:
624:
620:
616:
609:
607:
605:
601:
596:
592:
588:
586:9780128011577
582:
578:
571:
568:
557:
553:
547:
544:
532:
526:
523:
518:
516:9781402062414
512:
508:
504:
500:
493:
491:
487:
482:
478:
474:
470:
466:
462:
458:
454:
450:
446:
439:
437:
435:
433:
431:
429:
425:
414:
410:
403:
400:
395:
391:
387:
383:
379:
375:
371:
367:
360:
358:
354:
349:
345:
341:
339:9781607616283
335:
331:
327:
323:
316:
314:
312:
310:
308:
304:
299:
295:
291:
287:
283:
279:
275:
271:
264:
262:
260:
256:
249:
247:
240:
238:
236:
227:
225:
223:
214:
212:
208:
204:
200:
194:
192:
185:
183:
180:
176:
173:
169:
166:
162:
156:
154:
150:
146:
143:
139:
136:
132:
126:
124:
116:
109:
107:
105:
99:
97:
93:
88:
84:
81:
76:
75:primer dimers
72:
67:
63:
55:
48:
46:
42:
40:
36:
31:
26:
25:primer dimers
22:
18:
17:Hot start PCR
1493:Kjell Kleppe
1431:
1423:(nested PCR)
1390:Optimization
1300:Denaturation
1219:
1215:
1205:
1180:
1176:
1170:
1130:(1): 47–51.
1127:
1123:
1113:
1070:
1060:
1051:
1008:
1004:
994:
984:
964:
929:
925:
869:
865:
841:. Retrieved
839:. 2019-03-03
836:
809:. Retrieved
805:
782:
739:
735:
679:
675:
665:
625:(20): e131.
622:
618:
576:
570:
559:. Retrieved
555:
546:
535:. Retrieved
525:
498:
448:
444:
416:. Retrieved
412:
402:
369:
365:
321:
276:(5): 506–9.
273:
269:
244:
231:
218:
209:
205:
201:
198:
189:
181:
177:
174:
170:
167:
163:
161:Preheating:
160:
151:
147:
144:
140:
137:
133:
130:
121:
100:
89:
85:
60:
43:
35:denaturation
16:
15:
1498:Alice Chien
1488:Kary Mullis
1011:(21): e94.
241:Limitations
235:specificity
222:precipitate
138:Wax beads:
64:(PCR) is a
1481:and people
1457:(COLD-PCR)
1328:Polymerase
1319:Final hold
1310:Elongation
1281:techniques
843:2020-05-29
811:2020-05-29
561:2019-10-09
537:2019-10-03
418:2020-05-29
250:References
228:Advantages
168:Freezing:
49:Background
1305:Annealing
1287:Procedure
1144:0887-8013
1087:1364-548X
1025:0305-1048
948:0261-3166
886:0003-2700
756:2396-8923
696:0305-1048
639:1362-4962
595:960448665
386:1940-3402
92:cofactors
1535:Category
1439:(OE-PCR)
1411:(RT-PCR)
1395:variants
1238:25967906
1197:12219733
1162:11835531
1105:18188468
1043:11058144
956:18776352
904:19438248
774:32161793
714:14576300
657:18796527
394:29717052
348:20301005
80:reagents
1525:Biology
1478:History
1463:(dePCR)
1153:6808141
1096:3760149
895:2712722
765:6994073
648:2582603
481:4267296
473:8632804
453:Bibcode
298:2885453
290:7764710
149:react.
110:Methods
1511:Portal
1469:(RAPD)
1451:(MLPA)
1405:(qPCR)
1336:Klenow
1296:Cycle
1236:
1195:
1160:
1150:
1142:
1103:
1093:
1085:
1041:
1034:113163
1031:
1023:
976:
954:
946:
902:
892:
884:
772:
762:
754:
712:
705:275455
702:
694:
655:
645:
637:
593:
583:
513:
479:
471:
445:Nature
392:
384:
346:
336:
296:
288:
477:S2CID
294:S2CID
1393:and
1365:Vent
1343:and
1234:PMID
1193:PMID
1158:PMID
1140:ISSN
1101:PMID
1083:ISSN
1039:PMID
1021:ISSN
952:PMID
944:ISSN
900:PMID
882:ISSN
770:PMID
752:ISSN
710:PMID
692:ISSN
653:PMID
635:ISSN
591:OCLC
581:ISBN
511:ISBN
469:PMID
390:PMID
382:ISSN
370:2018
344:PMID
334:ISBN
286:PMID
1380:Tfu
1375:Tli
1370:Pwo
1360:Pfu
1355:Tth
1350:Taq
1224:doi
1185:doi
1148:PMC
1132:doi
1091:PMC
1075:doi
1029:PMC
1013:doi
934:doi
890:PMC
874:doi
760:PMC
744:doi
700:PMC
684:doi
643:PMC
627:doi
503:doi
461:doi
449:381
374:doi
326:doi
278:doi
1537::
1345:T7
1341:T4
1232:.
1220:61
1218:.
1214:.
1191:.
1181:16
1179:.
1156:.
1146:.
1138:.
1128:16
1126:.
1122:.
1099:.
1089:.
1081:.
1069:.
1037:.
1027:.
1019:.
1009:28
1007:.
1003:.
950:.
942:.
930:52
928:.
924:.
912:^
898:.
888:.
880:.
870:81
868:.
864:.
852:^
835:.
820:^
804:.
791:^
768:.
758:.
750:.
738:.
734:.
722:^
708:.
698:.
690:.
680:31
678:.
674:.
651:.
641:.
633:.
623:36
621:.
617:.
603:^
589:.
554:.
509:.
489:^
475:.
467:.
459:.
447:.
427:^
411:.
388:.
380:.
368:.
356:^
342:.
332:.
306:^
292:.
284:.
274:12
272:.
258:^
237:.
1513::
1271:e
1264:t
1257:v
1240:.
1226::
1199:.
1187::
1164:.
1134::
1107:.
1077::
1045:.
1015::
958:.
936::
906:.
876::
846:.
814:.
776:.
746::
740:2
716:.
686::
659:.
629::
597:.
564:.
540:.
519:.
505::
483:.
463::
455::
421:.
396:.
376::
350:.
328::
300:.
280::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.