1067:
42:
844:; also called Sdh5 in yeast and SdhE in bacteria) and by some of the Krebs cycle intermediates. Fumarate most strongly stimulates covalent flavinylation of SDHA. Through studies of the bacterial system, the mechanism of FAD attachment has been shown to involve a quinone:methide intermediate. In mitochondrial, but not bacterial, assembly, SDHA interacts with a second assembly factor called succinate dehydrogenase assembly factor 4 (SDHAF4; called Sdh8 in yeast) before it is inserted into the final complex.
905:
894:
4278:
1056:
1349:. Indeed, oxaloacetate is one of the most potent inhibitors of Complex II. Why a common TCA cycle intermediate would inhibit Complex II is not entirely understood, though it may exert a protective role in minimizing reverse-electron transfer mediated production of superoxide by Complex I. Atpenin 5a are highly potent Complex II inhibitors mimicking ubiquinone binding.
414:
857:
are being preformed in the mitochondrial matrix by protein complex ISU. The complex is also thought to be capable of inserting the iron-sulphur clusters in SDHB during its maturation. The studies suggest that Fe-S cluster insertion precedes SDHA-SDHB dimer forming. Such incorporation requires
1507:
Reduced levels of the mitochondrial enzyme succinate dehydrogenase (SDH), the main element of complex II, are observed post mortem in the brains of patients with
Huntington's Disease, and energy metabolism defects have been identified in both presymptomatic and symptomatic HD patients.
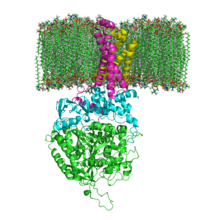
515:, are also found in the SdhC and SdhD subunits (not shown in the image). They serve to occupy the hydrophobic space below the heme b. These subunits are displayed in the attached image. SdhA is green, SdhB is teal, SdhC is fuchsia, and SdhD is yellow. Around SdhC and SdhD is a
475:
and SdhB contains three iron-sulfur clusters: , , and . The second two subunits are hydrophobic membrane anchor subunits, SdhC and SdhD. Human mitochondria contain two distinct isoforms of SdhA (Fp subunits type I and type II), these isoforms are also found in
2109:
Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, et al. (January 2008). "High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates".
938:
of the enzymes from multiple organisms shows that this is well poised for the proton transfer step. Thereafter, there are two possible elimination mechanisms: E2 or E1cb. In the E2 elimination, the mechanism is concerted. The basic
2254:
Dubos T, Pasquali M, Pogoda F, Casanova A, Hoffmann L, Beyer M (January 2013). "Differences between the succinate dehydrogenase sequences of isopyrazam sensitive
Zymoseptoria tritici and insensitive Fusarium graminearum strains".
2468:
Skillings EA, Morton AJ (2016). "Delayed Onset and
Reduced Cognitive Deficits through Pre-Conditioning with 3-Nitropropionic Acid is Dependent on Sex and CAG Repeat Length in the R6/2 Mouse Model of Huntington's Disease".
3769:
1631:
Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K (August 2003). "Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase)".
1767:"Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction"
2398:
Rijken JA, Niemeijer ND, Jonker MA, Eijkelenkamp K, Jansen JC, van Berkel A, et al. (January 2018). "The penetrance of paraganglioma and pheochromocytoma in SDHB germline mutation carriers".
2181:
Avenot HF, Michailides TJ (2010). "Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi".
2521:
885:
insertion and even its function. Heme b prosthetic group does not appear to be part of electron transporting pathway within the complex II. The cofactor rather maintains the anchor stability.
1329:
There are two distinct classes of inhibitors (SDHIs) of complex II: those that bind in the succinate pocket and those that bind in the ubiquinone pocket. Ubiquinone type inhibitors include
1670:
Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, et al. (January 2003). "Architecture of succinate dehydrogenase and reactive oxygen species generation".
2897:
2860:
870:(Sdh6) and SDHAF3 (Sdh7 in yeast), seem to be involved in SDHB maturation in way of protecting the subunit or dimer SDHA-SDHB from Fe-S cluster damage caused by ROS.
222:
3702:
2514:
1558:"The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies"
708:
of His207 of subunit B, Ser27 and Arg31 of subunit C, and Tyr83 of subunit D. The quinone ring is surrounded by Ile28 of subunit C and Pro160 of subunit B. These
291:
279:
2794:
241:
3736:
3378:
2507:
1489:. SDHB mutations are the most penetrant for paraganglioma and pheochromocytoms and both SDHD and SDHAF2 mutations have some maternal imprinting effects.
3879:
1527:
930:, which involves the transfer of a proton and a hydride. A combination of mutagenesis and structural analysis identifies Arg-286 of the SDHA subunit (
874:
3373:
3464:
3046:
2765:
3132:
362:. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.
2307:
2236:
2165:
1409:
2941:
3749:
3745:
2890:
3964:
3695:
1606:
1471:
3232:
3167:
2647:
2566:
862:
residues within active site of SDHB. Both reduced cysteine residues and already incorporated Fe-S clusters are highly susceptible to
3997:
3837:
3732:
3177:
3002:
1066:
3503:
3301:
2936:
2355:
Barletta JA, Hornick JL (July 2012). "Succinate dehydrogenase-deficient tumors: diagnostic advances and clinical implications".
234:
3670:
3498:
3162:
2883:
1281:. His207 and Asp82 most likely facilitate this process. Other studies claim that Tyr83 of subunit D is coordinated to a nearby
700:. Ubiquinone binding site Qp, which shows higher affinity to ubiquinone, is located in a gap composed of SdhB, SdhC, and SdhD.
3797:
3754:
3688:
3493:
3279:
3237:
2804:
2634:
2622:
2571:
1598:
1467:
1406:. Some agriculturally important fungi are not sensitive towards members of the new generation of ubiquinone type inhibitors.
161:
4153:
185:
3832:
2643:
2330:
1386:. More recently, other compounds with a broader spectrum against a range of plant pathogens have been developed including
4268:
3959:
3792:
3083:
3025:
2915:
2630:
983:. Further research is required to determine which elimination mechanism succinate undergoes in Succinate Dehydrogenase.
347:
3822:
3137:
2070:"The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b"
1018:
976:
830:
784:
753:
461:
3954:
3842:
3802:
3311:
3306:
3110:
2976:
4138:
3715:
4254:
4241:
4228:
4215:
4202:
4189:
4176:
3910:
3896:
3852:
3779:
3764:
3728:
3036:
2731:
4148:
2433:
Baysal BE (May 2013). "Mitochondrial complex II and genomic imprinting in inheritance of paraganglioma tumors".
41:
4102:
4045:
3719:
3227:
3105:
2726:
2686:
2561:
512:
359:
179:
72:
847:
2301:
4050:
3274:
2667:
1903:"The roles of SDHAF2 and dicarboxylate in covalent flavinylation of SDHA, the human complex II flavoprotein"
1425:
1334:
1187:
1175:
944:
863:
850:
833:
724:, which lies closer to inter-membrane space, is composed of SdhD only and has lower affinity to ubiquinone.
465:
166:
3915:
3269:
2926:
2710:
2659:
2626:
4071:
3990:
3817:
3643:
3383:
3259:
3095:
3073:
2604:
1413:
516:
246:
4143:
154:
3827:
3759:
3484:
3390:
3247:
3196:
3142:
2775:
2705:
2593:
2264:
2190:
2024:
1914:
1679:
456:(SdhB), form a hydrophilic head where enzymatic activity of the complex takes place. SdhA contains a
89:
4107:
2855:
1962:"How an assembly factor enhances covalent FAD attachment to the flavoprotein subunit of complex II"
1765:
Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, et al. (March 2006).
1300:
1214:
940:
904:
764:
709:
453:
398:
303:
84:
3680:
1374:
were found to have comparable activity to carboxin and a number of these were marketed, including
829:
into the mitochondrial matrix. Subsequently, one of the first steps is covalent attachment of the
182:
4040:
3905:
3414:
3182:
3056:
2746:
2380:
1703:
1537:
1202:
927:
106:
1155:
in succinate dehydrogenase is still being researched, some studies have asserted that the first
893:
712:, along with Il209, Trp163, and Trp164 of subunit B, and Ser27 (C atom) of subunit C, form the
4298:
3932:
3807:
3404:
3207:
2988:
2534:
2486:
2450:
2415:
2372:
2280:
2242:
2232:
2161:
2127:
2091:
2050:
1993:
1942:
1883:
1842:
1788:
1747:
1695:
1649:
1579:
935:
810:
806:
366:
355:
173:
4086:
4081:
4055:
3983:
3812:
3217:
3010:
2829:
2824:
2697:
2551:
2478:
2442:
2407:
2364:
2272:
2224:
2198:
2153:
2119:
2081:
2040:
2032:
1983:
1973:
1932:
1922:
1873:
1832:
1824:
1778:
1737:
1687:
1641:
1569:
1459:
1451:
1183:
802:. All edge-to-edge distances between the centers are less than the suggested 14 Å limit for
1261:
and is coordinated by His207 of subunit B, Arg31 of subunit C, and Asp82 of subunit D. The
4133:
4117:
4030:
3122:
3015:
1463:
437:
142:
2499:
2268:
2194:
2028:
1918:
1683:
118:
4282:
4171:
4112:
3711:
2045:
2012:
1988:
1961:
1937:
1902:
1837:
1812:
1482:
1475:
1399:
1254:
1109:
343:
217:
77:
17:
1878:
1861:
1356:
in agriculture since the 1960s. Carboxin was mainly used to control disease caused by
197:
4292:
4076:
4035:
2906:
1493:
1486:
1479:
1455:
1433:
1357:
1105:
1093:
803:
489:
429:
284:
192:
2384:
1707:
672:
648:
624:
617:
592:
585:
4025:
3539:
3514:
3172:
3051:
2964:
2770:
2756:
2678:
1429:
1395:
1346:
948:
733:
689:
686:
504:
485:
472:
449:
1610:
308:
296:
2446:
2368:
2276:
2228:
2202:
1828:
4249:
4184:
4020:
1862:"The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase"
1371:
1366:
1262:
1258:
1124:
1033:
991:
956:
821:
All subunits of human mitochondrial SDH are nuclear encoded. After translation,
745:
713:
705:
637:
508:
445:
441:
201:
4277:
2157:
2148:
Walter H (2016). "Fungicidal
Succinate-Dehydrogenase-Inhibiting Carboxamides".
2036:
1907:
Proceedings of the
National Academy of Sciences of the United States of America
1742:
1725:
1055:
3291:
2953:
2614:
2530:
1978:
1379:
1316:
1293:
1274:
1266:
1222:
1195:
1168:
1160:
1140:
1132:
1113:
1101:
1089:
1026:
780:
701:
500:
457:
390:
351:
4223:
4197:
3222:
2556:
1927:
1726:"Crystal structure of mitochondrial respiratory membrane protein complex II"
1691:
1436:
1391:
1375:
1361:
1353:
1297:
1282:
1278:
1246:
1206:
1136:
1097:
1022:
1014:
1011:
1007:
964:
960:
924:
921:
826:
776:
760:
741:
737:
469:
433:
394:
378:
374:
370:
340:
2490:
2454:
2419:
2376:
2284:
2246:
2131:
2095:
2086:
2069:
2054:
1997:
1946:
1846:
1792:
1783:
1766:
1751:
1699:
1653:
1583:
1574:
1557:
3368:
3242:
2875:
2588:
2068:
Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH (October 2006).
2013:"Crystal structure of an assembly intermediate of respiratory Complex II"
1887:
1403:
1387:
1383:
1338:
1330:
1308:
1286:
1230:
1179:
1156:
1128:
1117:
1082:
1044:
1029:
987:
984:
968:
859:
756:
749:
402:
382:
2150:
Bioactive
Carboxylic Compound Classes: Pharmaceuticals and Agrochemicals
1813:"Protein-mediated assembly of succinate dehydrogenase and its cofactors"
1645:
1221:. His207 of subunit B is in direct proximity to the cluster, the bound
3856:
3528:
3523:
3443:
2482:
2123:
1270:
1250:
1242:
995:
980:
972:
952:
799:
717:
554:
492:
149:
130:
2411:
1182:
sink. Its role is to prevent the interaction of the intermediate with
4236:
4006:
3947:
3942:
3937:
3920:
3783:
3633:
3628:
3598:
3593:
3473:
3453:
3448:
3438:
3433:
3428:
3423:
2537:
1497:
1444:
1440:
1342:
1312:
1289:
1085:
1041:
882:
867:
841:
837:
796:
792:
496:
336:
229:
125:
113:
101:
1021:, they tunnel along the relay until they reach the cluster. These
1724:
Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, et al. (July 2005).
413:
4210:
3925:
3653:
3648:
3638:
3623:
3618:
3613:
3608:
3603:
3588:
3583:
3578:
3573:
3568:
3563:
3558:
3553:
3548:
3344:
3331:
2839:
2834:
2223:. Advances in Applied Microbiology. Vol. 90. pp. 29–92.
1501:
1120:
783:-binding site are connected by a chain of redox centers including
666:
Succinate dehydrogenase cytochrome b small subunit, mitochondrial
547:
412:
386:
1901:
Sharma P, Maklashina E, Cecchini G, Iverson TM (September 2020).
1447:
has also been shown to be lethal at the embryonic stage in mice.
3884:
3874:
3869:
2819:
2814:
2809:
2799:
2789:
2582:
1532:
1522:
1517:
1504:
suppression; and, therefore, is the object of ongoing research.
1226:
1218:
1210:
1191:
1172:
1164:
1152:
1037:
878:
854:
822:
788:
669:
661:
645:
621:
614:
605:
589:
582:
573:
559:
137:
3979:
3684:
2879:
2503:
2011:
Sharma P, Maklashina E, Cecchini G, Iverson TM (January 2018).
1337:. Succinate-analogue inhibitors include the synthetic compound
1424:
The fundamental role of succinate-coenzyme Q reductase in the
1304:
642:
Succinate dehydrogenase cytochrome b560 subunit, mitochondrial
35:
succinate dehydrogenase (succinate-ubiquinone oxidoreductase)
579:
Succinate dehydrogenase flavoprotein subunit, mitochondrial
1131:
arrives from the cluster to provide full reduction of the
611:
Succinate dehydrogenase iron-sulfur subunit, mitochondrial
3975:
1811:
Van
Vranken JG, Na U, Winge DR, Rutter J (December 2014).
873:
Assembly of the hydrophobic anchor consisting of subunits
720:-binding pocket Qp. In contrast, ubiquinone binding site Q
1603:
1492:
Mammalian succinate dehydrogenase functions not only in
1217:
His207, which lies directly between the cluster and the
836:(covalent flavinylation). This process is enhanced by
1817:
Critical
Reviews in Biochemistry and Molecular Biology
1096:
interactions with Tyr83 of subunit D. The presence of
748:
Thr254, His354, and Arg399 of subunit A stabilize the
692:
can be recognized on mammalian SDH – matrix-proximal Q
519:
with the intermembrane space at the top of the image.
354:. It is the only enzyme that participates in both the
4266:
1960:
Maklashina E, Iverson TM, Cecchini G (October 2022).
1599:"Using Histochemistry to Determine Muscle Properties"
1104:
into a second orientation. This facilitates a second
1100:
in the iron sulphur cluster induces the movement of
3770:
Dihydroxymethyloxo-tetrahydroquinoline dehydrogenase
4162:
4126:
4095:
4064:
4013:
3895:
3851:
3778:
3727:
3537:
3512:
3482:
3462:
3412:
3403:
3357:
3320:
3288:
3256:
3204:
3195:
3151:
3119:
3092:
3070:
3033:
3024:
3001:
2973:
2950:
2923:
2914:
2848:
2739:
2723:
2695:
2676:
2657:
2612:
2603:
2544:
1116:and Ser27 of subunit C. Following the first single
795:clusters. This chain extends over 40 Å through the
537:
302:
290:
278:
270:
265:
260:
240:
228:
216:
211:
191:
172:
160:
148:
136:
124:
112:
100:
95:
83:
71:
66:
34:
1470:(GISTs). SDHA, SDHB, SDHD, and SDHAF1 can cause
975:intermediate is formed, shown in image 7, before
436:SQRs are composed of four structurally different
46:The structure of SQR in a phospholipid membrane.
2861:Electron-transferring-flavoprotein dehydrogenase
1450:All SDHx mutations can lead to tumorogenesis in
2766:Complex III/Coenzyme Q - cytochrome c reductase
1719:
1717:
1077:, iron-sulfur centers, heme b, and ubiquinone.
3991:
3696:
2891:
2515:
1412:has a working group for SDHIs and recommends
1352:Ubiquinone type inhibitors have been used as
8:
1213:from the cluster. A potential candidate is
1092:is oriented at the active site (image 4) by
1025:are subsequently transferred to an awaiting
1163:via may tunnel back and forth between the
3998:
3984:
3976:
3703:
3689:
3681:
3409:
3379:Mitochondrial permeability transition pore
3361:
3201:
3030:
2920:
2898:
2884:
2876:
2736:
2609:
2522:
2508:
2500:
1665:
1663:
1496:energy generation, but also has a role in
1073:Electron carriers of the SQR complex. FADH
208:
40:
2219:Lucas JA, Hawkins NJ, Fraaije BA (2015).
2143:
2141:
2085:
2044:
1987:
1977:
1936:
1926:
1877:
1836:
1782:
1741:
1573:
838:succinate dehydrogenase assembly factor 2
397:. This occurs in the inner mitochondrial
3374:Mitochondrial membrane transport protein
1454:, causing neuroendocrine tumors such as
1341:as well as the TCA cycle intermediates,
1311:, making it more suitable to donate its
1201:It has also been proposed that a gating
1065:
1054:
903:
892:
4273:
1548:
881:remains unclear. Especially in case of
866:damage. Two more SDH assembly factors,
3133:Cholesterol side-chain cleavage enzyme
1253:are needed. It has been argued that a
1127:radical species is formed. The second
1047:tunneling system is shown in image 9.
484:. The subunits form a membrane-bound
257:
31:
2308:Fungicide Resistance Action Committee
2257:Pesticide Biochemistry and Physiology
2221:The Evolution of Fungicide Resistance
1556:Oyedotun KS, Lemire BD (March 2004).
1273:delivered from HOH39, completing the
7:
3746:Enoyl-acyl carrier protein reductase
1806:
1804:
1802:
3965:3-oxo-5beta-steroid 4-dehydrogenase
3960:Isovaleryl coenzyme A dehydrogenase
3047:Coenzyme Q – cytochrome c reductase
2074:The Journal of Biological Chemistry
1966:The Journal of Biological Chemistry
1866:The Journal of Biological Chemistry
1771:The Journal of Biological Chemistry
1607:University of California, San Diego
1562:The Journal of Biological Chemistry
1472:Mitochondrial complex II deficiency
934:numbering) as the proton shuttle.
911:E1cb Succinate oxidation mechanism.
420:Subunits of succinate dehydrogenase
3233:Oxoglutarate dehydrogenase complex
3168:Glycerol-3-phosphate dehydrogenase
2752:Complex II/Succinate dehydrogenase
2648:Pyruvate dehydrogenase phosphatase
1233:flow between these redox centers.
1151:Although the functionality of the
25:
3838:Pyrroloquinoline-quinone synthase
3178:Carnitine palmitoyltransferase II
900:E2 Succinate oxidation mechanism.
767:, . This can be seen in image 5.
4276:
3302:Carbamoyl phosphate synthetase I
2942:Long-chain-fatty-acid—CoA ligase
2937:Carnitine palmitoyltransferase I
3163:Glutamate aspartate transporter
2776:Complex IV/Cytochrome c oxidase
2471:Journal of Huntington's Disease
2302:"SDHI Fungicides Working Group"
1209:from tunneling directly to the
1205:may be in place to prevent the
1171:intermediate. In this way, the
1143:reduction is shown in image 8.
1062:Ubiquinone reduction mechanism.
971:—refer to image 6. In E1cb, an
3798:Coproporphyrinogen III oxidase
3755:7-Dehydrocholesterol reductase
3280:Pyruvate dehydrogenase complex
3238:Succinyl coenzyme A synthetase
2623:Pyruvate dehydrogenase complex
2357:Advances in Anatomic Pathology
1468:Gastrointestinal stromal tumor
1370:diseases. In the 1980s simple
325:succinate-coenzyme Q reductase
1:
3833:Tryptophan alpha,beta-oxidase
2644:Pyruvate dehydrogenase kinase
2435:Biochimica et Biophysica Acta
1879:10.1016/S0021-9258(19)41598-6
3793:Dihydroorotate dehydrogenase
3084:Dihydroorotate dehydrogenase
2747:Complex I/NADH dehydrogenase
2447:10.1016/j.bbabio.2012.12.005
2369:10.1097/PAP.0b013e31825c6bc6
2277:10.1016/j.pestbp.2012.11.004
2229:10.1016/bs.aambs.2014.09.001
2203:10.1016/j.cropro.2010.02.019
1829:10.3109/10409238.2014.990556
813:is demonstrated in image 8.
523:Table of subunit composition
448:. The first two subunits, a
405:the two reactions together.
348:inner mitochondrial membrane
3823:Tetrahydroberberine oxidase
3138:Steroid 11-beta-hydroxylase
1108:interaction between the O4
990:, now loosely bound to the
825:subunit is translocated as
462:flavin adenine dinucleotide
4315:
3955:Glutaryl-CoA dehydrogenase
3843:L-galactonolactone oxidase
3803:Protoporphyrinogen oxidase
3312:N-Acetylglutamate synthase
3307:Ornithine transcarbamylase
3111:Glycerol phosphate shuttle
2977:monoamine neurotransmitter
2567:Oxoglutarate dehydrogenase
2331:"Recommendations for SDHI"
2158:10.1002/9783527693931.ch31
2037:10.1038/s41467-017-02713-8
1743:10.1016/j.cell.2005.05.025
562:family with Human protein
4154:Michaelis–Menten kinetics
3911:Butyryl-CoA dehydrogenase
3765:2,4 Dienoyl-CoA reductase
3666:
3364:
3340:
3037:oxidative phosphorylation
2732:oxidative phosphorylation
1979:10.1016/j.jbc.2022.102472
553:Protein description from
207:
39:
4046:Diffusion-limited enzyme
3228:Isocitrate dehydrogenase
3106:Malate-aspartate shuttle
2727:electron transport chain
2687:Methylmalonyl-CoA mutase
2562:Isocitrate dehydrogenase
1860:Kenney WC (April 1975).
1597:webmaster (2009-03-04).
1139:. This mechanism of the
920:Much is known about the
513:phosphatidylethanolamine
360:electron transport chain
3865:Succinate dehydrogenase
3275:Glutamate dehydrogenase
3062:Succinate dehydrogenase
2668:Glutamate dehydrogenase
2579:Succinate dehydrogenase
2572:Succinyl CoA synthetase
2112:The Biochemical Journal
1928:10.1073/pnas.2007391117
1692:10.1126/science.1079605
1634:Journal of Biochemistry
1432:makes it vital in most
1426:electron transfer chain
1335:thenoyltrifluoroacetone
1257:(HOH39) arrives at the
1198:, is shown in image 4.
1188:reactive oxygen species
817:Assembly and maturation
681:Ubiquinone binding site
339:complex, found in many
317:Succinate dehydrogenase
261:Succinate dehydrogenase
18:Resistance to fluopyram
3916:Acyl CoA dehydrogenase
3671:mitochondrial diseases
3270:Aspartate transaminase
2927:fatty acid degradation
2711:Aspartate transaminase
2087:10.1074/jbc.M607476200
1784:10.1074/jbc.M508173200
1575:10.1074/jbc.M311876200
1078:
1063:
994:, is free to exit the
951:, and FAD accepts the
912:
901:
728:Succinate binding site
482:Caenorhabditis elegans
421:
333:respiratory complex II
274:Respiratory complex II
4139:Eadie–Hofstee diagram
4072:Allosteric regulation
3818:Dihydrouracil oxidase
3716:CH–CH oxidoreductases
3384:Mitochondrial carrier
3260:anaplerotic reactions
3096:mitochondrial shuttle
3074:pyrimidine metabolism
2017:Nature Communications
1414:resistance management
1229:; and could modulate
1147:Heme prosthetic group
1069:
1058:
907:
896:
704:is stabilized by the
517:phospholipid membrane
416:
4149:Lineweaver–Burk plot
3828:Secologanin synthase
3760:Biliverdin reductase
3391:Translocator protein
3248:Malate dehydrogenase
3143:Aldosterone synthase
2706:Pyruvate carboxylase
2594:Malate dehydrogenase
2152:. pp. 405–425.
1241:To fully reduce the
1051:Ubiquinone reduction
765:iron-sulfur clusters
763:to the first of the
3750:Enoyl ACP reductase
3003:Intermembrane space
2856:Alternative oxidase
2660:α-ketoglutaric acid
2269:2013PBioP.105...28D
2195:2010CrPro..29..643A
2080:(43): 32310–32317.
2029:2018NatCo...9..274S
1919:2020PNAS..11723548S
1913:(38): 23548–23556.
1684:2003Sci...299..700Y
1194:group, relative to
916:Succinate oxidation
716:environment of the
696:and matrix-distal Q
503:-binding site. Two
454:iron-sulfur protein
4108:Enzyme superfamily
4041:Enzyme promiscuity
3906:Fumarate reductase
3358:Other/to be sorted
3323:alcohol metabolism
3183:Uncoupling protein
3057:NADH dehydrogenase
2483:10.3233/JHD-160189
2124:10.1042/BJ20071162
1538:Fumarate reductase
1474:which can lead to
1439:, removal of this
1285:as well as the O1
1079:
1064:
1002:Electron tunneling
936:Crystal structures
913:
902:
779:-binding site and
732:SdhA provides the
422:
4264:
4263:
3973:
3972:
3899:: Other acceptors
3808:Bilirubin oxidase
3678:
3677:
3662:
3661:
3405:Mitochondrial DNA
3399:
3398:
3353:
3352:
3208:citric acid cycle
3191:
3190:
2997:
2996:
2989:Monoamine oxidase
2873:
2872:
2869:
2868:
2719:
2718:
2535:Citric acid cycle
2412:10.1111/cge.13055
2400:Clinical Genetics
2238:978-0-12-802275-7
2167:978-3-527-33947-1
1777:(11): 7309–7316.
1678:(5607): 700–704.
1646:10.1093/jb/mvg144
1568:(10): 9424–9431.
1010:are derived from
947:deprotonates the
851:prosthetic groups
811:electron transfer
807:electron transfer
678:
677:
488:complex with six
367:citric acid cycle
365:In step 6 of the
356:citric acid cycle
314:
313:
256:
255:
252:
251:
155:metabolic pathway
16:(Redirected from
4306:
4281:
4280:
4272:
4144:Hanes–Woolf plot
4087:Enzyme activator
4082:Enzyme inhibitor
4056:Enzyme catalysis
4000:
3993:
3986:
3977:
3813:Acyl-CoA oxidase
3705:
3698:
3691:
3682:
3542:
3517:
3487:
3467:
3417:
3410:
3362:
3325:
3295:
3263:
3218:Citrate synthase
3211:
3202:
3156:
3126:
3099:
3077:
3040:
3031:
3011:Adenylate kinase
2982:
2958:
2930:
2921:
2900:
2893:
2886:
2877:
2737:
2698:oxaloacetic acid
2610:
2552:Citrate synthase
2524:
2517:
2510:
2501:
2495:
2494:
2465:
2459:
2458:
2430:
2424:
2423:
2395:
2389:
2388:
2352:
2346:
2345:
2343:
2342:
2327:
2321:
2320:
2318:
2317:
2298:
2292:
2288:
2250:
2213:
2207:
2206:
2178:
2172:
2171:
2145:
2136:
2135:
2106:
2100:
2099:
2089:
2065:
2059:
2058:
2048:
2008:
2002:
2001:
1991:
1981:
1957:
1951:
1950:
1940:
1930:
1898:
1892:
1891:
1881:
1872:(8): 3089–3094.
1857:
1851:
1850:
1840:
1808:
1797:
1796:
1786:
1762:
1756:
1755:
1745:
1736:(7): 1043–1057.
1721:
1712:
1711:
1667:
1658:
1657:
1628:
1622:
1621:
1619:
1618:
1609:. Archived from
1594:
1588:
1587:
1577:
1553:
1460:pheochromocytoma
1452:chromaffin cells
1184:molecular oxygen
759:and carries the
685:Two distinctive
538:
535:
534:
530:
258:
209:
61:
57:
53:
49:
44:
32:
21:
4314:
4313:
4309:
4308:
4307:
4305:
4304:
4303:
4289:
4288:
4287:
4275:
4267:
4265:
4260:
4172:Oxidoreductases
4158:
4134:Enzyme kinetics
4122:
4118:List of enzymes
4091:
4060:
4031:Catalytic triad
4009:
4004:
3974:
3969:
3891:
3847:
3774:
3723:
3712:Oxidoreductases
3709:
3679:
3674:
3658:
3538:
3533:
3513:
3508:
3483:
3478:
3463:
3458:
3413:
3395:
3349:
3336:
3321:
3316:
3289:
3284:
3257:
3252:
3205:
3187:
3152:
3147:
3123:steroidogenesis
3120:
3115:
3093:
3088:
3071:
3066:
3034:
3020:
3016:Creatine kinase
2993:
2979:
2974:
2969:
2951:
2946:
2924:
2910:
2904:
2874:
2865:
2844:
2786:
2760:
2730:
2725:
2715:
2691:
2672:
2653:
2599:
2540:
2528:
2498:
2467:
2466:
2462:
2432:
2431:
2427:
2397:
2396:
2392:
2354:
2353:
2349:
2340:
2338:
2329:
2328:
2324:
2315:
2313:
2300:
2299:
2295:
2291:
2253:
2239:
2218:
2214:
2210:
2183:Crop Protection
2180:
2179:
2175:
2168:
2147:
2146:
2139:
2108:
2107:
2103:
2067:
2066:
2062:
2010:
2009:
2005:
1959:
1958:
1954:
1900:
1899:
1895:
1859:
1858:
1854:
1810:
1809:
1800:
1764:
1763:
1759:
1723:
1722:
1715:
1669:
1668:
1661:
1630:
1629:
1625:
1616:
1614:
1596:
1595:
1591:
1555:
1554:
1550:
1546:
1514:
1464:renal carcinoma
1422:
1420:Role in disease
1327:
1315:to the reduced
1249:as well as two
1239:
1237:Proton transfer
1149:
1076:
1053:
1004:
918:
891:
853:of the subunit
819:
773:
730:
723:
699:
695:
683:
536:
532:
528:
526:
525:
507:molecules, one
495:containing one
427:
411:
280:OPM superfamily
62:
59:
55:
51:
47:
28:
23:
22:
15:
12:
11:
5:
4312:
4310:
4302:
4301:
4291:
4290:
4286:
4285:
4262:
4261:
4259:
4258:
4245:
4232:
4219:
4206:
4193:
4180:
4166:
4164:
4160:
4159:
4157:
4156:
4151:
4146:
4141:
4136:
4130:
4128:
4124:
4123:
4121:
4120:
4115:
4110:
4105:
4099:
4097:
4096:Classification
4093:
4092:
4090:
4089:
4084:
4079:
4074:
4068:
4066:
4062:
4061:
4059:
4058:
4053:
4048:
4043:
4038:
4033:
4028:
4023:
4017:
4015:
4011:
4010:
4005:
4003:
4002:
3995:
3988:
3980:
3971:
3970:
3968:
3967:
3962:
3957:
3952:
3951:
3950:
3945:
3940:
3930:
3929:
3928:
3923:
3913:
3908:
3902:
3900:
3893:
3892:
3890:
3889:
3888:
3887:
3882:
3877:
3872:
3861:
3859:
3849:
3848:
3846:
3845:
3840:
3835:
3830:
3825:
3820:
3815:
3810:
3805:
3800:
3795:
3789:
3787:
3776:
3775:
3773:
3772:
3767:
3762:
3757:
3752:
3742:
3740:
3725:
3724:
3710:
3708:
3707:
3700:
3693:
3685:
3676:
3675:
3667:
3664:
3663:
3660:
3659:
3657:
3656:
3651:
3646:
3641:
3636:
3631:
3626:
3621:
3616:
3611:
3606:
3601:
3596:
3591:
3586:
3581:
3576:
3571:
3566:
3561:
3556:
3551:
3545:
3543:
3535:
3534:
3532:
3531:
3526:
3520:
3518:
3510:
3509:
3507:
3506:
3501:
3496:
3490:
3488:
3480:
3479:
3477:
3476:
3470:
3468:
3460:
3459:
3457:
3456:
3451:
3446:
3441:
3436:
3431:
3426:
3420:
3418:
3407:
3401:
3400:
3397:
3396:
3394:
3393:
3388:
3387:
3386:
3381:
3371:
3365:
3359:
3355:
3354:
3351:
3350:
3348:
3347:
3341:
3338:
3337:
3335:
3334:
3328:
3326:
3318:
3317:
3315:
3314:
3309:
3304:
3298:
3296:
3286:
3285:
3283:
3282:
3277:
3272:
3266:
3264:
3254:
3253:
3251:
3250:
3245:
3240:
3235:
3230:
3225:
3220:
3214:
3212:
3199:
3193:
3192:
3189:
3188:
3186:
3185:
3180:
3175:
3170:
3165:
3159:
3157:
3149:
3148:
3146:
3145:
3140:
3135:
3129:
3127:
3117:
3116:
3114:
3113:
3108:
3102:
3100:
3090:
3089:
3087:
3086:
3080:
3078:
3068:
3067:
3065:
3064:
3059:
3054:
3049:
3043:
3041:
3028:
3026:Inner membrane
3022:
3021:
3019:
3018:
3013:
3007:
3005:
2999:
2998:
2995:
2994:
2992:
2991:
2985:
2983:
2971:
2970:
2968:
2967:
2961:
2959:
2948:
2947:
2945:
2944:
2939:
2933:
2931:
2918:
2916:Outer membrane
2912:
2911:
2905:
2903:
2902:
2895:
2888:
2880:
2871:
2870:
2867:
2866:
2864:
2863:
2858:
2852:
2850:
2846:
2845:
2843:
2842:
2837:
2832:
2827:
2822:
2817:
2812:
2807:
2802:
2797:
2792:
2784:
2779:
2778:
2773:
2768:
2763:
2758:
2754:
2749:
2743:
2741:
2734:
2721:
2720:
2717:
2716:
2714:
2713:
2708:
2702:
2700:
2693:
2692:
2690:
2689:
2683:
2681:
2674:
2673:
2671:
2670:
2664:
2662:
2655:
2654:
2652:
2651:
2642:(regulated by
2639:
2638:
2619:
2617:
2607:
2601:
2600:
2598:
2597:
2591:
2586:
2575:
2574:
2569:
2564:
2559:
2554:
2548:
2546:
2542:
2541:
2529:
2527:
2526:
2519:
2512:
2504:
2497:
2496:
2460:
2441:(5): 573–577.
2425:
2390:
2363:(4): 193–203.
2347:
2322:
2293:
2290:
2289:
2251:
2237:
2215:
2208:
2189:(7): 643–651.
2173:
2166:
2137:
2118:(2): 491–499.
2101:
2060:
2003:
1972:(10): 102472.
1952:
1893:
1852:
1823:(2): 168–180.
1798:
1757:
1713:
1659:
1640:(2): 191–195.
1623:
1589:
1547:
1545:
1542:
1541:
1540:
1535:
1530:
1525:
1520:
1513:
1510:
1483:encephalopathy
1476:Leigh syndrome
1421:
1418:
1400:pydiflumetofen
1358:basidiomycetes
1326:
1323:
1319:intermediate.
1303:decreases the
1255:water molecule
1238:
1235:
1148:
1145:
1110:carbonyl group
1074:
1052:
1049:
1003:
1000:
917:
914:
890:
887:
818:
815:
772:
769:
729:
726:
721:
697:
693:
682:
679:
676:
675:
667:
664:
659:
656:
652:
651:
643:
640:
635:
632:
628:
627:
612:
609:
603:
600:
596:
595:
580:
577:
571:
568:
564:
563:
557:
551:
545:
542:
524:
521:
452:(SdhA) and an
426:
423:
410:
407:
312:
311:
306:
300:
299:
294:
288:
287:
282:
276:
275:
272:
268:
267:
263:
262:
254:
253:
250:
249:
244:
238:
237:
232:
226:
225:
220:
214:
213:
205:
204:
195:
189:
188:
177:
170:
169:
164:
158:
157:
152:
146:
145:
140:
134:
133:
128:
122:
121:
116:
110:
109:
104:
98:
97:
93:
92:
87:
81:
80:
75:
69:
68:
64:
63:
45:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4311:
4300:
4297:
4296:
4294:
4284:
4279:
4274:
4270:
4256:
4252:
4251:
4246:
4243:
4239:
4238:
4233:
4230:
4226:
4225:
4220:
4217:
4213:
4212:
4207:
4204:
4200:
4199:
4194:
4191:
4187:
4186:
4181:
4178:
4174:
4173:
4168:
4167:
4165:
4161:
4155:
4152:
4150:
4147:
4145:
4142:
4140:
4137:
4135:
4132:
4131:
4129:
4125:
4119:
4116:
4114:
4113:Enzyme family
4111:
4109:
4106:
4104:
4101:
4100:
4098:
4094:
4088:
4085:
4083:
4080:
4078:
4077:Cooperativity
4075:
4073:
4070:
4069:
4067:
4063:
4057:
4054:
4052:
4049:
4047:
4044:
4042:
4039:
4037:
4036:Oxyanion hole
4034:
4032:
4029:
4027:
4024:
4022:
4019:
4018:
4016:
4012:
4008:
4001:
3996:
3994:
3989:
3987:
3982:
3981:
3978:
3966:
3963:
3961:
3958:
3956:
3953:
3949:
3946:
3944:
3941:
3939:
3936:
3935:
3934:
3931:
3927:
3924:
3922:
3919:
3918:
3917:
3914:
3912:
3909:
3907:
3904:
3903:
3901:
3898:
3894:
3886:
3883:
3881:
3878:
3876:
3873:
3871:
3868:
3867:
3866:
3863:
3862:
3860:
3858:
3854:
3850:
3844:
3841:
3839:
3836:
3834:
3831:
3829:
3826:
3824:
3821:
3819:
3816:
3814:
3811:
3809:
3806:
3804:
3801:
3799:
3796:
3794:
3791:
3790:
3788:
3785:
3781:
3777:
3771:
3768:
3766:
3763:
3761:
3758:
3756:
3753:
3751:
3747:
3744:
3743:
3741:
3738:
3734:
3730:
3726:
3721:
3717:
3713:
3706:
3701:
3699:
3694:
3692:
3687:
3686:
3683:
3673:
3672:
3665:
3655:
3652:
3650:
3647:
3645:
3642:
3640:
3637:
3635:
3632:
3630:
3627:
3625:
3622:
3620:
3617:
3615:
3612:
3610:
3607:
3605:
3602:
3600:
3597:
3595:
3592:
3590:
3587:
3585:
3582:
3580:
3577:
3575:
3572:
3570:
3567:
3565:
3562:
3560:
3557:
3555:
3552:
3550:
3547:
3546:
3544:
3541:
3536:
3530:
3527:
3525:
3522:
3521:
3519:
3516:
3511:
3505:
3502:
3500:
3497:
3495:
3492:
3491:
3489:
3486:
3481:
3475:
3472:
3471:
3469:
3466:
3461:
3455:
3452:
3450:
3447:
3445:
3442:
3440:
3437:
3435:
3432:
3430:
3427:
3425:
3422:
3421:
3419:
3416:
3411:
3408:
3406:
3402:
3392:
3389:
3385:
3382:
3380:
3377:
3376:
3375:
3372:
3370:
3367:
3366:
3363:
3360:
3356:
3346:
3343:
3342:
3339:
3333:
3330:
3329:
3327:
3324:
3319:
3313:
3310:
3308:
3305:
3303:
3300:
3299:
3297:
3294:
3293:
3287:
3281:
3278:
3276:
3273:
3271:
3268:
3267:
3265:
3262:
3261:
3255:
3249:
3246:
3244:
3241:
3239:
3236:
3234:
3231:
3229:
3226:
3224:
3221:
3219:
3216:
3215:
3213:
3210:
3209:
3203:
3200:
3198:
3194:
3184:
3181:
3179:
3176:
3174:
3171:
3169:
3166:
3164:
3161:
3160:
3158:
3155:
3150:
3144:
3141:
3139:
3136:
3134:
3131:
3130:
3128:
3125:
3124:
3118:
3112:
3109:
3107:
3104:
3103:
3101:
3098:
3097:
3091:
3085:
3082:
3081:
3079:
3076:
3075:
3069:
3063:
3060:
3058:
3055:
3053:
3050:
3048:
3045:
3044:
3042:
3039:
3038:
3032:
3029:
3027:
3023:
3017:
3014:
3012:
3009:
3008:
3006:
3004:
3000:
2990:
2987:
2986:
2984:
2981:
2978:
2972:
2966:
2963:
2962:
2960:
2957:
2955:
2949:
2943:
2940:
2938:
2935:
2934:
2932:
2929:
2928:
2922:
2919:
2917:
2913:
2908:
2907:Mitochondrial
2901:
2896:
2894:
2889:
2887:
2882:
2881:
2878:
2862:
2859:
2857:
2854:
2853:
2851:
2847:
2841:
2838:
2836:
2833:
2831:
2828:
2826:
2823:
2821:
2818:
2816:
2813:
2811:
2808:
2806:
2803:
2801:
2798:
2796:
2793:
2791:
2787:
2781:
2780:
2777:
2774:
2772:
2769:
2767:
2764:
2762:
2755:
2753:
2750:
2748:
2745:
2744:
2742:
2738:
2735:
2733:
2728:
2724:Mitochondrial
2722:
2712:
2709:
2707:
2704:
2703:
2701:
2699:
2694:
2688:
2685:
2684:
2682:
2680:
2675:
2669:
2666:
2665:
2663:
2661:
2656:
2649:
2645:
2641:
2640:
2636:
2632:
2628:
2624:
2621:
2620:
2618:
2616:
2611:
2608:
2606:
2602:
2595:
2592:
2590:
2587:
2584:
2580:
2577:
2576:
2573:
2570:
2568:
2565:
2563:
2560:
2558:
2555:
2553:
2550:
2549:
2547:
2543:
2539:
2536:
2532:
2525:
2520:
2518:
2513:
2511:
2506:
2505:
2502:
2492:
2488:
2484:
2480:
2476:
2472:
2464:
2461:
2456:
2452:
2448:
2444:
2440:
2436:
2429:
2426:
2421:
2417:
2413:
2409:
2405:
2401:
2394:
2391:
2386:
2382:
2378:
2374:
2370:
2366:
2362:
2358:
2351:
2348:
2336:
2332:
2326:
2323:
2311:
2309:
2303:
2297:
2294:
2286:
2282:
2278:
2274:
2270:
2266:
2262:
2258:
2252:
2248:
2244:
2240:
2234:
2230:
2226:
2222:
2217:
2216:
2212:
2209:
2204:
2200:
2196:
2192:
2188:
2184:
2177:
2174:
2169:
2163:
2159:
2155:
2151:
2144:
2142:
2138:
2133:
2129:
2125:
2121:
2117:
2113:
2105:
2102:
2097:
2093:
2088:
2083:
2079:
2075:
2071:
2064:
2061:
2056:
2052:
2047:
2042:
2038:
2034:
2030:
2026:
2022:
2018:
2014:
2007:
2004:
1999:
1995:
1990:
1985:
1980:
1975:
1971:
1967:
1963:
1956:
1953:
1948:
1944:
1939:
1934:
1929:
1924:
1920:
1916:
1912:
1908:
1904:
1897:
1894:
1889:
1885:
1880:
1875:
1871:
1867:
1863:
1856:
1853:
1848:
1844:
1839:
1834:
1830:
1826:
1822:
1818:
1814:
1807:
1805:
1803:
1799:
1794:
1790:
1785:
1780:
1776:
1772:
1768:
1761:
1758:
1753:
1749:
1744:
1739:
1735:
1731:
1727:
1720:
1718:
1714:
1709:
1705:
1701:
1697:
1693:
1689:
1685:
1681:
1677:
1673:
1666:
1664:
1660:
1655:
1651:
1647:
1643:
1639:
1635:
1627:
1624:
1613:on 2018-10-10
1612:
1608:
1604:
1600:
1593:
1590:
1585:
1581:
1576:
1571:
1567:
1563:
1559:
1552:
1549:
1543:
1539:
1536:
1534:
1531:
1529:
1526:
1524:
1521:
1519:
1516:
1515:
1511:
1509:
1505:
1503:
1499:
1495:
1494:mitochondrial
1490:
1488:
1487:optic atrophy
1484:
1481:
1480:mitochondrial
1477:
1473:
1469:
1465:
1461:
1457:
1456:paraganglioma
1453:
1448:
1446:
1442:
1438:
1435:
1434:multicellular
1431:
1427:
1419:
1417:
1415:
1411:
1407:
1405:
1401:
1397:
1393:
1389:
1385:
1381:
1377:
1373:
1369:
1368:
1363:
1359:
1355:
1350:
1348:
1344:
1340:
1336:
1332:
1324:
1322:
1320:
1318:
1314:
1310:
1306:
1302:
1299:
1295:
1291:
1288:
1284:
1280:
1277:reduction to
1276:
1272:
1268:
1264:
1260:
1256:
1252:
1248:
1244:
1236:
1234:
1232:
1228:
1224:
1220:
1216:
1212:
1208:
1204:
1199:
1197:
1193:
1189:
1185:
1181:
1177:
1174:
1170:
1166:
1162:
1159:delivered to
1158:
1154:
1146:
1144:
1142:
1138:
1134:
1130:
1126:
1122:
1119:
1115:
1111:
1107:
1106:hydrogen bond
1103:
1099:
1095:
1094:hydrogen bond
1091:
1087:
1084:
1072:
1068:
1061:
1057:
1050:
1048:
1046:
1043:
1039:
1035:
1031:
1028:
1024:
1020:
1016:
1013:
1009:
1001:
999:
997:
993:
989:
986:
982:
978:
974:
970:
966:
962:
958:
954:
950:
946:
942:
937:
933:
929:
926:
923:
915:
910:
906:
899:
895:
888:
886:
884:
880:
876:
871:
869:
865:
861:
858:reduction of
856:
852:
849:
845:
843:
839:
835:
832:
828:
824:
816:
814:
812:
808:
805:
804:physiological
801:
798:
794:
790:
786:
782:
778:
771:Redox centers
770:
768:
766:
762:
758:
755:
751:
747:
743:
739:
735:
727:
725:
719:
715:
711:
707:
703:
691:
690:binding sites
688:
680:
674:
671:
668:
665:
663:
660:
657:
654:
653:
650:
647:
644:
641:
639:
636:
633:
630:
629:
626:
623:
619:
616:
613:
610:
607:
604:
601:
598:
597:
594:
591:
587:
584:
581:
578:
575:
572:
569:
566:
565:
561:
558:
556:
552:
549:
546:
544:Subunit name
543:
540:
539:
531:
522:
520:
518:
514:
510:
506:
502:
498:
494:
491:
490:transmembrane
487:
483:
479:
474:
471:
467:
463:
459:
455:
451:
447:
443:
439:
435:
431:
430:Mitochondrial
424:
419:
415:
408:
406:
404:
400:
396:
392:
388:
384:
380:
376:
372:
368:
363:
361:
357:
353:
349:
345:
342:
338:
334:
330:
326:
322:
318:
310:
307:
305:
301:
298:
295:
293:
289:
286:
283:
281:
277:
273:
269:
264:
259:
248:
245:
243:
239:
236:
233:
231:
227:
224:
221:
219:
215:
210:
206:
203:
199:
196:
194:
193:Gene Ontology
190:
187:
184:
181:
178:
175:
171:
168:
165:
163:
159:
156:
153:
151:
147:
144:
141:
139:
135:
132:
131:NiceZyme view
129:
127:
123:
120:
117:
115:
111:
108:
105:
103:
99:
94:
91:
88:
86:
82:
79:
76:
74:
70:
65:
43:
38:
33:
30:
19:
4250:Translocases
4247:
4234:
4221:
4208:
4195:
4185:Transferases
4182:
4169:
4026:Binding site
3933:5α-reductase
3864:
3668:
3515:ATP synthase
3322:
3290:
3258:
3206:
3173:ATP synthase
3153:
3121:
3094:
3072:
3061:
3052:Cytochrome c
3035:
2975:
2965:Kynureninase
2952:
2925:
2782:
2771:Cytochrome c
2751:
2679:succinyl-CoA
2578:
2477:(1): 19–32.
2474:
2470:
2463:
2438:
2434:
2428:
2406:(1): 60–66.
2403:
2399:
2393:
2360:
2356:
2350:
2339:. Retrieved
2337:. March 2020
2334:
2325:
2314:. Retrieved
2312:. 2020-01-31
2305:
2296:
2263:(1): 28–35.
2260:
2256:
2220:
2211:
2186:
2182:
2176:
2149:
2115:
2111:
2104:
2077:
2073:
2063:
2020:
2016:
2006:
1969:
1965:
1955:
1910:
1906:
1896:
1869:
1865:
1855:
1820:
1816:
1774:
1770:
1760:
1733:
1729:
1675:
1671:
1637:
1633:
1626:
1615:. Retrieved
1611:the original
1602:
1592:
1565:
1561:
1551:
1506:
1500:sensing and
1491:
1449:
1430:mitochondria
1423:
1408:
1396:fluxapyroxad
1372:benzanilides
1365:
1351:
1347:oxaloacetate
1328:
1321:
1245:in SQR, two
1240:
1200:
1150:
1080:
1070:
1059:
1005:
979:accepts the
949:alpha carbon
931:
919:
908:
897:
872:
846:
820:
774:
734:binding site
731:
684:
505:phospholipid
499:group and a
486:cytochrome b
481:
478:Ascaris suum
477:
473:binding site
450:flavoprotein
428:
417:
364:
332:
328:
324:
320:
316:
315:
119:BRENDA entry
29:
4021:Active site
3465:Complex III
2788:synthesis:
2605:Anaplerotic
1416:practices.
1367:Rhizoctonia
1265:species is
1263:semiquinone
1259:active site
1190:(ROS). The
1186:to produce
1178:acts as an
1125:semiquinone
1034:active site
1032:within the
992:active site
957:beta carbon
746:side chains
714:hydrophobic
706:side chains
509:cardiolipin
446:hydrophobic
442:hydrophilic
346:and in the
292:OPM protein
266:Identifiers
107:IntEnz view
67:Identifiers
4224:Isomerases
4198:Hydrolases
4065:Regulation
3485:Complex IV
3292:urea cycle
2980:metabolism
2956:metabolism
2954:tryptophan
2783:Coenzyme Q
2757:Coenzyme Q
2615:acetyl-CoA
2531:Metabolism
2341:2022-07-05
2316:2022-07-05
2023:(1): 274.
1617:2017-12-27
1544:References
1380:flutolanil
1362:stem rusts
1354:fungicides
1325:Inhibitors
1317:ubiquinone
1294:ubiquinone
1275:ubiquinone
1267:protonated
1225:, and the
1223:ubiquinone
1196:ubiquinone
1169:ubiquinone
1161:ubiquinone
1141:ubiquinone
1133:ubiquinone
1114:ubiquinone
1102:ubiquinone
1090:ubiquinone
1027:ubiquinone
1006:After the
963:the bound
827:apoprotein
781:ubiquinone
702:Ubiquinone
687:ubiquinone
662:DHSD_HUMAN
638:C560_HUMAN
501:ubiquinone
458:covalently
391:ubiquinone
352:eukaryotes
304:Membranome
176:structures
143:KEGG entry
90:9028-11-9
4103:EC number
3669:see also
3415:Complex I
3223:Aconitase
2557:Aconitase
1443:from the
1437:organisms
1392:fluopyram
1376:benodanil
1298:histidine
1283:histidine
1279:ubiquinol
1247:electrons
1207:electrons
1203:mechanism
1137:ubiquinol
1121:reduction
1098:electrons
1071:Image 9:
1060:Image 8:
1023:electrons
1015:oxidation
1012:succinate
1008:electrons
965:succinate
961:oxidizing
955:from the
928:mechanism
925:oxidation
922:succinate
909:Image 7:
898:Image 6:
889:Mechanism
777:succinate
761:electrons
742:succinate
738:oxidation
470:succinate
460:attached
434:bacterial
432:and many
409:Structure
395:ubiquinol
387:reduction
385:with the
379:succinate
375:oxidation
371:catalyzes
341:bacterial
96:Databases
4299:EC 1.3.5
4293:Category
4127:Kinetics
4051:Cofactor
4014:Activity
3786:acceptor
3739:acceptor
3369:Frataxin
3243:Fumarase
2909:proteins
2589:Fumarase
2491:27031731
2455:23291190
2420:28503760
2385:32088940
2377:22692282
2285:24238287
2247:25596029
2132:17916065
2096:16950775
2055:29348404
1998:36089066
1947:32887801
1847:25488574
1793:16407191
1752:15989954
1708:29222766
1700:12560550
1654:12966066
1584:14672929
1512:See also
1404:sedaxane
1388:boscalid
1384:mepronil
1360:such as
1339:malonate
1331:carboxin
1309:tyrosine
1287:carbonyl
1231:electron
1180:electron
1176:cofactor
1167:and the
1157:electron
1129:electron
1123:step, a
1118:electron
1083:carbonyl
1045:electron
1030:molecule
988:fumarate
985:Oxidized
969:fumarate
945:cofactor
860:cysteine
834:cofactor
787:and the
757:oxidizes
750:molecule
736:for the
710:residues
673:PF05328
649:PF01127
625:PF13183
593:PF02910
550:protein
511:and one
468:and the
466:cofactor
444:and two
438:subunits
425:Subunits
418:Image 5:
403:coupling
399:membrane
383:fumarate
358:and the
247:proteins
235:articles
223:articles
180:RCSB PDB
4283:Biology
4237:Ligases
4007:Enzymes
3857:Quinone
3529:MT-ATP8
3524:MT-ATP6
3444:MT-ND4L
2761:(CoQ10)
2740:Primary
2596:and ETC
2538:enzymes
2265:Bibcode
2191:Bibcode
2046:5773532
2025:Bibcode
1989:9557727
1938:7519310
1915:Bibcode
1838:4653115
1680:Bibcode
1672:Science
1301:residue
1271:protons
1251:protons
1243:quinone
1215:residue
1081:The O1
996:protein
981:hydride
973:enolate
953:hydride
941:residue
932:E. coli
809:. This
800:monomer
718:quinone
618:PF13085
586:PF00890
555:UniProt
493:helices
202:QuickGO
167:profile
150:MetaCyc
85:CAS no.
78:1.3.5.1
4269:Portal
4211:Lyases
3948:SRD5A3
3943:SRD5A2
3938:SRD5A1
3921:ACADSB
3897:1.3.99
3784:Oxygen
3634:MT-TS2
3629:MT-TS1
3599:MT-TL2
3594:MT-TL1
3504:MT-CO3
3499:MT-CO2
3494:MT-CO1
3474:MT-CYB
3454:MT-ND6
3449:MT-ND5
3439:MT-ND4
3434:MT-ND3
3429:MT-ND2
3424:MT-ND1
3197:Matrix
2830:COQ10B
2825:COQ10A
2489:
2453:
2418:
2383:
2375:
2306:FRAC (
2283:
2245:
2235:
2164:
2130:
2094:
2053:
2043:
1996:
1986:
1945:
1935:
1888:235539
1886:
1845:
1835:
1791:
1750:
1706:
1698:
1652:
1582:
1498:oxygen
1485:, and
1466:, and
1445:genome
1441:enzyme
1343:malate
1313:proton
1296:. The
1290:oxygen
1086:oxygen
1042:Sulfur
1036:. The
883:heme b
868:SDHAF1
842:SDHAF2
797:enzyme
793:sulfur
752:while
744:. The
608:_HUMAN
576:_HUMAN
527:": -->
497:heme b
464:(FAD)
440:: two
369:, SQR
337:enzyme
335:is an
271:Symbol
230:PubMed
212:Search
198:AmiGO
186:PDBsum
126:ExPASy
114:BRENDA
102:IntEnz
73:EC no.
27:Enzyme
4163:Types
3926:ACADS
3853:1.3.5
3780:1.3.3
3729:1.3.1
3654:MT-TY
3649:MT-TW
3644:MT-TV
3639:MT-TT
3624:MT-TR
3619:MT-TQ
3614:MT-TP
3609:MT-TN
3604:MT-TM
3589:MT-TK
3584:MT-TI
3579:MT-TH
3574:MT-TG
3569:MT-TF
3564:MT-TE
3559:MT-TD
3554:MT-TC
3549:MT-TA
3345:PMPCB
3332:ALDH2
3154:other
2849:Other
2840:PDSS2
2835:PDSS1
2545:Cycle
2381:S2CID
1704:S2CID
1502:tumor
548:Human
344:cells
331:) or
323:) or
162:PRIAM
4255:list
4248:EC7
4242:list
4235:EC6
4229:list
4222:EC5
4216:list
4209:EC4
4203:list
4196:EC3
4190:list
4183:EC2
4177:list
4170:EC1
3885:SDHD
3880:SDHC
3875:SDHB
3870:SDHA
3737:NADP
3722:1.3)
3540:tRNA
2820:COQ9
2815:COQ7
2810:COQ6
2805:COQ5
2800:COQ4
2795:COQ3
2790:COQ2
2646:and
2583:SDHA
2487:PMID
2451:PMID
2439:1827
2416:PMID
2373:PMID
2335:FRAC
2281:PMID
2243:PMID
2233:ISBN
2162:ISBN
2128:PMID
2092:PMID
2051:PMID
1994:PMID
1943:PMID
1884:PMID
1843:PMID
1789:PMID
1748:PMID
1730:Cell
1696:PMID
1650:PMID
1580:PMID
1533:SDHD
1528:SDHC
1523:SDHB
1518:SDHA
1410:FRAC
1402:and
1382:and
1364:and
1345:and
1333:and
1227:heme
1219:heme
1211:heme
1192:heme
1173:heme
1165:heme
1153:heme
1038:Iron
1017:via
879:SDHD
877:and
875:SDHC
855:SDHB
848:Fe-S
823:SDHA
789:iron
775:The
670:Pfam
658:SdhD
646:Pfam
634:SdhC
622:Pfam
615:Pfam
606:SDHB
602:SdhB
590:Pfam
583:Pfam
574:SDHA
570:SdhA
560:Pfam
541:No.
529:edit
480:and
373:the
297:1zoy
242:NCBI
183:PDBe
138:KEGG
60:SdhD
58:and
56:SdhC
52:SdhB
48:SdhA
3733:NAD
2696:to
2677:to
2658:to
2613:to
2479:doi
2443:doi
2408:doi
2365:doi
2273:doi
2261:105
2225:doi
2199:doi
2154:doi
2120:doi
2116:409
2082:doi
2078:281
2041:PMC
2033:doi
1984:PMC
1974:doi
1970:298
1933:PMC
1923:doi
1911:117
1874:doi
1870:250
1833:PMC
1825:doi
1779:doi
1775:281
1738:doi
1734:121
1688:doi
1676:299
1642:doi
1638:134
1570:doi
1566:279
1428:of
1307:of
1305:pKa
1292:of
1269:by
1135:to
1112:of
1088:of
1019:FAD
977:FAD
967:to
943:or
864:ROS
831:FAD
785:FAD
754:FAD
740:of
588:,
401:by
393:to
389:of
381:to
377:of
350:of
329:SQR
321:SDH
309:656
218:PMC
174:PDB
4295::
3855::
3782::
3731::
3720:EC
3714::
2785:10
2759:10
2635:E3
2633:,
2631:E2
2629:,
2627:E1
2533::
2485:.
2473:.
2449:.
2437:.
2414:.
2404:93
2402:.
2379:.
2371:.
2361:19
2359:.
2333:.
2304:.
2279:.
2271:.
2259:.
2241:.
2231:.
2197:.
2187:29
2185:.
2160:.
2140:^
2126:.
2114:.
2090:.
2076:.
2072:.
2049:.
2039:.
2031:.
2019:.
2015:.
1992:.
1982:.
1968:.
1964:.
1941:.
1931:.
1921:.
1909:.
1905:.
1882:.
1868:.
1864:.
1841:.
1831:.
1821:50
1819:.
1815:.
1801:^
1787:.
1773:.
1769:.
1746:.
1732:.
1728:.
1716:^
1702:.
1694:.
1686:.
1674:.
1662:^
1648:.
1636:.
1605:.
1601:.
1578:.
1564:.
1560:.
1478:,
1462:,
1458:,
1398:,
1394:,
1390:,
1378:,
998:.
959:,
620:,
200:/
54:,
50:,
4271::
4257:)
4253:(
4244:)
4240:(
4231:)
4227:(
4218:)
4214:(
4205:)
4201:(
4192:)
4188:(
4179:)
4175:(
3999:e
3992:t
3985:v
3748:/
3735:/
3718:(
3704:e
3697:t
3690:v
2899:e
2892:t
2885:v
2729:/
2650:)
2637:)
2625:(
2585:)
2581:(
2523:e
2516:t
2509:v
2493:.
2481::
2475:5
2457:.
2445::
2422:.
2410::
2387:.
2367::
2344:.
2319:.
2310:)
2287:.
2275::
2267::
2249:.
2227::
2205:.
2201::
2193::
2170:.
2156::
2134:.
2122::
2098:.
2084::
2057:.
2035::
2027::
2021:9
2000:.
1976::
1949:.
1925::
1917::
1890:.
1876::
1849:.
1827::
1795:.
1781::
1754:.
1740::
1710:.
1690::
1682::
1656:.
1644::
1620:.
1586:.
1572::
1075:2
1040:-
840:(
791:-
722:D
698:D
694:P
655:4
631:3
599:2
567:1
533:]
327:(
319:(
285:3
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.