451:
varies between organisms, cell types, and even in individual cells. Cholesterol, a major component of plasma membranes, regulates the fluidity of the overall membrane, meaning that cholesterol controls the amount of movement of the various cell membrane components based on its concentrations. In high temperatures, cholesterol inhibits the movement of phospholipid fatty acid chains, causing a reduced permeability to small molecules and reduced membrane fluidity. The opposite is true for the role of cholesterol in cooler temperatures. Cholesterol production, and thus concentration, is up-regulated (increased) in response to cold temperature. At cold temperatures, cholesterol interferes with fatty acid chain interactions. Acting as antifreeze, cholesterol maintains the fluidity of the membrane. Cholesterol is more abundant in cold-weather animals than warm-weather animals. In plants, which lack cholesterol, related compounds called sterols perform the same function as cholesterol.
1279:, types of bacteria, share similar functions to mitochondria and blue-green algae (cyanobacteria) share similar functions to chloroplasts. Endosymbiotic theory proposes that through the course of evolution, a eukaryotic cell engulfed these two types of bacteria, leading to the formation of mitochondria and chloroplasts inside eukaryotic cells. This engulfment lead to the double-membranes systems of these organelles in which the outer membrane originated from the host's plasma membrane and the inner membrane was the endosymbiont's plasma membrane. Considering that mitochondria and chloroplasts both contain their own DNA is further support that both of these organelles evolved from engulfed bacteria that thrived inside a eukaryotic cell.
279:
significance of the cell membrane as it was acknowledged. Finally, two scientists Gorter and
Grendel (1925) made the discovery that the membrane is "lipid-based". From this, they furthered the idea that this structure would have to be in a formation that mimicked layers. Once studied further, it was found by comparing the sum of the cell surfaces and the surfaces of the lipids, a 2:1 ratio was estimated; thus, providing the first basis of the bilayer structure known today. This discovery initiated many new studies that arose globally within various fields of scientific studies, confirming that the structure and functions of the cell membrane are widely accepted.
832:: Endocytosis is the process in which cells absorb molecules by engulfing them. The plasma membrane creates a small deformation inward, called an invagination, in which the substance to be transported is captured. This invagination is caused by proteins on the outside on the cell membrane, acting as receptors and clustering into depressions that eventually promote accumulation of more proteins and lipids on the cytosolic side of the membrane. The deformation then pinches off from the membrane on the inside of the cell, creating a vesicle containing the captured substance. Endocytosis is a pathway for internalizing solid particles ("cell eating" or
793:), can move across the plasma membrane by diffusion, which is a passive transport process. Because the membrane acts as a barrier for certain molecules and ions, they can occur in different concentrations on the two sides of the membrane. Diffusion occurs when small molecules and ions move freely from high concentration to low concentration in order to equilibrate the membrane. It is considered a passive transport process because it does not require energy and is propelled by the concentration gradient created by each side of the membrane. Such a concentration gradient across a semipermeable membrane sets up an
847:: Just as material can be brought into the cell by invagination and formation of a vesicle, the membrane of a vesicle can be fused with the plasma membrane, extruding its contents to the surrounding medium. This is the process of exocytosis. Exocytosis occurs in various cells to remove undigested residues of substances brought in by endocytosis, to secrete substances such as hormones and enzymes, and to transport a substance completely across a cellular barrier. In the process of exocytosis, the undigested waste-containing food vacuole or the secretory vesicle budded from
665:
that are embedded in the lipid bilayer that allow protons to travel through the membrane by transferring from one amino acid side chain to another. Processes such as electron transport and generating ATP use proton pumps. A G-protein coupled receptor is a single polypeptide chain that crosses the lipid bilayer seven times responding to signal molecules (i.e. hormones and neurotransmitters). G-protein coupled receptors are used in processes such as cell to cell signaling, the regulation of the production of cAMP, and the regulation of ion channels.
229:
results of the initial experiment. Independently, the leptoscope was invented in order to measure very thin membranes by comparing the intensity of light reflected from a sample to the intensity of a membrane standard of known thickness. The instrument could resolve thicknesses that depended on pH measurements and the presence of membrane proteins that ranged from 8.6 to 23.2 nm, with the lower measurements supporting the lipid bilayer hypothesis. Later in the 1930s, the membrane structure model developed in general agreement to be the
200:. Initially it was believed that all cells contained a hard cell wall since only plant cells could be observed at the time. Microscopists focused on the cell wall for well over 150 years until advances in microscopy were made. In the early 19th century, cells were recognized as being separate entities, unconnected, and bound by individual cell walls after it was found that plant cells could be separated. This theory extended to include animal cells to suggest a universal mechanism for cell protection and development.
688:
33:
1083:
696:
1144:
208:. It was also inferred that cell membranes were not vital components to all cells. Many refuted the existence of a cell membrane still towards the end of the 19th century. In 1890, an update to the Cell Theory stated that cell membranes existed, but were merely secondary structures. It was not until later studies with osmosis and permeability that cell membranes gained more recognition. In 1895,
353:
1346:: Sarcolemma is the name given to the cell membrane of muscle cells. Although the sarcolemma is similar to other cell membranes, it has other functions that set it apart. For instance, the sarcolemma transmits synaptic signals, helps generate action potentials, and is very involved in muscle contraction. Unlike other cell membranes, the sarcolemma makes up small channels called
1357:: The oolemma of oocytes, (immature egg cells) are not consistent with a lipid bilayer as they lack a bilayer and do not consist of lipids. Rather, the structure has an inner layer, the fertilization envelope, and the exterior is made up of the vitelline layer, which is made up of glycoproteins; however, channels and proteins are still present for their functions in the membrane.
986:
45:
314:, notably lipids and proteins. Composition is not set, but constantly changing for fluidity and changes in the environment, even fluctuating during different stages of cell development. Specifically, the amount of cholesterol in human primary neuron cell membrane changes, and this change in composition affects fluidity throughout development stages.
851:, is first moved by cytoskeleton from the interior of the cell to the surface. The vesicle membrane comes in contact with the plasma membrane. The lipid molecules of the two bilayers rearrange themselves and the two membranes are, thus, fused. A passage is formed in the fused membrane and the vesicles discharges its contents outside the cell.
809:: Transmembrane proteins extend through the lipid bilayer of the membranes; they function on both sides of the membrane to transport molecules across it. Nutrients, such as sugars or amino acids, must enter the cell, and certain products of metabolism must leave the cell. Such molecules can diffuse passively through protein channels such as
1024:) are the major driving forces in the formation of lipid bilayers. An increase in interactions between hydrophobic molecules (causing clustering of hydrophobic regions) allows water molecules to bond more freely with each other, increasing the entropy of the system. This complex interaction can include noncovalent interactions such as
302:, 1867), plasmatic membrane (Pfeffer, 1900), plasma membrane, cytoplasmic membrane, cell envelope and cell membrane. Some authors who did not believe that there was a functional permeable boundary at the surface of the cell preferred to use the term plasmalemma (coined by Mast, 1924) for the external region of the cell.
1306:, its membrane will have more pores. The protein composition of the nucleus can vary greatly from the cytosol as many proteins are unable to cross through pores via diffusion. Within the nuclear membrane, the inner and outer membranes vary in protein composition, and only the outer membrane is continuous with the
1243:-based extensions. These extensions are ensheathed in membrane and project from the surface of the cell in order to sense the external environment and/or make contact with the substrate or other cells. The apical surfaces of epithelial cells are dense with actin-based finger-like projections known as
1067:
cells. One important role is to regulate the movement of materials into and out of cells. The phospholipid bilayer structure (fluid mosaic model) with specific membrane proteins accounts for the selective permeability of the membrane and passive and active transport mechanisms. In addition, membranes
664:
allow inorganic ions such as sodium, potassium, calcium, or chlorine to diffuse down their electrochemical gradient across the lipid bilayer through hydrophilic pores across the membrane. The electrical behavior of cells (i.e. nerve cells) is controlled by ion channels. Proton pumps are protein pumps
1031:
Lipid bilayers are generally impermeable to ions and polar molecules. The arrangement of hydrophilic heads and hydrophobic tails of the lipid bilayer prevent polar solutes (ex. amino acids, nucleic acids, carbohydrates, proteins, and ions) from diffusing across the membrane, but generally allows for
902:
and phospholipids forming the interior. The outer membrane typically has a porous quality due to its presence of membrane proteins, such as gram-negative porins, which are pore-forming proteins. The inner plasma membrane is also generally symmetric whereas the outer membrane is asymmetric because of
228:
In 1925 it was determined by Fricke that the thickness of erythrocyte and yeast cell membranes ranged between 3.3 and 4 nm, a thickness compatible with a lipid monolayer. The choice of the dielectric constant used in these studies was called into question but future tests could not disprove the
224:
lack both nuclei and cytoplasmic organelles, the plasma membrane is the only lipid-containing structure in the cell. Consequently, all of the lipids extracted from the cells can be assumed to have resided in the cells' plasma membranes. The ratio of the surface area of water covered by the extracted
1313:
The ER, which is part of the endomembrane system, which makes up a very large portion of the cell's total membrane content. The ER is an enclosed network of tubules and sacs, and its main functions include protein synthesis, and lipid metabolism. There are 2 types of ER, smooth and rough. The rough
797:
for the water. Osmosis, in biological systems involves a solvent, moving through a semipermeable membrane similarly to passive diffusion as the solvent still moves with the concentration gradient and requires no energy. While water is the most common solvent in cell, it can also be other liquids as
472:
from the inside of the vesicle to the ambient solution allows researchers to better understand membrane permeability. Vesicles can be formed with molecules and ions inside the vesicle by forming the vesicle with the desired molecule or ion present in the solution. Proteins can also be embedded into
450:
Cholesterol is normally found dispersed in varying degrees throughout cell membranes, in the irregular spaces between the hydrophobic tails of the membrane lipids, where it confers a stiffening and strengthening effect on the membrane. Additionally, the amount of cholesterol in biological membranes
245:
eggs. Since the surface tension values appeared to be much lower than would be expected for an oil–water interface, it was assumed that some substance was responsible for lowering the interfacial tensions in the surface of cells. It was suggested that a lipid bilayer was in between two thin protein
911:
was grown in 37C for 24h, the membrane exhibited a more fluid state instead of a gel-like state. This supports the concept that in higher temperatures, the membrane is more fluid than in colder temperatures. When the membrane is becoming more fluid and needs to become more stabilized, it will make
219:
and
Grendel, created speculation in the description of the cell membrane bilayer structure based on crystallographic studies and soap bubble observations. In an attempt to accept or reject the hypothesis, researchers measured membrane thickness. These researchers extracted the lipid from human red
203:
By the second half of the 19th century, microscopy was still not advanced enough to make a distinction between cell membranes and cell walls. However, some microscopists correctly identified at this time that while invisible, it could be inferred that cell membranes existed in animal cells due to
1321:
has two interconnected round Golgi cisternae. Compartments of the apparatus forms multiple tubular-reticular networks responsible for organization, stack connection and cargo transport that display a continuous grape-like stringed vesicles ranging from 50 to 60 nm. The apparatus consists of
407:
usually contain an even number of carbon atoms, typically between 16 and 20. The 16- and 18-carbon fatty acids are the most common. Fatty acids may be saturated or unsaturated, with the configuration of the double bonds nearly always "cis". The length and the degree of unsaturation of fatty acid
968:
in which lipid and protein molecules diffuse more or less easily. Although the lipid bilayers that form the basis of the membranes do indeed form two-dimensional liquids by themselves, the plasma membrane also contains a large quantity of proteins, which provide more structure. Examples of such
934:
out into periplasmic protrusions under stress conditions or upon virulence requirements while encountering a host target cell, and thus such blebs may work as virulence organelles. Bacterial cells provide numerous examples of the diverse ways in which prokaryotic cell membranes are adapted with
463:
are approximately spherical pockets that are enclosed by a lipid bilayer. These structures are used in laboratories to study the effects of chemicals in cells by delivering these chemicals directly to the cell, as well as getting more insight into cell membrane permeability. Lipid vesicles and
278:
For many centuries, the scientists cited disagreed with the significance of the structure they were seeing as the cell membrane. For almost two centuries, the membranes were seen but mostly disregarded as an important structure with cellular function. It was not until the 20th century that the
265:
has been modernized to detail contemporary discoveries, the basics have remained constant: the membrane is a lipid bilayer composed of hydrophilic exterior heads and a hydrophobic interior where proteins can interact with hydrophilic heads through polar interactions, but proteins that span the
825:, are usually quite specific, and they only recognize and transport a limited variety of chemical substances, often limited to a single substance. Another example of a transmembrane protein is a cell-surface receptor, which allow cell signaling molecules to communicate between cells.
1310:(ER) membrane. Like the ER, the outer membrane also possesses ribosomes responsible for producing and transporting proteins into the space between the two membranes. The nuclear membrane disassembles during the early stages of mitosis and reassembles in later stages of mitosis.
392:. The amount of each depends upon the type of cell, but in the majority of cases phospholipids are the most abundant, often contributing for over 50% of all lipids in plasma membranes. Glycolipids only account for a minute amount of about 2% and sterols make up the rest. In
581:, which interacts with internal molecules, a hydrophobic membrane-spanning domain that anchors it within the cell membrane, and a hydrophilic extracellular domain that interacts with external molecules. The hydrophobic domain consists of one, multiple, or a combination of
1015:
that spontaneously arrange so that the hydrophobic "tail" regions are isolated from the surrounding water while the hydrophilic "head" regions interact with the intracellular (cytosolic) and extracellular faces of the resulting bilayer. This forms a continuous, spherical
2696:
435:. It means the lipid molecules are free to diffuse and exhibit rapid lateral diffusion along the layer in which they are present. However, the exchange of phospholipid molecules between intracellular and extracellular leaflets of the bilayer is a very slow process.
1122:, and away from the lumen. Basolateral membrane is a compound phrase referring to the terms "basal (base) membrane" and "lateral (side) membrane", which, especially in epithelial cells, are identical in composition and activity. Proteins (such as ion channels and
1134:
join epithelial cells near their apical surface to prevent the migration of proteins from the basolateral membrane to the apical membrane. The basal and lateral surfaces thus remain roughly equivalent to one another, yet distinct from the apical surface.
628:
Attached to integral membrane proteins, or associated with peripheral regions of the lipid bilayer. These proteins tend to have only temporary interactions with biological membranes, and once reacted, the molecule dissociates to carry on its work in the
1219:
that extend from the cell. Indeed, cytoskeletal elements interact extensively and intimately with the cell membrane. Anchoring proteins restricts them to a particular cell surface — for example, the apical surface of epithelial cells that line the
1414:
of the molecule. Due to the cell membrane's hydrophobic nature, small electrically neutral molecules pass through the membrane more easily than charged, large ones. The inability of charged molecules to pass through the cell membrane results in
935:
structures that suit the organism's niche. For example, proteins on the surface of certain bacterial cells aid in their gliding motion. Many gram-negative bacteria have cell membranes which contain ATP-driven protein exporting systems.
672:, are present on the surface of the membrane. Functions of membrane proteins can also include cell–cell contact, surface recognition, cytoskeleton contact, signaling, enzymatic activity, or transporting substances across the membrane.
1263:, which contribute to the overall function of the cell. The origin, structure, and function of each organelle leads to a large variation in the cell composition due to the individual uniqueness associated with each organelle.
644:
The cell membrane has large content of proteins, typically around 50% of membrane volume These proteins are important for the cell because they are responsible for various biological activities. Approximately a third of the
774:. The cell membrane thus works as a selective filter that allows only certain things to come inside or go outside the cell. The cell employs a number of transport mechanisms that involve biological membranes:
477:
the desired proteins in the presence of detergents and attaching them to the phospholipids in which the liposome is formed. These provide researchers with a tool to examine various membrane protein functions.
343:
Although the concentration of membrane components in the aqueous phase is low (stable membrane components have low solubility in water), there is an exchange of molecules between the lipid and aqueous phases.
906:
Also, for the prokaryotic membranes, there are multiple things that can affect the fluidity. One of the major factors that can affect the fluidity is fatty acid composition. For example, when the bacteria
506:
in eukaryotes; they are located on the surface of the cell where they recognize host cells and share information. Viruses that bind to cells using these receptors cause an infection. For the most part, no
270:
not only provided an accurate representation of membrane mechanics, it enhanced the study of hydrophobic forces, which would later develop into an essential descriptive limitation to describe biological
668:
The cell membrane, being exposed to the outside environment, is an important site of cell–cell communication. As such, a large variety of protein receptors and identification proteins, such as
679:, which inserts the proteins into a lipid bilayer. Once inserted, the proteins are then transported to their final destination in vesicles, where the vesicle fuses with the target membrane.
660:
As shown in the adjacent table, integral proteins are amphipathic transmembrane proteins. Examples of integral proteins include ion channels, proton pumps, and g-protein coupled receptors.
1367:
of nerve cells that is responsible for the generation of the action potential. It consists of a granular, densely packed lipid bilayer that works closely with the cytoskeleton components
1350:
that pass through the entirety of muscle cells. It has also been found that the average sarcolemma is 10 nm thick as opposed to the 4 nm thickness of a general cell membrane.
443:-enriched microdomains in the cell membrane. Also, a fraction of the lipid in direct contact with integral membrane proteins, which is tightly bound to the protein surface is called
613:
Covalently bound to single or multiple lipid molecules; hydrophobically insert into the cell membrane and anchor the protein. The protein itself is not in contact with the membrane.
246:
layers. The paucimolecular model immediately became popular and it dominated cell membrane studies for the following 30 years, until it became rivaled by the fluid mosaic model of
396:
studies, 30% of the plasma membrane is lipid. However, for the majority of eukaryotic cells, the composition of plasma membranes is about half lipids and half proteins by weight.
1247:, which increase cell surface area and thereby increase the absorption rate of nutrients. Localized decoupling of the cytoskeleton and cell membrane results in formation of a
1332:
408:
chains have a profound effect on membrane fluidity as unsaturated lipids create a kink, preventing the fatty acids from packing together as tightly, thus decreasing the
225:
lipid to the surface area calculated for the red blood cells from which the lipid was 2:1(approx) and they concluded that the plasma membrane contains a lipid bilayer.
926:(amino acids and sugars). Some eukaryotic cells also have cell walls, but none that are made of peptidoglycan. The outer membrane of gram negative bacteria is rich in
1641:
1118:
or basolateral cell membrane of a polarized cell is the surface of the plasma membrane that forms its basal and lateral surfaces. It faces outwards, towards the
675:
Most membrane proteins must be inserted in some way into the membrane. For this to occur, an N-terminus "signal sequence" of amino acids directs proteins to the
3373:
894:
have only a plasma membrane. These two membranes differ in many aspects. The outer membrane of the gram-negative bacteria differs from other prokaryotes due to
3983:
1314:
ER has ribosomes attached to it used for protein synthesis, while the smooth ER is used more for the processing of toxins and calcium regulation in the cell.
1526:
340:
If a membrane is continuous with a tubular structure made of membrane material, then material from the tube can be drawn into the membrane continuously.
2407:
329:) not only excretes the contents of the vesicle but also incorporates the vesicle membrane's components into the cell membrane. The membrane may form
3802:
1744:
930:, which are combined poly- or oligosaccharide and carbohydrate lipid regions that stimulate the cell's natural immunity. The outer membrane can
2283:
1635:
1552:
220:
blood cells and measured the amount of surface area the lipid would cover when spread over the surface of the water. Since mature mammalian
4113:
1713:
1215:
is found underlying the cell membrane in the cytoplasm and provides a scaffolding for membrane proteins to anchor to, as well as forming
2881:
Whatley JM, John P, Whatley FR (April 1979). "From extracellular to intracellular: the establishment of mitochondria and chloroplasts".
1298:, providing the strict regulation of materials in to and out of the nucleus. Materials move between the cytosol and the nucleus through
3346:
2561:
2949:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "The
Transport of Molecules between the Nucleus and the Cytosol".
3366:
2685:"A structural model for virulence organellae of gram negative organisms with reference to Salmonella pathogenicity in chicken ileum"
2529:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "Transport into the Cell from the Plasma
Membrane: Endocytosis".
2186:
1982:
511:
occurs on membranes within the cell; rather generally glycosylation occurs on the extracellular surface of the plasma membrane. The
2055:
2041:
1032:
the passive diffusion of hydrophobic molecules. This affords the cell the ability to control the movement of these substances via
3976:
3435:
2587:"Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides"
1494:
188:
3321:
1227:— and limits how far they may diffuse within the bilayer. The cytoskeleton is able to form appendage-like organelles, such as
1907:
1068:
in prokaryotes and in the mitochondria and chloroplasts of eukaryotes facilitate the synthesis of ATP through chemiosmosis.
4133:
3537:
2536:
2515:
2339:
1625:
3336:
2975:
2456:
2313:
3795:
3359:
1595:
1474:
3969:
3880:
1997:
S J Singer and G L Nicolson."The fluid mosaic model of the structure of cell membranes." Science. (1972) 175. 720–731.
4081:
818:
806:
601:
520:
204:
intracellular movement of components internally but not externally and that membranes were not the equivalent of a
4177:
3871:
2204:"Assessment of Membrane Fluidity Fluctuations during Cellular Development Reveals Time and Cell Type Specificity"
3145:"Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol"
2838:
Doherty GJ, McMahon HT (2008). "Mediation, modulation, and consequences of membrane-cytoskeleton interactions".
4076:
3532:
1443:
1260:
883:
361:
3341:
1523:
1375:. These cytoskeleton components are able to bind to and interact with transmembrane proteins in the axolemma.
427:
interaction of hydrophobic tails, however the structure is quite fluid and not fixed rigidly in place. Under
266:
bilayer fully or partially have hydrophobic amino acids that interact with the non-polar lipid interior. The
261:, it remains the primary archetype for the cell membrane long after its inception in the 1970s. Although the
3788:
1385:
1303:
1192:
953:
428:
417:
247:
3497:
2400:
762:
of materials needed for survival. The movement of substances across the membrane can be achieved by either
464:
liposomes are formed by first suspending a lipid in an aqueous solution then agitating the mixture through
3884:
3866:
3472:
1479:
1295:
1291:
1224:
1126:) are free to move from the basal to the lateral surface of the cell or vice versa in accordance with the
997:
head groups separate the grey hydrophobic tails from the aqueous cytosolic and extracellular environments.
965:
879:
875:
871:
755:
608:
140:
4187:
4043:
4028:
3920:
3875:
3736:
3522:
3415:
3051:
Reed R, Wouston TW, Todd PM (July 1966). "Structure and function of the sarcolemma of skeletal muscle".
2932:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "The
Structure and Function of DNA".
1589:
1307:
1033:
961:
814:
676:
503:
469:
3326:
687:
3769:
3156:
3060:
3002:
2991:"The asymmetrical structure of Golgi apparatus membranes revealed by in situ atomic force microscope"
2890:
2771:
2762:
Singer SJ, Nicolson GL (February 1972). "The fluid mosaic model of the structure of cell membranes".
2215:
2140:
Plowe JQ (1931). "Membranes in the plant cell. I. Morphological membranes at protoplasmic surfaces".
1782:
1736:
1156:
720:
365:
230:
117:
2356:
Brandley BK, Schnaar RL (July 1986). "Cell-surface carbohydrates in cell recognition and response".
1771:"The effects of intra-membrane viscosity on lipid membrane morphology: complete analytical solution"
32:
4182:
4002:
3395:
1489:
1433:
1248:
1188:
1082:
1077:
1045:
1025:
931:
444:
357:
330:
89:
77:
1322:
three main compartments, a flat disc-shaped cisterna with tubular-reticular networks and vesicles.
695:
657:
consist of three main types: integral proteins, peripheral proteins, and lipid-anchored proteins.
4022:
3992:
3332:
Membrane homeostasis, tension regulation, mechanosensitive membrane exchange and membrane traffic
3084:
2914:
2863:
2795:
2665:
2381:
2157:
2053:
2039:
1705:
1127:
1123:
1053:
1021:
949:
927:
912:
longer fatty acid chains or saturated fatty acid chains in order to help stabilize the membrane.
771:
759:
636:
632:
623:
369:
299:
267:
262:
258:
136:
121:
3322:
Lipids, Membranes and
Vesicle Trafficking – The Virtual Library of Biochemistry and Cell Biology
4010:
3925:
3719:
3557:
3440:
3301:
3233:
3184:
3125:
3076:
3030:
2906:
2855:
2787:
2744:
2657:
2616:
2567:
2557:
2487:
2373:
2279:
2243:
2182:
1978:
1955:
1882:
1818:
1800:
1686:
1631:
1577:
1548:
1448:
1420:
1407:
994:
957:
763:
413:
322:
251:
105:
2851:
539:. Sialic acid carries a negative charge, providing an external barrier to charged particles.
4053:
4018:
3940:
3858:
3291:
3283:
3223:
3215:
3174:
3164:
3115:
3068:
3020:
3010:
2898:
2847:
2779:
2734:
2726:
2647:
2606:
2598:
2479:
2365:
2233:
2223:
2149:
2120:
1945:
1937:
1872:
1862:
1808:
1790:
1676:
1668:
1283:
1107:
794:
767:
724:
654:
590:
567:
317:
Material is incorporated into the membrane, or deleted from it, by a variety of mechanisms:
156:
148:
113:
109:
2025:
Pfeffer, W. 1877. Osmotische
Untersuchungen: Studien zur Zell Mechanik. Engelmann, Leipzig.
840:), and macromolecules. Endocytosis requires energy and is thus a form of active transport.
3935:
3420:
2585:
Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS (February 2011).
2059:
2045:
1911:
1530:
1484:
1438:
1403:
1318:
1143:
1103:
1099:
848:
770:, requiring the cell to expend energy in transporting it. The membrane also maintains the
578:
536:
291:
205:
176:
160:
3104:"Skeletal muscle basement membrane-sarcolemma-cytoskeleton interaction minireview series"
1267:
Mitochondria and chloroplasts are considered to have evolved from bacteria, known as the
3160:
3064:
3006:
2894:
2775:
2508:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "Membrane
Proteins".
2219:
2090:(ed. Deamer D.W., Kleinzeller A., Fambrough D.M.), pp. 1–18, Academic Press, San Diego,
2073:
1786:
524:
163:
and serve as the attachment surface for several extracellular structures, including the
3820:
3383:
3296:
3271:
3228:
3203:
3025:
2990:
2739:
2714:
2611:
2586:
2238:
2203:
1950:
1925:
1877:
1850:
1813:
1770:
1681:
1656:
1172:
1168:
1131:
969:
structures are protein-protein complexes, pickets and fences formed by the actin-based
653:
code specifically for them, and this number is even higher in multicellular organisms.
432:
393:
238:
221:
209:
85:
3351:
3179:
3144:
27:
Biological membrane that separates the interior of a cell from its outside environment
4171:
4154:
4108:
3945:
3905:
3828:
3666:
3626:
3601:
3517:
3430:
2684:
2272:
1268:
1176:
1017:
1012:
1005:
1001:
990:
923:
748:
712:
708:
508:
487:
412:(increasing the fluidity) of the membrane. The ability of some organisms to regulate
409:
283:
272:
152:
93:
2918:
2867:
2799:
2669:
2385:
2161:
282:
The structure has been variously referred to by different writers as the ectoplast (
151:
In addition, cell membranes are involved in a variety of cellular processes such as
4138:
4118:
3843:
3833:
3646:
3618:
3596:
3589:
3569:
3509:
3425:
3410:
3272:"The role of the axolemma in the initiation of traumatically induced axonal injury"
3088:
1740:
1709:
1504:
1416:
1299:
1287:
1212:
1119:
1098:
of a polarized cell is the surface of the plasma membrane that faces inward to the
970:
895:
833:
802:
716:
519:
with microvilli. Recent data suggest the glycocalyx participates in cell adhesion,
491:
474:
424:
400:
381:
193:
172:
124:
that loosely attach to the outer (peripheral) side of the cell membrane, acting as
97:
3780:
2483:
1904:
3015:
2783:
2652:
2635:
2556:(4th ed.). Galveston (TX): University of Texas Medical Branch at Galveston.
2228:
2176:
2091:
216:
4128:
4123:
4100:
4038:
3838:
3748:
3714:
3641:
3636:
3527:
3143:
Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM (March 2004).
1569:
1464:
1458:
1453:
1343:
1232:
1180:
1164:
1049:
1009:
899:
891:
837:
829:
661:
597:
582:
532:
499:
495:
440:
436:
377:
334:
311:
234:
197:
104:(a lipid component) interspersed between them, maintaining appropriate membrane
101:
53:
17:
3331:
3252:
3149:
Proceedings of the
National Academy of Sciences of the United States of America
2950:
2933:
2530:
2509:
2429:
2331:
2267:
2086:
Kleinzeller, A. 1999. Charles Ernest
Overton's concept of a cell membrane. In:
1795:
747:, which provides a mechanical support to the cell and precludes the passage of
4148:
4071:
3915:
3910:
3743:
3683:
3676:
3661:
3656:
3651:
3606:
3552:
3542:
3492:
3487:
3477:
2967:
2551:
2448:
2305:
1867:
1499:
1411:
1399:
1391:
1339:
1244:
1221:
1184:
1064:
1041:
974:
859:
844:
810:
586:
516:
512:
465:
404:
385:
352:
326:
242:
168:
129:
49:
3337:
3D structures of proteins associated with plasma membrane of eukaryotic cells
2470:
Kramer EM, Myers DR (April 2013). "Osmosis is not driven by water dilution".
1804:
1151:
Cell membrane can form different types of "supramembrane" structures such as
4066:
4061:
3764:
3726:
3699:
3671:
3579:
3564:
3547:
3482:
3455:
3387:
3169:
2813:
2428:
Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000).
2330:
Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000).
1395:
1236:
1216:
1200:
1196:
1060:
919:
887:
782:
744:
704:
616:
528:
241:(1935). This model was based on studies of surface tension between oils and
164:
81:
37:
3237:
3188:
3129:
3120:
3103:
3034:
2902:
2859:
2748:
2661:
2620:
2571:
2491:
2247:
1959:
1886:
1822:
1690:
1581:
1331:
The cell membrane has different lipid and protein compositions in distinct
1259:
The content of the cell, inside the cell membrane, is composed of numerous
758:
and able to regulate what enters and exits the cell, thus facilitating the
3305:
3287:
3080:
2791:
2377:
2125:
2104:
1851:"Once upon a time the cell membranes: 175 years of cell boundary research"
985:
3950:
3900:
3709:
3465:
3450:
3445:
2883:
Proceedings of the Royal Society of London. Series B, Biological Sciences
2602:
1941:
1469:
1368:
1360:
1347:
1160:
1152:
1037:
915:
867:
732:
460:
2910:
2369:
2007:
de Vries H (1885). "Plasmolytische Studien über die Wand der Vakuolen".
4091:
4033:
3731:
3631:
3584:
3460:
2153:
1354:
863:
778:
736:
669:
575:
257:
Despite the numerous models of the cell membrane proposed prior to the
3219:
2730:
2430:"Post-Translational Modifications and Quality Control in the Rough ER"
1672:
44:
3961:
3930:
3848:
3574:
3072:
1290:
from the cytoplasm of the cell. The nuclear membrane is formed by an
1111:
1048:, which carries a negative charge, on the inner membrane. Along with
389:
125:
989:
Diagram of the arrangement of amphipathic lipid molecules to form a
785:: Some substances (small molecules, ions) such as carbon dioxide (CO
171:, as well as the intracellular network of protein fibers called the
1926:"On Bimolecular Layers of Lipoids on the Chromocytes of the Blood"
1372:
1364:
1240:
1228:
984:
740:
728:
715:
environment. The cell membrane also plays a role in anchoring the
694:
650:
351:
43:
333:
around extracellular material that pinch off to become vesicles (
1398:
of molecules through the membrane. These molecules are known as
646:
3965:
3784:
3355:
1302:
in the nuclear membrane. If a cell's nucleus is more active in
3704:
1576:. 1 SIU School of Medicine 2 Baptist Regional Medical Center.
1335:
and may have therefore specific names for certain cell types.
356:
Examples of the major membrane phospholipids and glycolipids:
144:
3257:
Basic Neurochemistry: Molecular, Cellular and Medical Aspects
2399:
Jesse Gray; Shana Groeschler; Tony Le; Zara Gonzalez (2002).
1977:(6th ed.). US: John Wiley & Sons, Inc. p. 120.
298:(skin layer, Pfeffer, 1886; used with a different meaning by
1657:"Membrane assembly driven by a biomimetic coupling reaction"
3342:
Lipid composition and proteins of some eukariotic membranes
2332:"Biomembranes: Structural Organization and Basic Functions"
2268:"Biomembranes: Structural Organization and Basic Functions"
175:. In the field of synthetic biology, cell membranes can be
132:
embedded in the outer lipid layer serve a similar purpose.
92:(the extracellular space). The cell membrane consists of a
1008:. The cell membrane consists primarily of a thin layer of
2989:
Xu H, Su W, Cai M, Jiang J, Zeng X, Wang H (2013-04-16).
196:'s discovery of cells in 1665 led to the proposal of the
766:, occurring without the input of cellular energy, or by
2715:"Microcompartments and protein machines in prokaryotes"
2048:. Translated by A. J. Ewart from the 2nd German ed. of
431:
phospholipid molecules in the cell membrane are in the
128:
to facilitate interaction with the cell's environment.
719:
to provide shape to the cell, and in attaching to the
2278:(4th ed.). New York: Scientific American Books.
2088:
Membrane permeability: 100 years since Ernest Overton
108:
at various temperatures. The membrane also contains
3270:
Fitzpatrick MO, Maxwell WL, Graham DI (March 1998).
1235:-based extensions covered by the cell membrane, and
1110:, but also describes other polarized cells, such as
4147:
4099:
4090:
4052:
4009:
3893:
3857:
3819:
3757:
3692:
3617:
3508:
3394:
2719:
Journal of Molecular Microbiology and Biotechnology
2636:"Bacterial lipopolysaccharides and innate immunity"
2266:Lodish H, Berk A, Zipursky LS, et al. (2000).
1624:Alberts B, Johnson A, Lewis J, et al. (2002).
3276:Journal of Neurology, Neurosurgery, and Psychiatry
3204:"Cell surface changes in the egg at fertilization"
2271:
2181:. Amsterdam: Elsevier/Academic Press. p. 17.
1195:. They are composed of specific proteins, such as
215:The lipid bilayer hypothesis, proposed in 1925 by
212:proposed that cell membranes were made of lipids.
1899:Leray, C. Chronological history of lipid center.
515:is an important feature in all cells, especially
2351:
2349:
836:), small molecules and ions ("cell drinking" or
468:, resulting in a vesicle. Measuring the rate of
2503:
2501:
1545:Bacteria in Biology, Biotechnology and Medicine
1175:. These structures are usually responsible for
376:The cell membrane consists of three classes of
2325:
2323:
2299:
2297:
2295:
1844:
1842:
1840:
1838:
1836:
1834:
1832:
1706:"Chemists Synthesize Artificial Cell Membrane"
1568:Tom Herrmann; Sandeep Sharma (March 2, 2019).
1402:molecules. Permeability depends mainly on the
1020:. Hydrophobic interactions (also known as the
964:, biological membranes can be considered as a
723:and other cells to hold them together to form
502:). Carbohydrates are important in the role of
3977:
3796:
3367:
2708:
2706:
2178:Plant Cell Biology: From Astronomy to Zoology
1036:complexes such as pores, channels and gates.
8:
3046:
3044:
2202:Noutsi P, Gratton E, Chaieb S (2016-06-30).
1764:
1762:
1630:(4th ed.). New York: Garland Science.
1410:of the molecule and to a lesser extent the
1052:, this creates an extra barrier to charged
707:of living cells, physically separating the
535:, as the sugar backbone is modified in the
447:; it behaves as a part of protein complex.
4096:
4001:Mechanisms for chemical transport through
3984:
3970:
3962:
3803:
3789:
3781:
3374:
3360:
3352:
1737:"Chemists create artificial cell membrane"
3327:Cell membrane protein extraction protocol
3295:
3227:
3178:
3168:
3119:
3024:
3014:
2738:
2651:
2610:
2550:Salton MR, Kim KS (1996). Baron S (ed.).
2261:
2259:
2257:
2237:
2227:
2124:
1949:
1876:
1866:
1812:
1794:
1680:
1363:: The specialized plasma membrane on the
1147:Diagram of the cell membrane's structures
898:forming the exterior of the bilayer, and
798:well as supercritical liquids and gases.
699:Illustration depicting cellular diffusion
574:Span the membrane and have a hydrophilic
423:The entire membrane is held together via
2852:10.1146/annurev.biophys.37.032807.125912
1661:Journal of the American Chemical Society
1142:
1081:
821:. Protein channel proteins, also called
686:
546:
416:by altering lipid composition is called
31:
1516:
1271:. This theory arose from the idea that
862:are divided into two different groups,
691:A detailed diagram of the cell membrane
3208:Molecular Reproduction and Development
1619:
1617:
1615:
1613:
1611:
1609:
1607:
1605:
1587:
870:, with bacteria dividing further into
167:and the carbohydrate layer called the
72:, and historically referred to as the
2591:Antimicrobial Agents and Chemotherapy
1747:from the original on January 28, 2012
1716:from the original on January 29, 2012
1394:of a membrane is the rate of passive
1059:Membranes serve diverse functions in
903:proteins such as the aforementioned.
817:or are pumped across the membrane by
7:
4114:Non-specific, adsorptive pinocytosis
3102:Campbell KP, Stull JT (April 2003).
1930:The Journal of Experimental Medicine
1903:. Last updated on 11 November 2017.
1655:Budin I, Devaraj NK (January 2012).
1028:, electrostatic and hydrogen bonds.
455:Phospholipids forming lipid vesicles
414:the fluidity of their cell membranes
310:Cell membranes contain a variety of
116:that span the membrane and serve as
3202:Wessel GM, Wong JL (October 2009).
3108:The Journal of Biological Chemistry
1769:Zeidi, Mahdi; Kim, Chun IL (2018).
1461:, including damage to cell membrane
960:(1972), which replaced the earlier
882:have both a plasma membrane and an
137:controls the movement of substances
2634:Alexander C, Rietschel ET (2001).
2306:"Structure of the Plasma Membrane"
1924:Gorter E, Grendel F (March 1925).
1102:. This is particularly evident in
25:
2689:Indian Journal of Poultry Science
2535:(4th ed.). Garland Science.
1547:(5th ed.). New York: Wiley.
3347:Prokaryotic and Eukaryotic Cells
2978:from the original on 2017-10-03.
2955:(4th ed.). Garland Science.
2938:(4th ed.). Garland Science.
2699:from the original on 2014-11-07.
2539:from the original on 2018-06-05.
2518:from the original on 2018-06-05.
2459:from the original on 2018-06-05.
2342:from the original on 2018-06-05.
2316:from the original on 2017-09-19.
1644:from the original on 2017-12-20.
1353:Oolemma is the cell membrane in
703:The cell membrane surrounds the
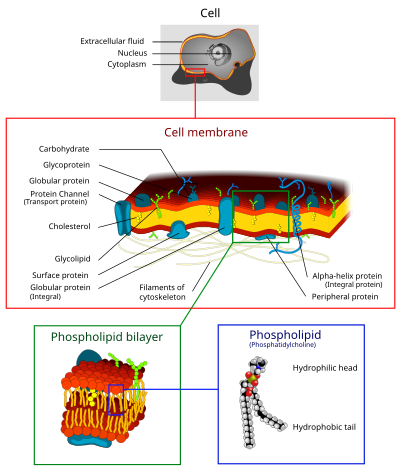
80:that separates and protects the
3253:"Characteristics of the Neuron"
2410:from the original on 2007-01-08
2077:. New York: McGraw Hill, p. 42.
1495:History of cell membrane theory
189:History of cell membrane theory
2972:The Cell: A Molecular Approach
2453:The Cell: A Molecular Approach
2449:"Transport of Small Molecules"
2310:The Cell: A Molecular Approach
1286:separates the contents of the
803:Transmembrane protein channels
486:Plasma membranes also contain
1:
4134:Receptor-mediated endocytosis
3538:Microtubule organizing center
2952:Molecular Biology of the Cell
2935:Molecular Biology of the Cell
2640:Journal of Endotoxin Research
2532:Molecular Biology of the Cell
2511:Molecular Biology of the Cell
2484:10.1016/j.tplants.2012.12.001
2105:"Structure and locomotion in
1627:Molecular Biology of the Cell
1419:of substances throughout the
1056:moving through the membrane.
494:, but with some glycolipids (
439:and caveolae are examples of
3016:10.1371/journal.pone.0061596
2784:10.1126/science.175.4023.720
2653:10.1177/09680519010070030101
2447:Cooper, Geoffrey M. (2000).
2358:Journal of Leukocyte Biology
2229:10.1371/journal.pone.0158313
1475:Elasticity of cell membranes
1187:. They can be visualized by
1004:form through the process of
962:model of Davson and Danielli
139:in and out of a cell, being
3881:Peripheral membrane protein
2968:"The Endoplasmic Reticulum"
2840:Annual Review of Biophysics
2814:"Basolateral cell membrane"
1849:Lombard J (December 2014).
96:, made up of two layers of
4204:
4082:Secondary active transport
3872:Integral membrane proteins
2062:. Clarendon Press, Oxford.
1975:Cell and Molecular Biology
1796:10.1038/s41598-018-31251-6
1735:Staff (January 26, 2012).
1704:Staff (January 25, 2012).
1383:
1075:
819:transmembrane transporters
602:G protein-coupled receptor
531:and the terminal sugar is
186:
3999:
1868:10.1186/s13062-014-0032-7
1282:In eukaryotic cells, the
1261:membrane-bound organelles
1171:, and different types of
918:are also surrounded by a
571:or transmembrane proteins
4077:Primary active transport
3533:Prokaryotic cytoskeleton
2074:Introduction To Cytology
2036:The Physiology of Plants
2034:Pfeffer, W., 1900–1906.
1594:: CS1 maint: location (
1444:Bacterial cell structure
433:liquid crystalline state
429:physiological conditions
362:phosphatidylethanolamine
321:Fusion of intracellular
177:artificially reassembled
3916:Lipid raft/microdomains
3170:10.1073/pnas.0307332101
2472:Trends in Plant Science
1524:Kimball's Biology pages
1386:Intestinal permeability
1255:Intracellular membranes
1193:fluorescence microscopy
1086:Alpha intercalated cell
609:Lipid anchored proteins
523:, and many others. The
418:homeoviscous adaptation
3921:Membrane contact sites
3885:Lipid-anchored protein
3867:Membrane glycoproteins
3121:10.1074/jbc.r300005200
2903:10.1098/rspb.1979.0020
2434:Molecular Cell Biology
2336:Molecular Cell Biology
2274:Molecular Cell Biology
1570:"Physiology, Membrane"
1480:Gram-positive bacteria
1148:
1087:
998:
966:two-dimensional liquid
880:Gram-negative bacteria
700:
692:
373:
57:
41:
4029:Facilitated diffusion
3876:transmembrane protein
3523:Intermediate filament
3416:Endoplasmic reticulum
3288:10.1136/jnnp.64.3.285
2126:10.1002/ar.1090290205
2071:Sharp, L. W. (1921).
1973:Karp, Gerald (2009).
1308:endoplasmic reticulum
1146:
1085:
1034:transmembrane protein
988:
909:Staphylococcus aureus
815:facilitated diffusion
756:selectively permeable
754:The cell membrane is
698:
690:
677:endoplasmic reticulum
504:cell-cell recognition
473:the membrane through
355:
141:selectively permeable
118:membrane transporters
47:
35:
4003:biological membranes
3901:Caveolae/Coated pits
3770:Extracellular matrix
2603:10.1128/AAC.00680-10
2553:Medical Microbiology
2406:. Davidson College.
2401:"Membrane Structure"
1942:10.1084/jem.41.4.439
1543:Singleton P (1999).
1512:Notes and references
1269:endosymbiotic theory
1157:postsynaptic density
1116:basolateral membrane
721:extracellular matrix
711:components from the
399:The fatty chains in
366:phosphatidylinositol
312:biological molecules
231:paucimolecular model
70:cytoplasmic membrane
3473:Cytoplasmic granule
3161:2004PNAS..101.4083M
3065:1966Natur.211..534R
3007:2013PLoSO...861596X
2895:1979RSPSB.204..165W
2776:1972Sci...175..720S
2683:YashRoy RC (1999).
2370:10.1002/jlb.40.1.97
2220:2016PLoSO..1158313N
2050:Pflanzenphysiologie
1787:2018NatSR...812845Z
1490:Membrane nanotubule
1434:Annular lipid shell
1189:electron microscopy
1139:Membrane structures
1078:Epithelial polarity
1046:phosphatidyl serine
928:lipopolysaccharides
624:Peripheral proteins
445:annular lipid shell
410:melting temperature
358:phosphatidylcholine
325:with the membrane (
122:peripheral proteins
90:outside environment
78:biological membrane
64:(also known as the
4023:mediated transport
3993:Membrane transport
3926:Membrane nanotubes
3811:Structures of the
3498:Weibel–Palade body
3382:Structures of the
2966:Cooper GM (2000).
2304:Cooper GM (2000).
2154:10.1007/BF01618716
2058:2018-06-01 at the
2044:2018-06-02 at the
1910:2017-10-13 at the
1775:Scientific Reports
1529:2009-01-25 at the
1421:fluid compartments
1149:
1128:fluid mosaic model
1088:
1022:hydrophobic effect
999:
973:, and potentially
950:fluid mosaic model
944:Fluid mosaic model
701:
693:
459:Lipid vesicles or
374:
370:phosphatidylserine
268:fluid mosaic model
263:fluid mosaic model
259:fluid mosaic model
149:organic molecules.
135:The cell membrane
58:
42:
36:Illustration of a
4165:
4164:
4161:
4160:
4011:Passive transport
3959:
3958:
3859:Membrane proteins
3778:
3777:
3558:Spindle pole body
3251:Raine CS (1999).
3220:10.1002/mrd.21090
3114:(15): 12599–600.
2731:10.1159/000351625
2713:Saier MH (2013).
2285:978-0-7167-3136-8
1901:Cyberlipid Center
1673:10.1021/ja2076873
1637:978-0-8153-3218-3
1554:978-0-471-98880-9
1449:Bangstad syndrome
1179:, communication,
1108:endothelial cells
1072:Membrane polarity
948:According to the
890:; however, other
764:passive transport
655:Membrane proteins
642:
641:
568:Integral proteins
521:lymphocyte homing
114:integral proteins
110:membrane proteins
16:(Redirected from
4195:
4178:Membrane biology
4097:
4054:Active transport
4019:Simple diffusion
3986:
3979:
3972:
3963:
3941:Nuclear envelope
3936:Nodes of Ranvier
3805:
3798:
3791:
3782:
3376:
3369:
3362:
3353:
3310:
3309:
3299:
3267:
3261:
3260:
3248:
3242:
3241:
3231:
3199:
3193:
3192:
3182:
3172:
3140:
3134:
3133:
3123:
3099:
3093:
3092:
3073:10.1038/211534b0
3048:
3039:
3038:
3028:
3018:
2986:
2980:
2979:
2974:(2nd ed.).
2963:
2957:
2956:
2946:
2940:
2939:
2929:
2923:
2922:
2889:(1155): 165–87.
2878:
2872:
2871:
2835:
2829:
2828:
2826:
2824:
2810:
2804:
2803:
2770:(4023): 720–31.
2759:
2753:
2752:
2742:
2710:
2701:
2700:
2680:
2674:
2673:
2655:
2631:
2625:
2624:
2614:
2582:
2576:
2575:
2547:
2541:
2540:
2526:
2520:
2519:
2514:(4th ed.).
2505:
2496:
2495:
2467:
2461:
2460:
2455:(2nd ed.).
2444:
2438:
2437:
2425:
2419:
2418:
2416:
2415:
2405:
2396:
2390:
2389:
2353:
2344:
2343:
2338:(4th ed.).
2327:
2318:
2317:
2312:(2nd ed.).
2301:
2290:
2289:
2277:
2263:
2252:
2251:
2241:
2231:
2199:
2193:
2192:
2175:Wayne R (2009).
2172:
2166:
2165:
2137:
2131:
2130:
2128:
2103:Mast SO (1924).
2100:
2094:
2084:
2078:
2069:
2063:
2032:
2026:
2023:
2017:
2016:
2009:Jahrb. Wiss. Bot
2004:
1998:
1995:
1989:
1988:
1970:
1964:
1963:
1953:
1921:
1915:
1897:
1891:
1890:
1880:
1870:
1846:
1827:
1826:
1816:
1798:
1766:
1757:
1756:
1754:
1752:
1732:
1726:
1725:
1723:
1721:
1701:
1695:
1694:
1684:
1652:
1646:
1645:
1621:
1600:
1599:
1593:
1585:
1565:
1559:
1558:
1540:
1534:
1533:, Cell Membranes
1521:
1284:nuclear membrane
1277:Rhodopseudomonas
1096:luminal membrane
768:active transport
749:larger molecules
547:
490:, predominantly
157:ion conductivity
48:Comparison of a
21:
18:Plasma membranes
4203:
4202:
4198:
4197:
4196:
4194:
4193:
4192:
4168:
4167:
4166:
4157:
4143:
4086:
4048:
4005:
3995:
3990:
3960:
3955:
3889:
3853:
3821:Membrane lipids
3815:
3809:
3779:
3774:
3753:
3688:
3613:
3504:
3421:Golgi apparatus
3397:
3390:
3380:
3318:
3313:
3269:
3268:
3264:
3259:(6th ed.).
3250:
3249:
3245:
3201:
3200:
3196:
3142:
3141:
3137:
3101:
3100:
3096:
3059:(5048): 534–6.
3050:
3049:
3042:
2988:
2987:
2983:
2965:
2964:
2960:
2948:
2947:
2943:
2931:
2930:
2926:
2880:
2879:
2875:
2837:
2836:
2832:
2822:
2820:
2818:www.uniprot.org
2812:
2811:
2807:
2761:
2760:
2756:
2725:(4–5): 243–69.
2712:
2711:
2704:
2682:
2681:
2677:
2633:
2632:
2628:
2584:
2583:
2579:
2564:
2549:
2548:
2544:
2528:
2527:
2523:
2507:
2506:
2499:
2469:
2468:
2464:
2446:
2445:
2441:
2436:(4th ed.).
2427:
2426:
2422:
2413:
2411:
2403:
2398:
2397:
2393:
2355:
2354:
2347:
2329:
2328:
2321:
2303:
2302:
2293:
2286:
2265:
2264:
2255:
2214:(6): e0158313.
2201:
2200:
2196:
2189:
2174:
2173:
2169:
2139:
2138:
2134:
2102:
2101:
2097:
2085:
2081:
2070:
2066:
2060:Wayback Machine
2046:Wayback Machine
2033:
2029:
2024:
2020:
2006:
2005:
2001:
1996:
1992:
1985:
1972:
1971:
1967:
1923:
1922:
1918:
1912:Wayback Machine
1898:
1894:
1848:
1847:
1830:
1768:
1767:
1760:
1750:
1748:
1734:
1733:
1729:
1719:
1717:
1703:
1702:
1698:
1654:
1653:
1649:
1638:
1623:
1622:
1603:
1586:
1567:
1566:
1562:
1555:
1542:
1541:
1537:
1531:Wayback Machine
1522:
1518:
1514:
1509:
1485:Membrane models
1439:Artificial cell
1429:
1404:electric charge
1388:
1382:
1329:
1319:Golgi apparatus
1257:
1209:
1141:
1132:Tight junctions
1092:apical membrane
1080:
1074:
983:
946:
941:
857:
849:Golgi apparatus
792:
789:) and oxygen (O
788:
685:
570:
545:
537:Golgi apparatus
484:
457:
350:
308:
294:, 1877, 1891),
222:red blood cells
206:plant cell wall
191:
185:
161:cell signalling
66:plasma membrane
28:
23:
22:
15:
12:
11:
5:
4201:
4199:
4191:
4190:
4185:
4180:
4170:
4169:
4163:
4162:
4159:
4158:
4153:
4151:
4145:
4144:
4142:
4141:
4136:
4131:
4126:
4121:
4116:
4111:
4105:
4103:
4094:
4088:
4087:
4085:
4084:
4079:
4074:
4069:
4064:
4058:
4056:
4050:
4049:
4047:
4046:
4041:
4036:
4031:
4026:
4015:
4013:
4007:
4006:
4000:
3997:
3996:
3991:
3989:
3988:
3981:
3974:
3966:
3957:
3956:
3954:
3953:
3948:
3946:Phycobilisomes
3943:
3938:
3933:
3928:
3923:
3918:
3913:
3908:
3906:Cell junctions
3903:
3897:
3895:
3891:
3890:
3888:
3887:
3878:
3869:
3863:
3861:
3855:
3854:
3852:
3851:
3846:
3841:
3836:
3831:
3825:
3823:
3817:
3816:
3810:
3808:
3807:
3800:
3793:
3785:
3776:
3775:
3773:
3772:
3767:
3761:
3759:
3755:
3754:
3752:
3751:
3746:
3741:
3740:
3739:
3734:
3724:
3723:
3722:
3717:
3712:
3702:
3696:
3694:
3693:Other internal
3690:
3689:
3687:
3686:
3681:
3680:
3679:
3674:
3669:
3664:
3659:
3654:
3649:
3644:
3639:
3629:
3623:
3621:
3615:
3614:
3612:
3611:
3610:
3609:
3604:
3594:
3593:
3592:
3587:
3582:
3577:
3567:
3562:
3561:
3560:
3555:
3550:
3545:
3535:
3530:
3525:
3520:
3514:
3512:
3506:
3505:
3503:
3502:
3501:
3500:
3495:
3490:
3485:
3480:
3470:
3469:
3468:
3463:
3458:
3453:
3448:
3443:
3433:
3428:
3423:
3418:
3413:
3408:
3402:
3400:
3392:
3391:
3381:
3379:
3378:
3371:
3364:
3356:
3350:
3349:
3344:
3339:
3334:
3329:
3324:
3317:
3316:External links
3314:
3312:
3311:
3262:
3243:
3214:(10): 942–53.
3194:
3155:(12): 4083–8.
3135:
3094:
3040:
2981:
2958:
2941:
2924:
2873:
2830:
2805:
2754:
2702:
2695:(2): 213–219.
2675:
2646:(3): 167–202.
2626:
2577:
2563:978-0963117212
2562:
2542:
2521:
2497:
2462:
2439:
2420:
2391:
2345:
2319:
2291:
2284:
2253:
2194:
2187:
2167:
2132:
2107:Amoeba proteus
2095:
2079:
2064:
2027:
2018:
1999:
1990:
1983:
1965:
1916:
1892:
1855:Biology Direct
1828:
1758:
1741:kurzweilai.net
1727:
1696:
1647:
1636:
1601:
1560:
1553:
1535:
1515:
1513:
1510:
1508:
1507:
1502:
1497:
1492:
1487:
1482:
1477:
1472:
1467:
1462:
1456:
1451:
1446:
1441:
1436:
1430:
1428:
1425:
1381:
1378:
1377:
1376:
1358:
1351:
1333:types of cells
1328:
1325:
1324:
1323:
1315:
1311:
1296:outer membrane
1280:
1256:
1253:
1208:
1205:
1173:cell junctions
1169:focal adhesion
1140:
1137:
1073:
1070:
1002:Lipid bilayers
982:
979:
958:G. L. Nicolson
945:
942:
940:
937:
884:outer membrane
856:
853:
790:
786:
772:cell potential
684:
681:
640:
639:
630:
626:
620:
619:
614:
611:
605:
604:
596:Ion channels,
594:
572:
564:
563:
558:
553:
544:
541:
483:
480:
456:
453:
394:red blood cell
349:
346:
345:
344:
341:
338:
307:
304:
290:(plasma skin,
273:macromolecules
210:Ernest Overton
187:Main article:
184:
181:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4200:
4189:
4186:
4184:
4181:
4179:
4176:
4175:
4173:
4156:
4155:Degranulation
4152:
4150:
4146:
4140:
4137:
4135:
4132:
4130:
4127:
4125:
4122:
4120:
4117:
4115:
4112:
4110:
4109:Efferocytosis
4107:
4106:
4104:
4102:
4098:
4095:
4093:
4089:
4083:
4080:
4078:
4075:
4073:
4070:
4068:
4065:
4063:
4060:
4059:
4057:
4055:
4051:
4045:
4042:
4040:
4037:
4035:
4032:
4030:
4027:
4024:
4020:
4017:
4016:
4014:
4012:
4008:
4004:
3998:
3994:
3987:
3982:
3980:
3975:
3973:
3968:
3967:
3964:
3952:
3949:
3947:
3944:
3942:
3939:
3937:
3934:
3932:
3931:Myelin sheath
3929:
3927:
3924:
3922:
3919:
3917:
3914:
3912:
3909:
3907:
3904:
3902:
3899:
3898:
3896:
3892:
3886:
3882:
3879:
3877:
3873:
3870:
3868:
3865:
3864:
3862:
3860:
3856:
3850:
3847:
3845:
3844:Sphingolipids
3842:
3840:
3837:
3835:
3834:Phospholipids
3832:
3830:
3829:Lipid bilayer
3827:
3826:
3824:
3822:
3818:
3814:
3813:cell membrane
3806:
3801:
3799:
3794:
3792:
3787:
3786:
3783:
3771:
3768:
3766:
3763:
3762:
3760:
3756:
3750:
3747:
3745:
3742:
3738:
3735:
3733:
3730:
3729:
3728:
3725:
3721:
3718:
3716:
3713:
3711:
3708:
3707:
3706:
3703:
3701:
3698:
3697:
3695:
3691:
3685:
3682:
3678:
3675:
3673:
3670:
3668:
3667:Proteinoplast
3665:
3663:
3660:
3658:
3655:
3653:
3650:
3648:
3645:
3643:
3640:
3638:
3635:
3634:
3633:
3630:
3628:
3627:Mitochondrion
3625:
3624:
3622:
3620:
3619:Endosymbionts
3616:
3608:
3605:
3603:
3602:Lamellipodium
3600:
3599:
3598:
3595:
3591:
3588:
3586:
3583:
3581:
3578:
3576:
3573:
3572:
3571:
3568:
3566:
3563:
3559:
3556:
3554:
3551:
3549:
3546:
3544:
3541:
3540:
3539:
3536:
3534:
3531:
3529:
3526:
3524:
3521:
3519:
3518:Microfilament
3516:
3515:
3513:
3511:
3507:
3499:
3496:
3494:
3491:
3489:
3486:
3484:
3481:
3479:
3476:
3475:
3474:
3471:
3467:
3464:
3462:
3459:
3457:
3454:
3452:
3449:
3447:
3444:
3442:
3439:
3438:
3437:
3434:
3432:
3431:Autophagosome
3429:
3427:
3424:
3422:
3419:
3417:
3414:
3412:
3409:
3407:
3406:Cell membrane
3404:
3403:
3401:
3399:
3396:Endomembrane
3393:
3389:
3385:
3377:
3372:
3370:
3365:
3363:
3358:
3357:
3354:
3348:
3345:
3343:
3340:
3338:
3335:
3333:
3330:
3328:
3325:
3323:
3320:
3319:
3315:
3307:
3303:
3298:
3293:
3289:
3285:
3281:
3277:
3273:
3266:
3263:
3258:
3254:
3247:
3244:
3239:
3235:
3230:
3225:
3221:
3217:
3213:
3209:
3205:
3198:
3195:
3190:
3186:
3181:
3176:
3171:
3166:
3162:
3158:
3154:
3150:
3146:
3139:
3136:
3131:
3127:
3122:
3117:
3113:
3109:
3105:
3098:
3095:
3090:
3086:
3082:
3078:
3074:
3070:
3066:
3062:
3058:
3054:
3047:
3045:
3041:
3036:
3032:
3027:
3022:
3017:
3012:
3008:
3004:
3001:(4): e61596.
3000:
2996:
2992:
2985:
2982:
2977:
2973:
2969:
2962:
2959:
2954:
2953:
2945:
2942:
2937:
2936:
2928:
2925:
2920:
2916:
2912:
2908:
2904:
2900:
2896:
2892:
2888:
2884:
2877:
2874:
2869:
2865:
2861:
2857:
2853:
2849:
2845:
2841:
2834:
2831:
2819:
2815:
2809:
2806:
2801:
2797:
2793:
2789:
2785:
2781:
2777:
2773:
2769:
2765:
2758:
2755:
2750:
2746:
2741:
2736:
2732:
2728:
2724:
2720:
2716:
2709:
2707:
2703:
2698:
2694:
2690:
2686:
2679:
2676:
2671:
2667:
2663:
2659:
2654:
2649:
2645:
2641:
2637:
2630:
2627:
2622:
2618:
2613:
2608:
2604:
2600:
2597:(2): 526–31.
2596:
2592:
2588:
2581:
2578:
2573:
2569:
2565:
2559:
2555:
2554:
2546:
2543:
2538:
2534:
2533:
2525:
2522:
2517:
2513:
2512:
2504:
2502:
2498:
2493:
2489:
2485:
2481:
2477:
2473:
2466:
2463:
2458:
2454:
2450:
2443:
2440:
2435:
2431:
2424:
2421:
2409:
2402:
2395:
2392:
2387:
2383:
2379:
2375:
2371:
2367:
2364:(1): 97–111.
2363:
2359:
2352:
2350:
2346:
2341:
2337:
2333:
2326:
2324:
2320:
2315:
2311:
2307:
2300:
2298:
2296:
2292:
2287:
2281:
2276:
2275:
2269:
2262:
2260:
2258:
2254:
2249:
2245:
2240:
2235:
2230:
2225:
2221:
2217:
2213:
2209:
2205:
2198:
2195:
2190:
2188:9780080921273
2184:
2180:
2179:
2171:
2168:
2163:
2159:
2155:
2151:
2147:
2143:
2136:
2133:
2127:
2122:
2118:
2114:
2110:
2108:
2099:
2096:
2092:
2089:
2083:
2080:
2076:
2075:
2068:
2065:
2061:
2057:
2054:
2052:, 1897–1904,
2051:
2047:
2043:
2040:
2037:
2031:
2028:
2022:
2019:
2014:
2010:
2003:
2000:
1994:
1991:
1986:
1984:9780470483374
1980:
1976:
1969:
1966:
1961:
1957:
1952:
1947:
1943:
1939:
1936:(4): 439–43.
1935:
1931:
1927:
1920:
1917:
1913:
1909:
1906:
1902:
1896:
1893:
1888:
1884:
1879:
1874:
1869:
1864:
1860:
1856:
1852:
1845:
1843:
1841:
1839:
1837:
1835:
1833:
1829:
1824:
1820:
1815:
1810:
1806:
1802:
1797:
1792:
1788:
1784:
1780:
1776:
1772:
1765:
1763:
1759:
1746:
1742:
1738:
1731:
1728:
1715:
1711:
1707:
1700:
1697:
1692:
1688:
1683:
1678:
1674:
1670:
1666:
1662:
1658:
1651:
1648:
1643:
1639:
1633:
1629:
1628:
1620:
1618:
1616:
1614:
1612:
1610:
1608:
1606:
1602:
1597:
1591:
1583:
1579:
1575:
1571:
1564:
1561:
1556:
1550:
1546:
1539:
1536:
1532:
1528:
1525:
1520:
1517:
1511:
1506:
1503:
1501:
1498:
1496:
1493:
1491:
1488:
1486:
1483:
1481:
1478:
1476:
1473:
1471:
1468:
1466:
1463:
1460:
1457:
1455:
1452:
1450:
1447:
1445:
1442:
1440:
1437:
1435:
1432:
1431:
1426:
1424:
1423:of the body.
1422:
1418:
1413:
1409:
1405:
1401:
1397:
1393:
1387:
1379:
1374:
1370:
1366:
1362:
1359:
1356:
1352:
1349:
1345:
1341:
1338:
1337:
1336:
1334:
1326:
1320:
1316:
1312:
1309:
1305:
1304:transcription
1301:
1300:nuclear pores
1297:
1293:
1289:
1285:
1281:
1278:
1274:
1270:
1266:
1265:
1264:
1262:
1254:
1252:
1250:
1246:
1242:
1238:
1234:
1230:
1226:
1223:
1218:
1214:
1206:
1204:
1202:
1198:
1194:
1190:
1186:
1182:
1178:
1177:cell adhesion
1174:
1170:
1166:
1162:
1158:
1154:
1145:
1138:
1136:
1133:
1129:
1125:
1121:
1117:
1113:
1109:
1105:
1101:
1097:
1093:
1084:
1079:
1071:
1069:
1066:
1062:
1057:
1055:
1051:
1047:
1043:
1039:
1035:
1029:
1027:
1026:van der Waals
1023:
1019:
1018:lipid bilayer
1014:
1013:phospholipids
1011:
1007:
1006:self-assembly
1003:
996:
993:. The yellow
992:
991:lipid bilayer
987:
981:Lipid bilayer
980:
978:
976:
972:
967:
963:
959:
955:
951:
943:
938:
936:
933:
929:
925:
924:peptidoglycan
921:
917:
913:
910:
904:
901:
897:
896:phospholipids
893:
889:
886:separated by
885:
881:
877:
876:gram-negative
873:
872:gram-positive
869:
865:
861:
854:
852:
850:
846:
841:
839:
835:
831:
826:
824:
820:
816:
812:
808:
804:
799:
796:
784:
780:
775:
773:
769:
765:
761:
757:
752:
750:
746:
742:
738:
734:
730:
726:
722:
718:
714:
713:extracellular
710:
709:intracellular
706:
697:
689:
682:
680:
678:
673:
671:
666:
663:
658:
656:
652:
648:
638:
637:some hormones
634:
631:
627:
625:
622:
621:
618:
615:
612:
610:
607:
606:
603:
599:
595:
592:
588:
584:
580:
577:
573:
569:
566:
565:
562:
559:
557:
554:
552:
549:
548:
542:
540:
538:
534:
530:
526:
522:
518:
514:
510:
509:glycosylation
505:
501:
497:
493:
492:glycoproteins
489:
488:carbohydrates
482:Carbohydrates
481:
479:
476:
471:
467:
462:
454:
452:
448:
446:
442:
438:
434:
430:
426:
421:
419:
415:
411:
406:
402:
401:phospholipids
397:
395:
391:
387:
383:
382:phospholipids
379:
371:
367:
363:
359:
354:
347:
342:
339:
336:
332:
328:
324:
320:
319:
318:
315:
313:
305:
303:
301:
297:
293:
289:
285:
280:
276:
274:
269:
264:
260:
255:
253:
249:
244:
240:
236:
232:
226:
223:
218:
213:
211:
207:
201:
199:
195:
190:
182:
180:
178:
174:
170:
166:
162:
158:
154:
153:cell adhesion
150:
146:
142:
138:
133:
131:
127:
123:
119:
115:
111:
107:
103:
99:
98:phospholipids
95:
94:lipid bilayer
91:
87:
83:
79:
75:
71:
67:
63:
62:cell membrane
56:cell membrane
55:
51:
46:
40:cell membrane
39:
34:
30:
19:
4188:Cell anatomy
4139:Transcytosis
4119:Phagocytosis
3839:Lipoproteins
3812:
3647:Gerontoplast
3597:Pseudopodium
3590:Radial spoke
3570:Undulipodium
3510:Cytoskeleton
3426:Parenthesome
3405:
3282:(3): 285–7.
3279:
3275:
3265:
3256:
3246:
3211:
3207:
3197:
3152:
3148:
3138:
3111:
3107:
3097:
3056:
3052:
2998:
2994:
2984:
2971:
2961:
2951:
2944:
2934:
2927:
2886:
2882:
2876:
2843:
2839:
2833:
2821:. Retrieved
2817:
2808:
2767:
2763:
2757:
2722:
2718:
2692:
2688:
2678:
2643:
2639:
2629:
2594:
2590:
2580:
2552:
2545:
2531:
2524:
2510:
2478:(4): 195–7.
2475:
2471:
2465:
2452:
2442:
2433:
2423:
2412:. Retrieved
2394:
2361:
2357:
2335:
2309:
2273:
2211:
2207:
2197:
2177:
2170:
2145:
2141:
2135:
2116:
2112:
2106:
2098:
2087:
2082:
2072:
2067:
2049:
2035:
2030:
2021:
2012:
2008:
2002:
1993:
1974:
1968:
1933:
1929:
1919:
1900:
1895:
1858:
1854:
1781:(1): 12845.
1778:
1774:
1751:February 18,
1749:. Retrieved
1730:
1720:February 18,
1718:. Retrieved
1710:ScienceDaily
1699:
1667:(2): 751–3.
1664:
1660:
1650:
1626:
1590:cite journal
1573:
1563:
1544:
1538:
1519:
1505:Trogocytosis
1417:pH partition
1392:permeability
1389:
1380:Permeability
1344:muscle cells
1330:
1276:
1272:
1258:
1239:, which are
1231:, which are
1213:cytoskeleton
1210:
1207:Cytoskeleton
1150:
1120:interstitium
1115:
1095:
1091:
1089:
1058:
1044:concentrate
1030:
1000:
971:cytoskeleton
954:S. J. Singer
947:
922:composed of
914:
908:
905:
900:lipoproteins
858:
842:
834:phagocytosis
827:
822:
807:transporters
800:
795:osmotic flow
776:
753:
743:also have a
717:cytoskeleton
702:
674:
667:
662:Ion channels
659:
643:
633:Some enzymes
598:proton pumps
560:
555:
550:
500:gangliosides
496:cerebrosides
485:
475:solubilizing
458:
449:
425:non-covalent
422:
398:
375:
316:
309:
295:
287:
281:
277:
256:
227:
214:
202:
194:Robert Hooke
192:
173:cytoskeleton
134:
112:, including
102:cholesterols
73:
69:
65:
61:
59:
29:
4129:Potocytosis
4124:Pinocytosis
4101:Endocytosis
3749:Magnetosome
3715:Spliceosome
3642:Chromoplast
3637:Chloroplast
3528:Microtubule
2148:: 196–220.
2142:Protoplasma
1465:Cell theory
1459:Cell damage
1454:Cell cortex
1233:microtubule
1181:endocytosis
1165:invadopodia
1065:prokaryotic
1042:scramblases
1010:amphipathic
975:lipid rafts
892:prokaryotes
860:Prokaryotes
855:Prokaryotes
838:pinocytosis
830:Endocytosis
777:1. Passive
556:Description
533:sialic acid
525:penultimate
441:cholesterol
437:Lipid rafts
405:glycolipids
386:glycolipids
378:amphipathic
335:endocytosis
306:Composition
296:Hautschicht
198:cell theory
130:Glycolipids
74:plasmalemma
54:prokaryotic
4183:Organelles
4172:Categories
4149:Exocytosis
4072:Antiporter
3911:Glycocalyx
3744:Proteasome
3737:Inclusions
3684:Nitroplast
3677:Apicoplast
3662:Elaioplast
3657:Amyloplast
3652:Leucoplast
3607:Filopodium
3553:Basal body
3543:Centrosome
3493:Peroxisome
3488:Glyoxysome
3478:Melanosome
3388:organelles
2414:2007-01-11
2015:: 465–598.
1574:StatPearls
1500:Lipid raft
1412:molar mass
1384:See also:
1340:Sarcolemma
1327:Variations
1273:Paracoccus
1245:microvilli
1222:vertebrate
1217:organelles
1185:exocytosis
1104:epithelial
1076:See also:
1061:eukaryotic
939:Structures
845:Exocytosis
811:aquaporins
629:cytoplasm.
617:G proteins
513:glycocalyx
466:sonication
368:(PtdIns),
364:(PtdEtn),
360:(PtdCho),
327:exocytosis
300:Hofmeister
288:Plasmahaut
243:echinoderm
169:glycocalyx
50:eukaryotic
38:eukaryotic
4067:Symporter
4062:Uniporter
3951:Porosomes
3765:Cell wall
3727:Cytoplasm
3700:Nucleolus
3672:Tannosome
3580:Flagellum
3565:Myofibril
3548:Centriole
3483:Microbody
3456:Phagosome
2846:: 65–95.
2119:(2): 88.
2113:Anat. Rec
1805:2045-2322
1396:diffusion
1348:T-tubules
1237:filopodia
1201:cadherins
1197:integrins
1161:podosomes
1038:Flippases
920:cell wall
888:periplasm
823:permeases
783:diffusion
760:transport
745:cell wall
705:cytoplasm
583:α-helices
576:cytosolic
529:galactose
527:sugar is
517:epithelia
461:liposomes
372:(PtdSer).
286:, 1885),
165:cell wall
88:from the
4044:Carriers
4039:Channels
4021:(or non-
3758:External
3710:Ribosome
3466:Acrosome
3451:Endosome
3446:Lysosome
3238:19658159
3189:15016920
3130:12556456
3035:23613878
2995:PLOS ONE
2976:Archived
2919:42398067
2868:17352662
2860:18573073
2800:83851531
2749:23920489
2697:Archived
2670:86224757
2662:11581570
2621:21115796
2572:21413343
2537:Archived
2516:Archived
2492:23298880
2457:Archived
2408:Archived
2386:45528175
2340:Archived
2314:Archived
2248:27362860
2208:PLOS ONE
2162:32248784
2056:Archived
2042:Archived
1960:19868999
1908:Archived
1887:25522740
1823:30150612
1745:Archived
1714:Archived
1691:22239722
1642:Archived
1582:30855799
1527:Archived
1470:Cytoneme
1427:See also
1408:polarity
1400:permeant
1369:spectrin
1361:Axolemma
1153:caveolae
1054:moieties
916:Bacteria
868:Bacteria
733:bacteria
683:Function
670:antigens
589:protein
561:Examples
543:Proteins
380:lipids:
323:vesicles
284:de Vries
254:(1972).
252:Nicolson
239:Danielli
106:fluidity
82:interior
4092:Cytosis
4034:Osmosis
3849:Sterols
3732:Cytosol
3632:Plastid
3585:Axoneme
3461:Vacuole
3441:Exosome
3436:Vesicle
3411:Nucleus
3306:9527135
3297:2169978
3229:2842880
3157:Bibcode
3089:4183025
3081:5967498
3061:Bibcode
3026:3628984
3003:Bibcode
2891:Bibcode
2823:15 June
2792:4333397
2772:Bibcode
2764:Science
2740:3832201
2612:3028772
2378:3011937
2239:4928918
2216:Bibcode
1951:2130960
1878:4304622
1814:6110749
1783:Bibcode
1682:3262119
1355:oocytes
1288:nucleus
1112:neurons
864:Archaea
779:osmosis
737:archaea
735:, most
725:tissues
587:β sheet
390:sterols
292:Pfeffer
183:History
126:enzymes
76:) is a
3575:Cilium
3398:system
3304:
3294:
3236:
3226:
3187:
3180:384699
3177:
3128:
3087:
3079:
3053:Nature
3033:
3023:
2917:
2909:
2866:
2858:
2798:
2790:
2747:
2737:
2668:
2660:
2619:
2609:
2570:
2560:
2490:
2384:
2376:
2282:
2246:
2236:
2185:
2160:
1981:
1958:
1948:
1885:
1875:
1861:: 32.
1821:
1811:
1803:
1689:
1679:
1634:
1580:
1551:
1114:. The
741:plants
739:, and
591:motifs
579:domain
470:efflux
388:, and
348:Lipids
248:Singer
235:Davson
217:Gorter
159:, and
120:, and
52:vs. a
3894:Other
3720:Vault
3085:S2CID
2915:S2CID
2911:36620
2864:S2CID
2796:S2CID
2666:S2CID
2404:(SWF)
2382:S2CID
2158:S2CID
1373:actin
1365:axons
1292:inner
1241:actin
1229:cilia
1124:pumps
1100:lumen
995:polar
729:Fungi
651:yeast
647:genes
331:blebs
100:with
84:of a
3384:cell
3302:PMID
3234:PMID
3185:PMID
3126:PMID
3077:PMID
3031:PMID
2907:PMID
2856:PMID
2825:2023
2788:PMID
2745:PMID
2658:PMID
2617:PMID
2568:PMID
2558:ISBN
2488:PMID
2374:PMID
2280:ISBN
2244:PMID
2183:ISBN
1979:ISBN
1956:PMID
1905:link
1883:PMID
1819:PMID
1801:ISSN
1753:2012
1722:2012
1687:PMID
1632:ISBN
1596:link
1578:PMID
1549:ISBN
1406:and
1390:The
1371:and
1317:The
1294:and
1275:and
1249:bleb
1211:The
1199:and
1183:and
1106:and
1090:The
1063:and
1050:NANA
1040:and
956:and
932:bleb
874:and
866:and
805:and
781:and
585:and
551:Type
498:and
403:and
250:and
237:and
147:and
145:ions
86:cell
60:The
3705:RNA
3292:PMC
3284:doi
3224:PMC
3216:doi
3175:PMC
3165:doi
3153:101
3116:doi
3112:278
3069:doi
3057:211
3021:PMC
3011:doi
2899:doi
2887:204
2848:doi
2780:doi
2768:175
2735:PMC
2727:doi
2648:doi
2607:PMC
2599:doi
2480:doi
2366:doi
2234:PMC
2224:doi
2150:doi
2121:doi
1946:PMC
1938:doi
1873:PMC
1863:doi
1809:PMC
1791:doi
1677:PMC
1669:doi
1665:134
1342:in
1225:gut
1191:or
1094:or
952:of
843:4.
828:3.
813:in
801:2.
649:in
233:of
143:to
68:or
4174::
3386:/
3300:.
3290:.
3280:64
3278:.
3274:.
3255:.
3232:.
3222:.
3212:76
3210:.
3206:.
3183:.
3173:.
3163:.
3151:.
3147:.
3124:.
3110:.
3106:.
3083:.
3075:.
3067:.
3055:.
3043:^
3029:.
3019:.
3009:.
2997:.
2993:.
2970:.
2913:.
2905:.
2897:.
2885:.
2862:.
2854:.
2844:37
2842:.
2816:.
2794:.
2786:.
2778:.
2766:.
2743:.
2733:.
2723:23
2721:.
2717:.
2705:^
2693:34
2691:.
2687:.
2664:.
2656:.
2642:.
2638:.
2615:.
2605:.
2595:55
2593:.
2589:.
2566:.
2500:^
2486:.
2476:18
2474:.
2451:.
2432:.
2380:.
2372:.
2362:40
2360:.
2348:^
2334:.
2322:^
2308:.
2294:^
2270:.
2256:^
2242:.
2232:.
2222:.
2212:11
2210:.
2206:.
2156:.
2146:12
2144:.
2117:29
2115:.
2111:.
2038:,
2013:16
2011:.
1954:.
1944:.
1934:41
1932:.
1928:.
1881:.
1871:.
1857:.
1853:.
1831:^
1817:.
1807:.
1799:.
1789:.
1777:.
1773:.
1761:^
1743:.
1739:.
1712:.
1708:.
1685:.
1675:.
1663:.
1659:.
1640:.
1604:^
1592:}}
1588:{{
1572:.
1251:.
1203:.
1167:,
1163:,
1159:,
1155:,
1130:.
977:.
878:.
751:.
731:,
727:.
635:,
600:,
420:.
384:,
337:).
275:.
179:.
155:,
4025:)
3985:e
3978:t
3971:v
3883:/
3874:/
3804:e
3797:t
3790:v
3375:e
3368:t
3361:v
3308:.
3286::
3240:.
3218::
3191:.
3167::
3159::
3132:.
3118::
3091:.
3071::
3063::
3037:.
3013::
3005::
2999:8
2921:.
2901::
2893::
2870:.
2850::
2827:.
2802:.
2782::
2774::
2751:.
2729::
2672:.
2650::
2644:7
2623:.
2601::
2574:.
2494:.
2482::
2417:.
2388:.
2368::
2288:.
2250:.
2226::
2218::
2191:.
2164:.
2152::
2129:.
2123::
2109:"
2093:.
1987:.
1962:.
1940::
1914:.
1889:.
1865::
1859:9
1825:.
1793::
1785::
1779:8
1755:.
1724:.
1693:.
1671::
1598:)
1584:.
1557:.
791:2
787:2
593:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.