86:
72:
YxxL/I). However, it is worth noting that in various sources, this consensus sequence differs, mainly in the number of amino acids between individual signatures. Apart from ITAMs which have the structure described above, there is also a variety of proteins containing ITAM-like motifs, which have a
499:
Suzuki-Inoue, Katsue; Fuller, Gemma L. J.; García, Angel; Eble, Johannes A.; Pöhlmann, Stefan; Inoue, Osamu; Gartner, T. Kent; Hughan, Sascha C.; Pearce, Andrew C.; Laing, Gavin D.; Theakston, R. David G. (2006-01-15). "A novel Syk-dependent mechanism of platelet activation by the C-type lectin
145:, resulting mostly in the activation of given cell. Paradoxically, in some cases, ITAMs and ITAM-like motifs do not have an activating effect, but rather an inhibitory one. Exact mechanisms of this phenomenon are as of yet not elucidated.
392:
Rogers, Neil C.; Slack, Emma C.; Edwards, Alexander D.; Nolte, Martijn A.; Schulz, Oliver; Schweighoffer, Edina; Williams, David L.; Gordon, Siamon; Tybulewicz, Victor L.; Brown, Gordon D.; Reis e Sousa, Caetano (April 2005).
597:
Pasquier, Benoit; Launay, Pierre; Kanamaru, Yutaka; Moura, Ivan C.; Pfirsch, Séverine; Ruffié, Claude; Hénin, Dominique; Benhamou, Marc; Pretolani, Marina; Blank, Ulrich; Monteiro, Renato C. (January 2005).
180:
Examples shown below list both proteins that contain the ITAM themselves and proteins that use ITAM-based signalling with the help of associated proteins which contain the motif.
137:, usually two domains in tandem, inducing a signaling cascade mediated by Syk family kinases (which are the primary proteins that bind to phosphorylated ITAMs), namely either
647:
O’Neill, Shannon K.; Getahun, Andrew; Gauld, Stephen B.; Merrell, Kevin T.; Tamir, Idan; Smith, Mia J.; Dal Porto, Joseph M.; Li, Quan-Zhen; Cambier, John C. (2011-11-23).
704:
Pfirsch-Maisonnas, Séverine; Aloulou, Meryem; Xu, Ting; Claver, Julien; Kanamaru, Yutaka; Tiwari, Meetu; Launay, Pierre; Monteiro, Renato C.; Blank, Ulrich (2011-04-19).
275:
Humphrey, Mary Beth; Daws, Michael R.; Spusta, Steve C.; Niemi, Eréne C.; Torchia, James A.; Lanier, Lewis L.; Seaman, William E.; Nakamura, Mary C. (February 2006).
29:
149:
333:
Paloneva, Juha; Mandelin, Jami; Kiialainen, Anna; Böhling, Tom; Prudlo, Johannes; Hakola, Panu; Haltia, Matti; Konttinen, Yrjö T.; Peltonen, Leena (2003-08-18).
191:
68:, giving the signature YxxL/I. Two of these signatures are typically separated by between 6 and 8 amino acids in the cytoplasmic tail of the molecule (YxxL/Ix
172:
Rare human genetic mutations are catalogued in the human genetic variation databases which can reportedly result in creation or deletion of ITIM and ITAMs.
164:. This serves not only for inhibition and regulation of signalling pathways related to ITAM-based signalling, but also for termination of signalling.
649:"Monophosphorylation of CD79a and b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy"
260:
101:
ITAMs are important for signal transduction, mainly in immune cells. They are found in the cytoplasmic tails of non-catalytic tyrosine-
40:
pathway and subsequently the activation of immune cells, although different functions have been described, for example an
707:"Inhibitory ITAM Signaling Traps Activating Receptors with the Phosphatase SHP-1 to Form Polarized "Inhibisome" Clusters"
152:) that, when phosphorylated, results in the inhibition of the signaling pathway via recruitment of phosphatases, namely
138:
600:"Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM"
395:"Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins"
1158:
1163:
554:
Dushek O, Goyette J, van der Merwe PA (November 2012). "Non-catalytic tyrosine-phosphorylated receptors".
444:"Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production"
276:
1108:
1051:
945:
1038:
Cummings BB, Karczewski KJ, Kosmicki JA, Seaby EG, Watts NA, Singer-Berk M, et al. (May 2020).
914:
743:
579:
533:
335:"DAP12/TREM2 Deficiency Results in Impaired Osteoclast Differentiation and Osteoporotic Features"
315:
97:
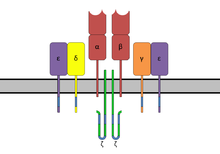
and ζ-chain accessory molecules. ITAMs are represented in blue on the tails of the CD3 subunits.
1134:
1077:
1020:
971:
906:
898:
859:
841:
800:
782:
735:
727:
686:
668:
629:
621:
571:
525:
517:
481:
463:
424:
416:
374:
356:
307:
299:
256:
126:
1124:
1116:
1067:
1059:
1010:
1002:
961:
953:
932:
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. (October 2015).
890:
849:
831:
790:
774:
719:
676:
660:
611:
563:
509:
471:
455:
442:
Underhill, David M.; Rossnagle, Eddie; Lowell, Clifford A.; Simmons, Randi M. (2005-10-01).
406:
364:
346:
291:
33:
1097:"TraPS-VarI: Identifying genetic variants altering phosphotyrosine based signalling motifs"
989:
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (January 2001).
820:"Termination of Immune Activation: An Essential Component of Healthy Host Immune Responses"
148:
Other non-catalytic tyrosine-phosphorylated receptors carry a conserved inhibitory motif (
114:
102:
1112:
1055:
949:
706:
1129:
1096:
1072:
1039:
966:
933:
854:
819:
795:
762:
681:
648:
476:
443:
369:
334:
277:"TREM2, a DAP12-associated receptor, regulates osteoclast differentiation and function"
203:
37:
818:
Kane, Barry A.; Bryant, Katherine J.; McNeil, H. Patrick; Tedla, Nicodemus T. (2014).
1152:
1015:
990:
894:
778:
747:
918:
319:
583:
106:
94:
36:. Its major role is being an integral component for the initiation of a variety of
537:
664:
616:
599:
411:
394:
122:
1120:
513:
459:
1063:
723:
134:
133:. Phosphorylated ITAMs serve as docking sites for other proteins containing a
110:
65:
61:
41:
25:
1040:"Transcript expression-aware annotation improves rare variant interpretation"
902:
845:
786:
731:
672:
625:
521:
467:
420:
360:
303:
878:
1138:
1081:
1024:
1006:
975:
910:
863:
804:
739:
690:
633:
575:
529:
485:
428:
378:
311:
351:
53:
957:
295:
125:. The tyrosine residues within these motifs become phosphorylated by
85:
57:
879:"Physiological mechanisms of signal termination in biological systems"
836:
567:
219:
215:
195:
161:
157:
130:
74:
121:-alpha and -beta chains of the B cell receptor complex, and certain
763:"Negative signaling by inhibitory receptors: the NK cell paradigm"
153:
142:
84:
227:
223:
211:
207:
199:
187:
183:
118:
253:
Basic
Immunology: Functions and Disorders of the Immune System
129:
following interaction of the receptor molecules with their
877:
Ligeti, E.; Csépányi-Kömi, R.; Hunyady, L. (April 2012).
73:
very similar structure and function (for example in
28:that is repeated twice in the cytoplasmic tails of
705:
934:"A global reference for human genetic variation"
991:"dbSNP: the NCBI database of genetic variation"
30:non-catalytic tyrosine-phosphorylated receptors
18:immunoreceptor tyrosine-based activation motif
8:
549:
547:
255:(3 ed.), Philadelphia, PA: Saunders,
1128:
1071:
1014:
965:
853:
835:
794:
680:
615:
475:
410:
368:
350:
32:, cell-surface proteins found mainly on
246:
244:
242:
238:
7:
284:Journal of Bone and Mineral Research
24:) is a conserved sequence of four
14:
895:10.1111/j.1748-1716.2012.02414.x
779:10.1111/j.1600-065X.2008.00660.x
339:Journal of Experimental Medicine
251:Abbas AK, Lichtman AH (2009),
1:
761:Long, Eric O. (August 2008).
93:with TCR-α and TCR-β chains,
665:10.1016/j.immuni.2011.10.011
617:10.1016/j.immuni.2004.11.017
412:10.1016/j.immuni.2005.03.004
1095:Ulaganathan VK (May 2020).
1180:
1121:10.1038/s41598-020-65146-2
824:Journal of Innate Immunity
514:10.1182/blood-2005-05-1994
460:10.1182/blood-2005-03-1239
1064:10.1038/s41586-020-2329-2
724:10.1126/scisignal.2001309
91:T-cell receptor complex
995:Nucleic Acids Research
105:receptors such as the
98:
767:Immunological Reviews
556:Immunological Reviews
88:
52:The motif contains a
1007:10.1093/nar/29.1.308
352:10.1084/jem.20030027
1113:2020NatSR..10.8453U
1056:2020Natur.581..452C
958:10.1038/nature15393
950:2015Natur.526...68T
296:10.1359/JBMR.051016
1101:Scientific Reports
500:receptor CLEC-2".
168:Genetic variations
127:Src family kinases
99:
1050:(7809): 452–458.
883:Acta Physiologica
837:10.1159/000363449
712:Science Signaling
568:10.1111/imr.12008
262:978-1-4160-4688-2
64:by any two other
56:separated from a
1171:
1143:
1142:
1132:
1092:
1086:
1085:
1075:
1035:
1029:
1028:
1018:
986:
980:
979:
969:
929:
923:
922:
874:
868:
867:
857:
839:
815:
809:
808:
798:
758:
752:
751:
709:
701:
695:
694:
684:
644:
638:
637:
619:
594:
588:
587:
551:
542:
541:
496:
490:
489:
479:
454:(7): 2543–2550.
439:
433:
432:
414:
389:
383:
382:
372:
354:
330:
324:
323:
281:
272:
266:
265:
248:
1179:
1178:
1174:
1173:
1172:
1170:
1169:
1168:
1149:
1148:
1147:
1146:
1094:
1093:
1089:
1037:
1036:
1032:
988:
987:
983:
944:(7571): 68–74.
931:
930:
926:
876:
875:
871:
817:
816:
812:
760:
759:
755:
703:
702:
698:
646:
645:
641:
596:
595:
591:
553:
552:
545:
498:
497:
493:
441:
440:
436:
391:
390:
386:
332:
331:
327:
279:
274:
273:
269:
263:
250:
249:
240:
235:
178:
170:
115:T cell receptor
83:
71:
50:
12:
11:
5:
1177:
1175:
1167:
1166:
1161:
1159:Cell signaling
1151:
1150:
1145:
1144:
1087:
1030:
981:
924:
889:(4): 469–478.
869:
830:(6): 727–738.
810:
753:
696:
659:(5): 746–756.
639:
589:
543:
508:(2): 542–549.
491:
434:
405:(4): 507–517.
384:
345:(4): 669–675.
325:
290:(2): 237–245.
267:
261:
237:
236:
234:
231:
177:
174:
169:
166:
103:phosphorylated
82:
79:
69:
49:
46:
13:
10:
9:
6:
4:
3:
2:
1176:
1165:
1164:Immune system
1162:
1160:
1157:
1156:
1154:
1140:
1136:
1131:
1126:
1122:
1118:
1114:
1110:
1106:
1102:
1098:
1091:
1088:
1083:
1079:
1074:
1069:
1065:
1061:
1057:
1053:
1049:
1045:
1041:
1034:
1031:
1026:
1022:
1017:
1012:
1008:
1004:
1001:(1): 308–11.
1000:
996:
992:
985:
982:
977:
973:
968:
963:
959:
955:
951:
947:
943:
939:
935:
928:
925:
920:
916:
912:
908:
904:
900:
896:
892:
888:
884:
880:
873:
870:
865:
861:
856:
851:
847:
843:
838:
833:
829:
825:
821:
814:
811:
806:
802:
797:
792:
788:
784:
780:
776:
772:
768:
764:
757:
754:
749:
745:
741:
737:
733:
729:
725:
721:
718:(169): ra24.
717:
713:
708:
700:
697:
692:
688:
683:
678:
674:
670:
666:
662:
658:
654:
650:
643:
640:
635:
631:
627:
623:
618:
613:
609:
605:
601:
593:
590:
585:
581:
577:
573:
569:
565:
562:(1): 258–76.
561:
557:
550:
548:
544:
539:
535:
531:
527:
523:
519:
515:
511:
507:
503:
495:
492:
487:
483:
478:
473:
469:
465:
461:
457:
453:
449:
445:
438:
435:
430:
426:
422:
418:
413:
408:
404:
400:
396:
388:
385:
380:
376:
371:
366:
362:
358:
353:
348:
344:
340:
336:
329:
326:
321:
317:
313:
309:
305:
301:
297:
293:
289:
285:
278:
271:
268:
264:
258:
254:
247:
245:
243:
239:
232:
230:
229:
225:
221:
217:
213:
209:
205:
201:
197:
193:
189:
185:
181:
175:
173:
167:
165:
163:
159:
155:
151:
146:
144:
140:
136:
132:
128:
124:
120:
117:complex, the
116:
112:
108:
104:
96:
92:
87:
80:
78:
76:
67:
63:
59:
55:
47:
45:
43:
39:
35:
31:
27:
23:
19:
1104:
1100:
1090:
1047:
1043:
1033:
998:
994:
984:
941:
937:
927:
886:
882:
872:
827:
823:
813:
770:
766:
756:
715:
711:
699:
656:
652:
642:
610:(1): 31–42.
607:
603:
592:
559:
555:
505:
501:
494:
451:
447:
437:
402:
398:
387:
342:
338:
328:
287:
283:
270:
252:
182:
179:
171:
147:
123:Fc receptors
100:
90:
51:
44:maturation.
34:immune cells
21:
17:
15:
1107:(1): 8453.
66:amino acids
26:amino acids
1153:Categories
233:References
135:SH2 domain
77:protein).
62:isoleucine
42:osteoclast
903:1748-1716
846:1662-811X
787:1600-065X
773:: 70–84.
748:206670699
732:1945-0877
673:1074-7613
626:1074-7613
522:0006-4971
468:0006-4971
421:1074-7613
361:0022-1007
304:0884-0431
198:(DAP12),
48:Structure
38:signaling
1139:32439998
1082:32461655
1025:11125122
976:26432245
919:13455093
911:22260256
864:25033984
805:18759921
740:21505186
691:22078222
653:Immunity
634:15664157
604:Immunity
576:23046135
530:16174766
486:15956283
429:15845454
399:Immunity
379:12925681
320:34957715
312:16418779
216:Dectin-1
176:Examples
111:ζ-chains
81:Function
75:Dectin-1
54:tyrosine
1130:7242328
1109:Bibcode
1073:7334198
1052:Bibcode
967:4750478
946:Bibcode
855:6741560
796:2587243
682:3232011
584:1549902
477:1895265
370:2194176
212:FcγRIII
131:ligands
113:of the
58:leucine
1137:
1127:
1080:
1070:
1044:Nature
1023:
1013:
974:
964:
938:Nature
917:
909:
901:
862:
852:
844:
803:
793:
785:
746:
738:
730:
689:
679:
671:
632:
624:
582:
574:
538:168505
536:
528:
520:
484:
474:
466:
427:
419:
377:
367:
359:
318:
310:
302:
259:
220:CLEC-1
208:FcγRII
196:TYROBP
143:ZAP-70
1016:29783
915:S2CID
744:S2CID
580:S2CID
534:S2CID
502:Blood
448:Blood
316:S2CID
280:(PDF)
204:FcγRI
200:FcαRI
162:SHIP1
158:SHP-2
154:SHP-1
70:(6-8)
1135:PMID
1078:PMID
1021:PMID
972:PMID
907:PMID
899:ISSN
860:PMID
842:ISSN
801:PMID
783:ISSN
736:PMID
728:ISSN
687:PMID
669:ISSN
630:PMID
622:ISSN
572:PMID
526:PMID
518:ISSN
482:PMID
464:ISSN
425:PMID
417:ISSN
375:PMID
357:ISSN
308:PMID
300:ISSN
257:ISBN
228:CD72
224:CD28
192:CD3ε
188:CD3δ
184:CD3γ
160:and
150:ITIM
119:CD79
109:and
89:The
22:ITAM
1125:PMC
1117:doi
1068:PMC
1060:doi
1048:581
1011:PMC
1003:doi
962:PMC
954:doi
942:526
891:doi
887:204
850:PMC
832:doi
791:PMC
775:doi
771:224
720:doi
677:PMC
661:doi
612:doi
564:doi
560:250
510:doi
506:107
472:PMC
456:doi
452:106
407:doi
365:PMC
347:doi
343:198
292:doi
141:or
139:Syk
107:CD3
95:CD3
60:or
16:An
1155::
1133:.
1123:.
1115:.
1105:10
1103:.
1099:.
1076:.
1066:.
1058:.
1046:.
1042:.
1019:.
1009:.
999:29
997:.
993:.
970:.
960:.
952:.
940:.
936:.
913:.
905:.
897:.
885:.
881:.
858:.
848:.
840:.
826:.
822:.
799:.
789:.
781:.
769:.
765:.
742:.
734:.
726:.
714:.
710:.
685:.
675:.
667:.
657:35
655:.
651:.
628:.
620:.
608:22
606:.
602:.
578:.
570:.
558:.
546:^
532:.
524:.
516:.
504:.
480:.
470:.
462:.
450:.
446:.
423:.
415:.
403:22
401:.
397:.
373:.
363:.
355:.
341:.
337:.
314:.
306:.
298:.
288:21
286:.
282:.
241:^
226:,
222:,
218:,
214:,
210:,
206:,
202:,
194:,
190:,
186:,
156:,
1141:.
1119::
1111::
1084:.
1062::
1054::
1027:.
1005::
978:.
956::
948::
921:.
893::
866:.
834::
828:6
807:.
777::
750:.
722::
716:4
693:.
663::
636:.
614::
586:.
566::
540:.
512::
488:.
458::
431:.
409::
381:.
349::
322:.
294::
20:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.