164:
polymerizing activity also at lower temperatures, which can cause DNA synthesis from primers after annealing to each other. Several methods have been developed to prevent PDs formation until the reaction reaches working temperature (60-70 °C), and these include initial inhibition of the DNA polymerase, or physical separation of reaction components reaction until the reaction mixture reaches the higher temperatures. These methods are referred to as
579:
296::also known as SAMRS, eliminating primer dimers by introducing nucleotide analogues T*, A*, G* and C* into the primer. The SAMRS DNA could bind to natural DNA, but not to other members of the same SAMRS species. For example, T* could bind to A but not A*, and A* could bind to T but not T*. Thus, through careful design, primers build from SAMRS could avoid primer-primer interactions and allowing sensitive SNP detection as well as multiplex PCR.
290:(rhPCR), utilizes a thermostable RNase HII to remove a blocking group from the PCR primers at high temperature. This RNase HII enzyme displays almost no activity at low temperature, making the removal of the block only occur at high temperature. The enzyme also possess inherent primer:template mismatch discrimination, resulting in additional selection against primer-dimers.
64:
314:: used when working with nonspecific dyes, such as SYBR Green I. It is based on the different length, and hence, different melting temperature of the PDs and the target sequence. In this method the signal is acquired below the melting temperature of the target sequence, but above the melting temperature of the PDs.
163:
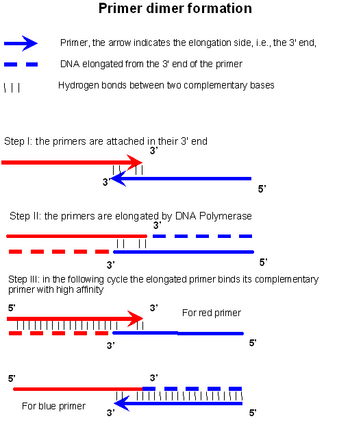
Because primers are designed to have low complementarity to each other, they may anneal (step I in the figure) only at low temperature, e.g. room temperature, such as during the preparation of the reaction mixture. Although DNA polymerases used in PCR are most active around 70 °C, they have some
131:
and temperature of the reaction. This method is somewhat limited by the physical-chemical characteristics that also determine the efficiency of amplification of the target sequence in the PCR. Therefore, reducing PDs formation may also result in reduced PCR efficiency. To overcome this limitation,
144:
formation and annealing of primers to itself or within primer pairs. Physical parameters that are taken into account by the software are potential self-complementarity and GC content of the primers; similar melting temperatures of the primers; and absence of secondary structures, such as
280:. The melting temperature of a chimeric sequence with another chimeric sequence is lower than that of chimeric sequence with DNA. This difference enables setting the annealing temperature such that the primer will anneal to its target sequence, but not to other chimeric primers.
111:, a nonspecific dye for detection of double-stranded DNA. Because they usually consist of short sequences, the PDs denature at lower temperature than the target sequence and hence can be distinguished by their melting-curve characteristics.
518:
496:
485:
474:
507:
79:
at the 3' ends and length of the overlap. The third step occurs in the next cycle, when a single strand of the product of step II is used as a template to which fresh primers anneal leading to synthesis of more PD product.
328:
probes generate signal only in the presence of their target (complementary) sequence, and this enhanced specificity precludes signal acquisition (but not possible inhibitory effects on product accumulation) from PDs.
265:
ystem): a nucleotide tail, complementary to the 3' end of the primer is added to the 5' end of the primer. Because of the close proximity of the 5' tail it anneals to the 3' end of the primer. The result is a
180:: DNA polymerase requires magnesium ions for activity, so the magnesium is chemically separated from the reaction by binding to a chemical compound, and is released into the solution only at high temperature
230:
of the DNA polymerase. The small molecule is released from the enzyme by incubation of the reaction mixture for 10–15 minutes at 95 °C. Once the small molecule is released, the enzyme is activated.
96:-stained gels are typically seen as a 30-50 base-pair (bp) band or smear of moderate to high intensity and distinguishable from the band of the target sequence, which is typically longer than 50 bp.
75:
will bind and extend the primers according to the complementary sequence (step II in the figure). An important factor contributing to the stability of the construct in step I is a high
71:
A primer dimer is formed and amplified in three steps. In the first step, two primers anneal at their respective 3' ends (step I in the figure). If this construct is stable enough, the
202:
bound to the enzyme at low temperature and inhibit its activity. After an incubation of 1–5 minutes at 95 °C, the inhibitor is released and the reaction starts.
239:
Another approach to prevent or reduce PD formation is by modifying the primers so that annealing with themselves or each other does not cause extension.
270:
primer that excludes annealing involving shorter overlaps, but permits annealing of the primer to its fully complementary sequence in the target.
174:: in this method the enzyme is spatially separated from the reaction mixture by wax that melts when the reaction reaches high temperature.
37:
119:
One approach to prevent PDs consists of physical-chemical optimization of the PCR system, i.e. changing the concentrations of primers,
811:
132:
other methods aim to reduce the formation of PDs only, including primer design, and use of different PCR enzyme systems or reagents.
601:"RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers"
816:
530:
Brownie, Jannine; Shawcross, Susan; Theaker, Jane; Whitcombe, David; Ferrie, Richard; Newton, Clive; Little, Stephen (1997).
308:. This approach is useful as long as there are few PDs formed and their inhibitory effect on product accumulation is minor.
304:
While the methods above are designed to reduce PD formation, another approach aims to minimize signal generated from PDs in
770:
801:
29:
25:
47:
amplifies the PD, leading to competition for PCR reagents, thus potentially inhibiting amplification of the
806:
104:
762:
701:"Elimilating primer dimers and improving SNP detection using self-avoiding molecular recognition systems"
287:
440:(2004). "Critical role of magnesium ions in DNA polymerase beta's closing and active site assembly".
389:"Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications"
199:
141:
89:
33:
785:
120:
730:
681:
632:
561:
457:
418:
720:
712:
671:
663:
622:
612:
551:
543:
449:
408:
400:
325:
305:
100:
93:
63:
52:
747:
725:
700:
676:
651:
627:
600:
128:
72:
44:
599:
Dobosy JR, Rose SD, Beltz KR, Rupp SM, Powers KM, Behlke MA, Walder JA (August 2011).
556:
531:
413:
388:
795:
437:
387:
Chou, Quin; Russell, Marion; Birch, David E.; Raymond, Jonathan; Bloch, Will (1992).
215:
158:
108:
28:(PCR), a common biotechnological method. As its name implies, a PD consists of two
716:
699:
Yang ZY, Le JT, Hutter D, Bradley KM, Overton BR, McLendon C, Benner SA (2020).
584:
227:
124:
777:
223:
219:
76:
40:
547:
617:
404:
267:
146:
734:
685:
667:
636:
461:
208:: is a modified DNA polymerase with almost no activity at low temperature.
140:
Primer-design software uses algorithms that check for the potential of DNA
652:"Artificial Generic Systems: Self-Avoiding ENA in PCR and Multiplexed PCR"
565:
422:
360:
191:
781:
276:: some DNA bases in the primer are replaced with RNA bases, creating a
195:
187:
453:
436:
Yang, Linjing; Arora, Karunesh; Beard, William A.; Wilson, Samuel H.;
321:
580:"Chimeric primers for improved nucleic acid amplification reactions"
62:
48:
67:
Primer dimer is formed and amplified in a three-step process
361:
The primer design page of Leiden
University Medical Center
374:
Polymerase Chain
Reaction: Techniques and Applications
532:"The elimination of primer-dimer accumulation in PCR"
350:(6th ed.). Garland Science. pp. 708–711.
55:, PDs may interfere with accurate quantification.
300:Preventing signal acquisition from primer dimers
650:Hoshika S, Chen F, Leal NA, Benner SA (2020).
763:"Online software for primer dimer prediction"
8:
294:Self-Avoiding molecular recognition systems
51:sequence targeted for PCR amplification. In
778:"Primer design. What is the primer-dimer?"
724:
675:
626:
616:
555:
475:US Patent application number 2007/0254327
412:
442:Journal of the American Chemical Society
338:
36:) to each other because of strings of
376:. Scientific Press. pp. 595–599.
214:: in this method a small molecule is
7:
235:Structural modifications of primers
88:Primer dimers may be visible after
24:) is a potential by-product in the
14:
184:Non-covalent binding of inhibitor
115:Preventing primer-dimer formation
107:with intercalating dyes, such as
43:in the primers. As a result, the
788:from the original on 2021-12-20.
149:, in the DNA target sequence.
32:molecules that have attached (
1:
705:Biology Methods and Protocols
348:Molecular Biology of the Cell
346:Alberts; et al. (2017).
206:Cold-sensitive Taq polymerase
771:Integrated DNA Technologies
92:of the PCR product. PDs in
833:
717:10.1093/biomethods/bpaa004
156:
812:Polymerase chain reaction
284:Blocked-cleavable primers
178:Slow release of magnesium
103:, PDs may be detected by
26:polymerase chain reaction
784:video. 3 November 2013.
519:US Patent number 5677152
508:US Patent number 6214557
497:US Patent number 6183967
486:US Patent number 5338671
318:Sequence-specific probes
618:10.1186/1472-6750-11-80
668:10.1002/ange.201001977
548:10.1093/nar/25.16.3235
536:Nucleic Acids Research
393:Nucleic Acids Research
136:Primer-design software
105:melting curve analysis
68:
59:Mechanism of formation
817:Laboratory techniques
405:10.1093/nar/20.7.1717
372:Patel, Ewing (2008).
288:RNase H-dependent PCR
212:Chemical modification
66:
286:: a method known as
186:: in this method a
142:secondary structure
90:gel electrophoresis
121:magnesium chloride
69:
802:Molecular biology
767:OligoAnalyzer 3.1
662:(32): 5686–5689.
605:BMC Biotechnology
454:10.1021/ja049412o
278:chimeric sequence
824:
789:
774:
750:
745:
739:
738:
728:
696:
690:
689:
679:
647:
641:
640:
630:
620:
596:
590:
589:
576:
570:
569:
559:
527:
521:
516:
510:
505:
499:
494:
488:
483:
477:
472:
466:
465:
433:
427:
426:
416:
384:
378:
377:
369:
363:
358:
352:
351:
343:
326:molecular beacon
306:quantitative PCR
274:Chimeric primers
216:covalently bound
101:quantitative PCR
94:ethidium bromide
53:quantitative PCR
832:
831:
827:
826:
825:
823:
822:
821:
792:
791:
776:
775:
761:
759:
754:
753:
746:
742:
698:
697:
693:
649:
648:
644:
598:
597:
593:
578:
577:
573:
542:(16): 3235–41.
529:
528:
524:
517:
513:
506:
502:
495:
491:
484:
480:
473:
469:
448:(27): 8441–53.
435:
434:
430:
386:
385:
381:
371:
370:
366:
359:
355:
345:
344:
340:
335:
302:
237:
161:
155:
138:
117:
86:
61:
12:
11:
5:
830:
828:
820:
819:
814:
809:
804:
794:
793:
758:
757:External links
755:
752:
751:
748:Four steps PCR
740:
711:(1): bpaa004.
691:
642:
591:
571:
522:
511:
500:
489:
478:
467:
438:Schlick, Tamar
428:
399:(7): 1717–23.
379:
364:
353:
337:
336:
334:
331:
312:Four steps PCR
301:
298:
236:
233:
200:non-covalently
157:Main article:
154:
151:
137:
134:
129:ionic strength
116:
113:
85:
82:
73:DNA polymerase
60:
57:
45:DNA polymerase
13:
10:
9:
6:
4:
3:
2:
829:
818:
815:
813:
810:
808:
807:Biotechnology
805:
803:
800:
799:
797:
790:
787:
783:
779:
772:
768:
764:
756:
749:
744:
741:
736:
732:
727:
722:
718:
714:
710:
706:
702:
695:
692:
687:
683:
678:
673:
669:
665:
661:
657:
653:
646:
643:
638:
634:
629:
624:
619:
614:
610:
606:
602:
595:
592:
587:
586:
581:
575:
572:
567:
563:
558:
553:
549:
545:
541:
537:
533:
526:
523:
520:
515:
512:
509:
504:
501:
498:
493:
490:
487:
482:
479:
476:
471:
468:
463:
459:
455:
451:
447:
443:
439:
432:
429:
424:
420:
415:
410:
406:
402:
398:
394:
390:
383:
380:
375:
368:
365:
362:
357:
354:
349:
342:
339:
332:
330:
327:
323:
319:
315:
313:
309:
307:
299:
297:
295:
291:
289:
285:
281:
279:
275:
271:
269:
264:
260:
256:
252:
248:
244:
240:
234:
232:
229:
225:
221:
217:
213:
209:
207:
203:
201:
197:
193:
189:
185:
181:
179:
175:
173:
169:
167:
166:hot-start PCR
160:
159:Hot start PCR
153:Hot-start PCR
152:
150:
148:
143:
135:
133:
130:
126:
122:
114:
112:
110:
106:
102:
97:
95:
91:
83:
81:
78:
74:
65:
58:
56:
54:
50:
46:
42:
39:
38:complementary
35:
31:
27:
23:
19:
766:
760:
743:
708:
704:
694:
659:
655:
645:
608:
604:
594:
583:
574:
539:
535:
525:
514:
503:
492:
481:
470:
445:
441:
431:
396:
392:
382:
373:
367:
356:
347:
341:
317:
316:
311:
310:
303:
293:
292:
283:
282:
277:
273:
272:
262:
258:
254:
250:
246:
242:
241:
238:
211:
210:
205:
204:
183:
182:
177:
176:
171:
170:
165:
162:
139:
118:
109:SYBR Green I
98:
87:
70:
21:
18:primer dimer
17:
15:
656:Angew. Chem
585:Patent Lens
228:active site
125:nucleotides
796:Categories
333:References
224:amino acid
220:side chain
147:stem-loops
77:GC-content
34:hybridized
268:stem-loop
84:Detection
786:Archived
735:32395633
686:20586087
637:21831278
462:15238001
253:ssisted
249:omo-Tag
192:antibody
782:YouTube
726:7200914
677:6027612
628:3224242
566:9241236
423:1579465
226:in the
218:to the
196:aptamer
188:peptide
733:
723:
684:
674:
635:
625:
611:: 80.
564:
557:146890
554:
460:
421:
414:312262
411:
322:TaqMan
222:of an
30:primer
261:imer
243:HANDS
41:bases
731:PMID
682:PMID
633:PMID
562:PMID
458:PMID
419:PMID
324:and
198:are
721:PMC
713:doi
672:PMC
664:doi
660:122
623:PMC
613:doi
552:PMC
544:doi
450:doi
446:126
409:PMC
401:doi
257:on-
194:or
172:Wax
99:In
49:DNA
798::
780:.
769:.
765:.
729:.
719:.
707:.
703:.
680:.
670:.
658:.
654:.
631:.
621:.
609:11
607:.
603:.
582:.
560:.
550:.
540:25
538:.
534:.
456:.
444:.
417:.
407:.
397:20
395:.
391:.
320::
190:,
168:.
127:,
123:,
22:PD
16:A
773:.
737:.
715::
709:5
688:.
666::
639:.
615::
588:.
568:.
546::
464:.
452::
425:.
403::
263:S
259:D
255:N
251:A
247:H
245:(
20:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.