185:
124:
33:
330:" phenomenon. Rather, bacterial retrons confer some protection from phage infection to bacterial hosts. Several retrons are located in DNA regions next to certain protein effector-coding genes. When their expression is activated, most of these effectors and their associated retrons function together to block phage infection.
567:
Bobonis, Jacob; Mitosch, Karin; Mateus, André; Karcher, Nicolai; Kritikos, George; Selkrig, Joel; Zietek, Matylda; Monzon, Vivian; Pfalz, Birgit; Garcia-Santamarina, Sarela; Galardini, Marco; Sueki, Anna; Kobayashi, Callie; Stein, Frank; Bateman, Alex (2022-09-01).
301:: Group II introns are the best characterized bacterial retroelement and the only type known to exhibit autonomous mobility; they consist of an RT encoded within a catalytic, self-splicing RNA structure. Group II intron mobility is mediated by a
756:
749:
742:
514:
Liu M, Gingery M, Doulatov SR, Liu Y, Hodes A, Baker S, Davis P, Simmonds M, Churcher C, Mungall K, Quail MA, Preston A, Harvill ET, Maskell DJ, Eiserling FA, Parkhill J, Miller JF (2004).
164:
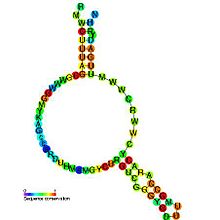
produced by retron elements and is the immediate precursor to the synthesis of msDNA. The retron msr RNA folds into a characteristic secondary structure that contains a conserved
311:(DGRs). The DGRs are not mobile, but function to diversify DNA sequences. For example, DGRs mediate the switch between pathogenic and free-living phases of
1312:
1245:
176:
end of the DNA chain via a 2′–5′ phosphodiester linkage that occurs from the 2′ position of the conserved internal guanosine residue.
1302:
308:
295:
Retrons were the first family of retroelement discovered in bacteria; the other two families of known bacterial retroelements are:
1307:
1230:
1218:
153:
244:
and other types of retroelements. Like other reverse transcriptases, the retron RT contains seven regions of conserved
188:
The retron operon carries a promoter sequence P that controls the synthesis of an RNA transcript carrying three loci:
172:
which is composed of small single-stranded DNA linked to small single-stranded RNA. The RNA strand is joined to the
623:
Millman A, Bernheim A, Stokar-Avihail A, Fedorenko T, Voichek M, Leavitt A, Oppenheimer-Shaanan Y, Sorek R (2020).
168:
residue at the end of a stem loop. Synthesis of DNA by the retron-encoded reverse transcriptase (RT) results in a
1235:
280:
For many years after their discovery in animal viruses, reverse transcriptases were believed to be absent from
1372:
1324:
468:
879:
149:
42:
111:
1334:
734:
728:
473:
909:
334:
38:
1203:
1125:
605:
385:
169:
1181:
1097:
999:
769:
697:
646:
597:
589:
549:
496:
437:
377:
302:
118:
106:
687:
677:
636:
581:
539:
531:
486:
478:
427:
419:
369:
94:
1041:
298:
1090:
1055:
1048:
1027:
1006:
983:
863:
692:
665:
491:
456:
432:
407:
161:
544:
515:
326:
Since retrons are not mobile, their appearance in diverse bacterial species is not a "
1366:
1329:
1289:
1257:
1150:
1118:
1104:
1083:
1069:
1020:
1013:
932:
805:
609:
569:
261:
257:
535:
389:
1346:
1279:
1143:
1111:
1076:
1062:
1034:
960:
939:
520:
Bacteriophages
Encoding Reverse Transcriptase-Mediated Tropism-Switching Cassettes"
354:
288:
228:, that are involved in msDNA synthesis. The DNA portion of msDNA is encoded by the
184:
1208:
953:
327:
641:
624:
585:
1339:
1286:
873:
786:
423:
313:
281:
245:
241:
593:
570:"Bacterial retrons encode phage-defending tripartite toxin–antitoxin systems"
946:
925:
827:
719:
165:
701:
650:
601:
553:
500:
441:
381:
1319:
1264:
851:
835:
800:
682:
482:
249:
145:
99:
1351:
1269:
1250:
810:
795:
253:
373:
173:
1240:
1225:
1175:
905:
213:
141:
17:
731:, on: EurekAlert!, 5 Nov 2020. Source: WEIZMANN INSTITUTE OF SCIENCE
216:
controlling the synthesis of an RNA transcript carrying three loci,
457:"A diversity of uncharacterized reverse transcriptases in bacteria"
32:
1274:
901:
845:
840:
183:
69:
1213:
817:
765:
723:
87:
64:
738:
305:
comprising an intron lariat bound to two intron-coded proteins.
240:
gene is a reverse transcriptase similar to the RTs produced by
137:
81:
291:, have been found in a wide variety of different bacteria:
212:
Retron elements are about 2 kb long. They contain a single
248:(labeled 1–7 in the figure), including a highly conserved
264:(YADD) sequence associated with the catalytic core. The
729:
Mystery molecule in bacteria is revealed to be a guard
666:"Retrons and their applications in genome engineering"
204:
gene product, a reverse transcriptase, processes the
1194:
1168:
1135:
991:
977:
970:
917:
900:
893:
861:
826:
784:
777:
152:and a unique single-stranded DNA/RNA hybrid called
117:
105:
93:
80:
75:
63:
55:
50:
25:
625:"Bacterial Retrons Function In Anti-Phage Defense"
664:Simon AJ, Ellington AD, Finkelstein IJ (2019).
268:gene product is responsible for processing the
401:
399:
750:
8:
284:. Currently, however, RT-encoding elements,
1313:Reverse transcriptase-related cellular gene
974:
914:
897:
781:
757:
743:
735:
355:"Retrons, msDNA, and the bacterial genome"
272:portion of the RNA transcript into msDNA.
691:
681:
640:
543:
490:
472:
431:
208:portion of the RNA transcript into msDNA.
232:gene, the RNA portion is encoded by the
1294:Retroelements not elsewhere classified
353:Lampson BC, Inouye M, Inouye S (2005).
345:
22:
7:
408:"Diversity-Generating Retroelements"
1246:Integrative and conjugative element
309:diversity-generating retroelements
14:
1303:Diversity-generating retroelement
516:"Genomic and Genetic Analysis of
333:Retrons are being developed into
1308:Telomerase reverse transcriptase
880:Microbes with highly unusual DNA
31:
536:10.1128/JB.186.5.1503-1517.2004
412:Current Opinion in Microbiology
236:gene, while the product of the
1231:Defective interfering particle
1:
1219:Clonally transmissible cancer
455:Simon DM, Zimmerly S (2008).
406:Medhekar B, Mille JF (2007).
276:Classification and occurrence
154:multicopy single-stranded DNA
772:, and comparable structures
1389:
642:10.1016/j.cell.2020.09.065
586:10.1038/s41586-022-05091-4
15:
424:10.1016/j.mib.2007.06.004
30:
1236:Endogenous viral element
16:Not to be confused with
720:Page for Retron msr RNA
148:species that codes for
670:Nucleic Acids Research
209:
180:Sequence and structure
140:sequence found in the
187:
150:reverse transcriptase
43:sequence conservation
1335:Transposable element
1325:Spiegelman's Monster
362:Cytogenet Genome Res
335:genome-editing tools
676:(21): 11007–11019.
39:secondary structure
1204:Bio-like structure
1126:Tolecusatellitidae
683:10.1093/nar/gkz865
483:10.1093/nar/gkn867
210:
1360:
1359:
1190:
1189:
1164:
1163:
1160:
1159:
1098:Portogloboviridae
1000:Alphasatellitidae
894:Non-cellular life
889:
888:
770:non-cellular life
580:(7925): 144–150.
467:(22): 7219–7229.
461:Nucleic Acids Res
374:10.1159/000084982
303:ribonucleoprotein
130:
129:
1380:
975:
915:
898:
782:
759:
752:
745:
736:
706:
705:
695:
685:
661:
655:
654:
644:
635:(6): 1551–1561.
620:
614:
613:
564:
558:
557:
547:
530:(5): 1503–1517.
511:
505:
504:
494:
476:
452:
446:
445:
435:
403:
394:
393:
368:(1–4): 491–499.
359:
350:
299:group II introns
35:
23:
1388:
1387:
1383:
1382:
1381:
1379:
1378:
1377:
1363:
1362:
1361:
1356:
1196:
1186:
1156:
1131:
1042:Finnlakeviridae
987:
966:
908:
904:
885:
857:
822:
773:
763:
714:
709:
663:
662:
658:
622:
621:
617:
566:
565:
561:
513:
512:
508:
474:10.1.1.358.8390
454:
453:
449:
405:
404:
397:
357:
352:
351:
347:
343:
324:
278:
182:
170:DNA/RNA chimera
46:
21:
12:
11:
5:
1386:
1384:
1376:
1375:
1373:Non-coding RNA
1365:
1364:
1358:
1357:
1355:
1354:
1349:
1344:
1343:
1342:
1332:
1327:
1322:
1317:
1316:
1315:
1310:
1305:
1300:
1292:
1284:
1283:
1282:
1272:
1267:
1262:
1253:
1248:
1243:
1238:
1233:
1228:
1223:
1222:
1221:
1216:
1206:
1200:
1198:
1192:
1191:
1188:
1187:
1185:
1184:
1179:
1172:
1170:
1166:
1165:
1162:
1161:
1158:
1157:
1155:
1154:
1147:
1139:
1137:
1133:
1132:
1130:
1129:
1122:
1115:
1108:
1101:
1094:
1091:Polydnaviridae
1087:
1080:
1073:
1066:
1059:
1056:Globuloviridae
1052:
1049:Fuselloviridae
1045:
1038:
1031:
1028:Bicaudaviridae
1024:
1017:
1010:
1007:Ampullaviridae
1003:
995:
993:
989:
988:
984:Naldaviricetes
981:
979:
972:
968:
967:
965:
964:
957:
950:
943:
936:
929:
921:
919:
912:
895:
891:
890:
887:
886:
884:
883:
877:
869:
867:
864:Incertae sedis
859:
858:
856:
855:
848:
843:
838:
832:
830:
824:
823:
821:
820:
815:
814:
813:
808:
798:
792:
790:
779:
775:
774:
764:
762:
761:
754:
747:
739:
733:
732:
726:
717:
713:
712:External links
710:
708:
707:
656:
615:
559:
506:
447:
418:(4): 388–395.
395:
344:
342:
339:
323:
320:
319:
318:
306:
296:
277:
274:
181:
178:
162:non-coding RNA
158:Retron msr RNA
136:is a distinct
128:
127:
122:
115:
114:
109:
103:
102:
97:
91:
90:
85:
78:
77:
73:
72:
67:
61:
60:
57:
53:
52:
48:
47:
36:
28:
27:
26:Retron msr RNA
13:
10:
9:
6:
4:
3:
2:
1385:
1374:
1371:
1370:
1368:
1353:
1350:
1348:
1345:
1341:
1338:
1337:
1336:
1333:
1331:
1330:Tandem repeat
1328:
1326:
1323:
1321:
1318:
1314:
1311:
1309:
1306:
1304:
1301:
1299:
1296:
1295:
1293:
1291:
1288:
1285:
1281:
1278:
1277:
1276:
1273:
1271:
1268:
1266:
1263:
1260:
1259:
1258:Nanobacterium
1254:
1252:
1249:
1247:
1244:
1242:
1239:
1237:
1234:
1232:
1229:
1227:
1224:
1220:
1217:
1215:
1212:
1211:
1210:
1207:
1205:
1202:
1201:
1199:
1193:
1183:
1180:
1177:
1174:
1173:
1171:
1167:
1153:
1152:
1151:Rhizidiovirus
1148:
1146:
1145:
1141:
1140:
1138:
1134:
1128:
1127:
1123:
1121:
1120:
1119:Thaspiviridae
1116:
1114:
1113:
1109:
1107:
1106:
1105:Pospiviroidae
1102:
1100:
1099:
1095:
1093:
1092:
1088:
1086:
1085:
1084:Plasmaviridae
1081:
1079:
1078:
1074:
1072:
1071:
1070:Halspiviridae
1067:
1065:
1064:
1060:
1058:
1057:
1053:
1051:
1050:
1046:
1044:
1043:
1039:
1037:
1036:
1032:
1030:
1029:
1025:
1023:
1022:
1021:Avsunviroidae
1018:
1016:
1015:
1014:Anelloviridae
1011:
1009:
1008:
1004:
1002:
1001:
997:
996:
994:
990:
986:
985:
980:
976:
973:
969:
963:
962:
958:
956:
955:
951:
949:
948:
944:
942:
941:
937:
935:
934:
933:Duplodnaviria
930:
928:
927:
923:
922:
920:
916:
913:
911:
907:
903:
899:
896:
892:
881:
878:
876:
875:
871:
870:
868:
866:
865:
860:
853:
849:
847:
844:
842:
839:
837:
834:
833:
831:
829:
825:
819:
816:
812:
809:
807:
806:Mitochondrion
804:
803:
802:
799:
797:
794:
793:
791:
788:
783:
780:
778:Cellular life
776:
771:
767:
760:
755:
753:
748:
746:
741:
740:
737:
730:
727:
725:
721:
718:
716:
715:
711:
703:
699:
694:
689:
684:
679:
675:
671:
667:
660:
657:
652:
648:
643:
638:
634:
630:
626:
619:
616:
611:
607:
603:
599:
595:
591:
587:
583:
579:
575:
571:
563:
560:
555:
551:
546:
541:
537:
533:
529:
525:
521:
519:
510:
507:
502:
498:
493:
488:
484:
480:
475:
470:
466:
462:
458:
451:
448:
443:
439:
434:
429:
425:
421:
417:
413:
409:
402:
400:
396:
391:
387:
383:
379:
375:
371:
367:
363:
356:
349:
346:
340:
338:
336:
331:
329:
321:
317:
315:
310:
307:
304:
300:
297:
294:
293:
292:
290:
289:retroelements
287:
283:
275:
273:
271:
267:
263:
259:
255:
251:
247:
243:
239:
235:
231:
227:
223:
219:
215:
207:
203:
199:
195:
191:
186:
179:
177:
175:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
126:
123:
120:
116:
113:
110:
108:
104:
101:
98:
96:
92:
89:
86:
83:
79:
74:
71:
68:
66:
62:
58:
54:
49:
44:
40:
34:
29:
24:
19:
1347:Transpoviron
1297:
1280:Fungal prion
1256:
1149:
1144:Dinodnavirus
1142:
1124:
1117:
1112:Spiraviridae
1110:
1103:
1096:
1089:
1082:
1077:Ovaliviridae
1075:
1068:
1063:Guttaviridae
1061:
1054:
1047:
1040:
1035:Clavaviridae
1033:
1026:
1019:
1012:
1005:
998:
982:
961:Varidnaviria
959:
952:
945:
940:Monodnaviria
938:
931:
924:
872:
862:
673:
669:
659:
632:
628:
618:
577:
573:
562:
527:
524:J. Bacteriol
523:
517:
509:
464:
460:
450:
415:
411:
365:
361:
348:
332:
325:
312:
285:
279:
269:
265:
242:retroviruses
237:
233:
229:
225:
221:
217:
211:
205:
201:
197:
193:
189:
157:
133:
131:
1290:microsphere
1209:Cancer cell
954:Ribozyviria
328:selfish DNA
282:prokaryotes
246:amino acids
51:Identifiers
1340:Retroposon
1287:Proteinoid
1197:structures
1195:Comparable
971:Unassigned
874:Parakaryon
787:Prokaryota
518:Bordetella
341:References
314:Bordetella
121:structures
112:SO:0000233
76:Other data
37:Predicted
947:Riboviria
926:Adnaviria
910:Satellite
828:Eukaryota
610:250643138
594:0028-0836
469:CiteSeerX
166:guanosine
156:(msDNA).
95:Domain(s)
1367:Category
1320:Ribozyme
1265:Phagemid
992:Families
852:Protista
836:Animalia
801:Bacteria
702:31598685
651:33157039
602:35850148
554:14973019
501:19004871
442:17703991
390:24854188
382:16093702
322:Function
146:bacteria
144:of many
100:Bacteria
1352:Xenobot
1270:Plasmid
1251:Jeewanu
1182:Obelisk
978:Classes
846:Plantae
811:Plastid
796:Archaea
693:6868368
492:2602772
433:2703298
270:msd/msr
206:msd/msr
160:is the
70:RF00170
1298:Retron
1241:Fosmid
1226:Cosmid
1176:Nanobe
1136:Genera
918:Realms
906:Viroid
700:
690:
649:
608:
600:
592:
574:Nature
552:
545:344406
542:
499:
489:
471:
440:
430:
388:
380:
224:, and
214:operon
200:. The
196:, and
142:genome
134:retron
56:Symbol
45:of msr
18:RetroN
1275:Prion
1169:Other
902:Virus
841:Fungi
606:S2CID
386:S2CID
358:(PDF)
1214:HeLa
818:LUCA
766:Life
724:Rfam
698:PMID
647:PMID
629:Cell
598:PMID
590:ISSN
550:PMID
497:PMID
438:PMID
378:PMID
286:i.e.
125:PDBe
88:Gene
84:type
65:Rfam
41:and
1178:(?)
882:(?)
722:at
688:PMC
678:doi
637:doi
633:183
582:doi
578:609
540:PMC
532:doi
528:186
487:PMC
479:doi
428:PMC
420:doi
370:doi
366:110
266:ret
262:asp
258:asp
254:ala
250:tyr
238:ret
234:msr
230:msd
226:ret
222:msd
218:msr
202:ret
198:ret
194:msd
190:msr
138:DNA
119:PDB
82:RNA
59:msr
1369::
768:,
696:.
686:.
674:47
672:.
668:.
645:.
631:.
627:.
604:.
596:.
588:.
576:.
572:.
548:.
538:.
526:.
522:.
495:.
485:.
477:.
465:36
463:.
459:.
436:.
426:.
416:10
414:.
410:.
398:^
384:.
376:.
364:.
360:.
337:.
220:,
192:,
174:5′
132:A
107:SO
1261:"
1255:"
854:'
850:'
789:"
785:"
758:e
751:t
744:v
704:.
680::
653:.
639::
612:.
584::
556:.
534::
503:.
481::
444:.
422::
392:.
372::
316:.
260:-
256:-
252:-
20:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.