107:
20:
271:
Conventional (up to 10 different oligonucleotides) existing molecule library serves as a starting point for the subsequent SELEX process. Selection, separation, and amplification using the mirror image of the target molecule is performed.
228:
420:
Wlotzka, Britta; Leva, Susanne; Eschgfäller, Bernd; Burmeister, Jens; Kleinjung, Frank; Kaduk, Christine; Muhn, Peter; Hess-Stumpp, Holger; Klussmann, Sven (June 2002).
326:-RNA aptamers has recently been demonstrated in diabetic nephropathy patients. They can also be used as diagnostic agents.
251:. In the case of peptides and small proteins that are produced synthetically, an enantiomer is made using synthetic
422:"In vivo properties of an anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class"
484:
Klussmann S, Nolte A, Bald R, Erdmann VA, Fürste JP (September 1996). "Mirror-image RNA that binds D-adenosine".
236:
315:
176:-RNA aptamers to their target molecules often lies in the pico to nanomolar range and is thus comparable to
133:
129:
543:
533:
137:
141:
433:
322:. They are currently in preclinical or clinical development. Proof-of-concept for an anti-CCL2/MCP-1
509:
194:-RNA aptamers have high stability in blood serum, since they are less susceptible to be cleaved
93:-RNA aptamers are considered potential drugs and are currently being tested in clinical trials.
259:. If the target is a larger protein molecule, beyond synthetic abilities, the enantiomer of an
538:
501:
461:
402:
493:
451:
441:
392:
381:"Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics"
356:
145:
66:
343:
Helmling, S.H.; Eulberg, D.E.; Maasch, C.M.; Buchner, K.B.; Klussmann, S.K. (July 2003).
437:
280:
The sequence of the oligonucleotide selected using SELEX is determined with the help of
281:
239:(PCR), used in SELEX. Therefore, the selection is done with mirrored target molecules.
456:
421:
284:. This information is used for the synthesis of the oligonucleotide's enantiomer, the
527:
513:
361:
344:
187:
202:
in a short time due to their low molar mass (which is below the renal threshold).
397:
380:
345:"RNA-Spiegelmers: a new substance class to efficiently inhibit peptide hormones"
426:
Proceedings of the
National Academy of Sciences of the United States of America
304:
256:
248:
195:
177:
149:
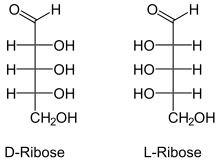
120:-ribose, are the enantiomers of natural oligonucleotides, which are made with
70:
209:
86:
465:
446:
406:
505:
497:
169:
165:
319:
260:
199:
78:
106:
311:
62:
19:
105:
18:
307:
235:-nucleic acids are not amenable to enzymatic methods, such as
54:
172:, and substances of low molecular weight. The affinity of
85:-nucleotides, they are highly resistant to degradation by
229:
systematic evolution of ligands by exponential enrichment
208:-RNA aptamers modified with a higher molar mass, such as
215:-RNA aptamers, show a prolonged plasma half-life.
247:The first step is the production of the target's
164:-RNA aptamers are able to bind molecules such as
374:
372:
479:
477:
475:
8:
227:-RNA aptamers are not directly made using
124:-ribose. Nucleic acid aptamers, including
455:
445:
396:
360:
303:-RNA aptamers have been obtained for the
335:
379:Vater A, Klussmann S (January 2015).
198:by enzymes. They are excreted by the
7:
186:-RNA aptamers themselves have low
14:
243:Reflection of the target molecule
190:. In contrast to other aptamers,
349:European Journal of Biochemistry
362:10.1111/j.1474-3833.2004.4119.x
1:
73:of natural oligonucleotides.
398:10.1016/j.drudis.2014.09.004
314:, the complement components
77:-RNA aptamers are a form of
116:-RNA aptamers, built using
65:units. It is an artificial
23:3-D structural model of an
560:
156:Biological characteristics
57:-like molecule built from
237:polymerase chain reaction
38:-ribonucleic acid aptamer
276:Sequencing and synthesis
128:-RNA aptamers, contain
223:Unlike other aptamers,
134:guanosine monophosphate
130:adenosine monophosphate
447:10.1073/pnas.132067399
138:cytidine monophosphate
110:
28:
160:Like other aptamers,
142:uridine monophosphate
109:
27:-RNA aptamer fragment
22:
498:10.1038/nbt0996-1112
385:Drug Discovery Today
288:-RNA aptamer, using
152:and a ribose sugar.
438:2002PNAS...99.8898W
102:Chemical properties
111:
69:named for being a
29:
325:
302:
291:
287:
254:
234:
226:
214:
207:
193:
185:
175:
163:
127:
123:
119:
115:
92:
84:
76:
60:
45:
36:
26:
16:RNA-like molecule
551:
518:
517:
481:
470:
469:
459:
449:
432:(13): 8898–902.
417:
411:
410:
400:
376:
367:
366:
364:
340:
323:
300:
289:
285:
252:
232:
224:
212:
205:
191:
183:
173:
161:
125:
121:
117:
113:
90:
82:
74:
58:
41:
32:
24:
559:
558:
554:
553:
552:
550:
549:
548:
524:
523:
522:
521:
486:Nat. Biotechnol
483:
482:
473:
419:
418:
414:
378:
377:
370:
342:
341:
337:
332:
298:
278:
269:
245:
221:
158:
146:phosphate group
104:
99:
81:. Due to their
67:oligonucleotide
17:
12:
11:
5:
557:
555:
547:
546:
541:
536:
526:
525:
520:
519:
471:
412:
391:(1): 147–155.
368:
334:
333:
331:
328:
297:
294:
292:-nucleotides.
282:DNA sequencing
277:
274:
268:
265:
244:
241:
220:
217:
196:hydrolytically
157:
154:
103:
100:
98:
95:
15:
13:
10:
9:
6:
4:
3:
2:
556:
545:
544:Biotechnology
542:
540:
537:
535:
534:Nucleic acids
532:
531:
529:
515:
511:
507:
503:
499:
495:
492:(9): 1112–5.
491:
487:
480:
478:
476:
472:
467:
463:
458:
453:
448:
443:
439:
435:
431:
427:
423:
416:
413:
408:
404:
399:
394:
390:
386:
382:
375:
373:
369:
363:
358:
354:
350:
346:
339:
336:
329:
327:
321:
317:
313:
309:
306:
295:
293:
283:
275:
273:
266:
264:
263:is produced.
262:
258:
250:
242:
240:
238:
230:
218:
216:
211:
203:
201:
197:
189:
181:
179:
171:
167:
155:
153:
151:
147:
143:
139:
135:
131:
108:
101:
96:
94:
88:
80:
72:
68:
64:
56:
52:
49:, trade name
48:
44:
39:
35:
21:
489:
485:
429:
425:
415:
388:
384:
352:
348:
338:
299:
279:
270:
246:
231:(SELEX), as
222:
204:
188:antigenicity
182:
159:
112:
71:mirror image
50:
47:-RNA aptamer
46:
42:
37:
33:
30:
257:amino acids
528:Categories
330:References
305:chemokines
249:enantiomer
219:Production
178:antibodies
150:nucleobase
51:Spiegelmer
210:PEGylated
87:nucleases
539:Peptides
514:41395593
466:12070349
407:25236655
170:proteins
166:peptides
97:Features
79:aptamers
53:) is an
506:9631061
434:Bibcode
320:ghrelin
261:epitope
200:kidneys
512:
504:
464:
457:124395
454:
405:
355:(S1).
312:CXCL12
63:ribose
510:S2CID
267:SELEX
502:PMID
462:PMID
403:PMID
318:and
310:and
308:CCL2
148:, a
144:, a
494:doi
452:PMC
442:doi
393:doi
357:doi
353:271
316:C5a
296:Use
55:RNA
31:An
530::
508:.
500:.
490:14
488:.
474:^
460:.
450:.
440:.
430:99
428:.
424:.
401:.
389:20
387:.
383:.
371:^
351:.
347:.
180:.
168:,
140:,
136:,
132:,
89:.
516:.
496::
468:.
444::
436::
409:.
395::
365:.
359::
324:L
301:L
290:L
286:L
255:-
253:D
233:L
225:L
213:L
206:L
192:L
184:L
174:L
162:L
126:L
122:D
118:L
114:L
91:L
83:L
75:L
61:-
59:L
43:L
40:(
34:L
25:L
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.