837:
40:
3338:
341:
218:
864:
102. These three key amino acids each play an essential role in the cleaving ability of the proteases. While the amino acid members of the triad are located far from one another on the sequence of the protein, due to folding, they will be very close to one another in the heart of the enzyme. The
1419:
Due to their catalytic activity, some serine proteases possess potent antimicrobial properties. Several in vitro studies have demonstrated the efficacy of some proteases in reducing virulence by cleaving viral surface proteins. Viral entry into host cells is mediated by the interaction of these
1108:
Zymogens are large, inactive structures, which have the ability to break apart or change into the smaller activated enzymes. The difference between zymogens and the activated enzymes lies in the fact that the active site for catalysis of the zymogens is distorted. As a result, the substrate
876:
binds (in this case, the polypeptide being cleaved), a product is released (the C-terminus "half" of the peptide with amino group visible), another substrate binds (in this case, water), and another product is released (the N-terminus "half" of the peptide with carboxyl group visible).
777:. They have been found to have roles in coagulation and digestion as well as in the pathophysiology of neurodegenerative disorders such as Alzheimer's and Parkinson's induced dementia. Many highly-toxic thrombin-like serine protease isoforms are found in snake venoms.
1237:
that resemble the tetrahedral intermediate, and thus fill up the active site, preventing the enzyme from working properly. Trypsin, a powerful digestive enzyme, is generated in the pancreas. Inhibitors prevent self-digestion of the pancreas itself.
1420:
surface proteins with the host cell. When these proteins are fragmented or inactivated on the viral surface, the viral entry is impaired, leading to a reduction in infectivity of a broad spectrum of pathologically relevant microorganisms like
1152:
secretions from the duodenal mucosa cleave the lysine 15 - isoleucine 16 peptide bond of the zymogen. As a result, the zymogen trypsinogen breaks down into trypsin. Recall that trypsin is also responsible for cleaving
865:
particular geometry of the triad members are highly characteristic to their specific function: it was shown that the position of just four points of the triad characterize the function of the containing enzyme.
2001:
Lopes BR, da Silva GS, de Lima
Menezes G, de Oliveira J, Watanabe AS, Porto BN, et al. (May 2022). "Serine proteases in neutrophil extracellular traps exhibit anti-Respiratory Syncytial Virus activity".
1002:
carbon. Once again, the electrons from the double bond move to the oxygen making it negative, as the bond between the oxygen of the water and the carbon is formed. This is coordinated by the nitrogen of the
1157:
peptide bonds, and thus, once a small amount of trypsin is generated, it participates in cleavage of its own zymogen, generating even more trypsin. The process of trypsin activation can thus be called
1958:
Breugelmans B, Simonet G, van Hoef V, Van Soest S, Vanden Broeck J (March 2009). "Pacifastin-related peptides: structural and functional characteristics of a family of serine peptidase inhibitors".
954:
substrate binds to the surface of the serine protease enzyme such that the scissile bond is inserted into the active site of the enzyme, with the carbonyl carbon of this bond positioned near the
844:
The main player in the catalytic mechanism in the serine proteases is the catalytic triad. The triad is located in the active site of the enzyme, where catalysis occurs, and is preserved in all
1252:, including synthetic chemical inhibitors for research or therapeutic purposes, and also natural proteinaceous inhibitors. One family of natural inhibitors called "serpins" (abbreviated from
1086:
Host organisms must ensure that the activity of serine proteases is adequately regulated. This is achieved by a requirement for initial protease activation, and the secretion of inhibitors.
1327:
Mutations may lead to decreased or increased activity of enzymes. This may have different consequences, depending on the normal function of the serine protease. For example, mutations in
1097:
are the usually inactive precursors of an enzyme. If the digestive enzymes were active when synthesized, they would immediately start chewing up the synthesizing organs and tissues.
749:
The S1 pocket of chymotrypsin-like enzymes is more hydrophobic than in trypsin-like proteases. This results in a specificity for medium to large sized hydrophobic residues, such as
2992:
2852:
181:
1217:. Therefore, it is essential that this activation does not occur prematurely. There are several protective measures taken by the organism to prevent self-digestion:
2592:
1933:
1344:
1101:
is such a condition, in which there is premature activation of the digestive enzymes in the pancreas, resulting in self-digestion (autolysis). It also complicates
504:
200:
352:
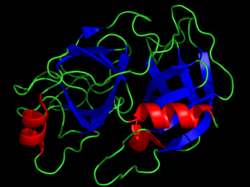
Hinge prediction are colored in blue (residues 23:28) and red (residues 175:182). The green colored region is the active site. Motion is generated using hdANM.
1347:
fusion). Exogenous snake venom serine proteases cause a vast array of coagulopathies when injected in a host due to the lack of regulation of their activity.
983:
The bond joining the nitrogen and the carbon in the peptide bond is now broken. The covalent electrons creating this bond move to attack the hydrogen of the
868:
In the event of catalysis, an ordered mechanism occurs in which several intermediates are generated. The catalysis of the peptide cleavage can be seen as a
1339:. Also, some proteases play a vital role in host cell-virus fusion activation by priming virus's Spike protein to show the protein named "fusion protein" (
713:
Serine proteases are characterised by a distinctive structure, consisting of two beta-barrel domains that converge at the catalytic active site. These
836:
1113:
does not occur. Only after activation, during which the conformation and structure of the zymogen change and the active site is opened, can
1593:
Madala PK, Tyndall JD, Nall T, Fairlie DP (June 2010). "Update 1 of: Proteases universally recognize beta strands in their active sites".
785:
Elastase-like proteases have a much smaller S1 cleft than either trypsin- or chymotrypsin-like proteases. Consequently, residues such as
2585:
2518:
2148:
2097:
1734:
Ovaere P, Lippens S, Vandenabeele P, Declercq W (September 2009). "The emerging roles of serine protease cascades in the epidermis".
3057:
1505:
2503:
193:
579:
733:). This specificity is driven by the residue which lies at the base of the enzyme's S1 pocket (generally a negatively charged
2578:
1176:
After the Arg 15 - Ile 16 bond in the chymotrypsinogen zymogen is cleaved by trypsin, the newly generated structure called a
160:
136:
3213:
717:
can be further categorised based on their substrate specificity as either trypsin-like, chymotrypsin-like or elastase-like.
2434:
2349:
3328:
1937:
2812:
2367:
1510:
1888:
Iván G, Szabadka Z, Ordög R, Grolmusz V, Náray-Szabó G (June 2009). "Four spatial points that define enzyme families".
812:. Subtilisin is evolutionarily unrelated to the chymotrypsin-clan, but shares the same catalytic mechanism utilising a
1224:
The zymogens are stored in zymogen granules, capsules that have walls that are thought to be resistant to proteolysis.
2570:
2039:"The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses"
991:
oxygen double bond move back from the negative oxygen to recreate the bond, generating an acyl-enzyme intermediate.
3198:
3314:
3301:
3288:
3275:
3262:
3249:
3236:
2727:
2712:
2612:
2550:
2466:
2389:
2359:
2341:
2294:
2259:
2187:
2174:
1495:
1384:
1380:
1253:
3208:
1078:
of the reaction. This "preferential binding" is responsible for much of the catalytic efficiency of the enzyme.
154:
3162:
3105:
2609:
2329:
2165:
2120:
1102:
1063:
873:
59:
44:
Crystal structure of bovine chymotrypsin. The catalytic residues are shown as yellow sticks. Rendered from PDB
2508:
425:
141:
3110:
3002:
2967:
2962:
2498:
2324:
2225:
2215:
2141:
1395:
604:
2312:
2200:
2110:
1897:
1500:
584:
390:
683:
205:
3131:
3050:
2957:
2807:
2319:
2220:
1070:
double bond, fits perfectly into the oxyanion hole. In effect, serine proteases preferentially bind the
405:
221:
129:
3203:
2947:
2493:
2302:
1782:
1690:
1475:
1373:
1332:
821:
311:
1902:
1066:
of step 1 and step 3 are generated, the negative oxygen ion, having accepted the electrons from the
3167:
2847:
2837:
2654:
2649:
2452:
2414:
2398:
1490:
1269:
1181:
845:
362:
326:
315:
295:
157:
1007:, which accepts a proton from the water. Overall, this generates another tetrahedral intermediate.
81:
3363:
3358:
3100:
2488:
2429:
2419:
2409:
2134:
1983:
1800:
1453:
1359:
1098:
643:
2037:
Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, et al. (May 2014). Dermody TS (ed.).
525:
2829:
2483:
2376:
2068:
2019:
1975:
1915:
1870:
1818:
1751:
1716:
1659:
1610:
1575:
1458:
1448:
1075:
976:
accepts the hydrogen from the -OH of the and a pair of electrons from the double bond of the
688:
599:
148:
45:
1355:
Determination of serine protease levels may be useful in the context of particular diseases.
3146:
3141:
3115:
3043:
2621:
2533:
2528:
2404:
2116:
2058:
2050:
2011:
1967:
1907:
1860:
1852:
1808:
1790:
1743:
1706:
1698:
1649:
1641:
1602:
1567:
1438:
1249:
1242:
1234:
1071:
668:
648:
509:
489:
450:
445:
430:
2094:
1677:
Khade PM, Scaramozzino D, Kumar A, Lacidogna G, Carpinteri A, Jernigan RL (November 2021).
117:
3193:
3177:
3090:
2862:
2857:
2629:
2538:
2523:
2101:
1485:
1463:
1369:
1149:
1145:
939:
813:
484:
303:
93:
1786:
1694:
64:
3342:
3231:
3172:
2559:
2195:
2063:
2038:
1865:
1840:
1711:
1678:
1654:
1629:
1365:
931:
725:
Trypsin-like proteases cleave peptide bonds following a positively charged amino acid (
369:
334:
299:
176:
3352:
3136:
3095:
3007:
2880:
2869:
2601:
2470:
2272:
1839:
Oliveira AL, Viegas MF, da Silva SL, Soares AM, Ramos MJ, Fernandes PA (2022-06-10).
1813:
1770:
1316:
1209:
is essential, because it activates its own reaction, as well as the reaction of both
1158:
927:
897:
861:
848:
of serine protease enzymes. The triad is a coordinated structure consisting of three
754:
738:
734:
663:
3085:
2903:
2797:
2639:
2178:
1987:
1971:
1537:
1265:
1171:
955:
916:
623:
564:
544:
307:
275:
256:
244:
39:
333:
class families, S: purely serine proteases. superfamily. Within each superfamily,
2015:
980:
oxygen moves to the oxygen. As a result, a tetrahedral intermediate is generated.
3309:
3244:
3080:
3022:
3017:
2666:
2424:
2371:
2307:
2267:
2126:
1480:
1399:
1362:
levels may be required in the diagnosis of hemorrhagic or thrombotic conditions.
1273:
1114:
1110:
951:
889:
849:
809:
559:
539:
470:
330:
271:
263:
17:
3337:
1911:
1856:
1775:
Proceedings of the
National Academy of Sciences of the United States of America
1747:
2984:
2926:
2707:
2702:
2682:
1702:
1645:
1532:
1527:
1391:
1336:
1304:
1277:
1027:
995:
805:
758:
628:
410:
385:
279:
267:
259:
3283:
3257:
3012:
2998:
2817:
2697:
2687:
2157:
2106:
1795:
1771:"Substrate specificity of trypsin investigated by using a genetic selection"
1421:
1407:
1403:
1328:
1300:
1281:
1019:
1004:
984:
973:
935:
904:
869:
853:
825:
466:
274:. Serine proteases fall into two broad categories based on their structure:
2072:
2023:
1979:
1919:
1874:
1755:
1720:
1663:
1614:
1579:
1105:, as the pancreas often digests itself before it can be assessed visually.
1822:
1410:, meaning it remains in the system for a clinically useful length of time.
1288:, respectively. Artificial irreversible small molecule inhibitors include
2898:
2893:
2842:
2692:
2677:
2644:
2161:
2054:
1558:
Hedstrom L (December 2002). "Serine protease mechanism and specificity".
1443:
1368:
is employed to determine the exocrine activity of the pancreas, e.g., in
1312:
1260:
bond with the serine protease, inhibiting its function. The best-studied
1257:
1194:
1067:
1049:
1023:
1015:
999:
988:
977:
969:
923:
908:
893:
770:
750:
730:
349:
987:, breaking the connection. The electrons that previously moved from the
105:
2952:
2942:
2888:
2802:
2634:
1804:
1518:- database of protease specificity, substrates, products and inhibitors
1515:
1470:
1340:
1139:
1094:
880:
Each amino acid in the triad performs a specific task in this process:
790:
786:
774:
613:
319:
248:
225:
124:
1630:"Characterizing and Predicting Protein Hinges for Mechanistic Insight"
1606:
1571:
337:
are designated by their catalytic nucleophile, (S: serine proteases).
3296:
3066:
2979:
2921:
2247:
2240:
2235:
2090:
1521:
1308:
1154:
1011:
965:
958:
912:
885:
857:
817:
794:
726:
714:
344:
Hinge motion in disordered activation domain in
Trypsinogen (PDB ID:
291:
252:
240:
188:
100:
88:
76:
524:
Phage K1F endosialidase CIMCD self-cleaving protein (Enterobacteria
345:
1010:
In a final reaction, the bond formed in the first step between the
3270:
2988:
2974:
2913:
2787:
2782:
2777:
2772:
2767:
2762:
2282:
2277:
2210:
2205:
1289:
1026:
carbon re-forms the double bond with the oxygen. As a result, the
339:
216:
2757:
2752:
2747:
2742:
2737:
2732:
2722:
2717:
1425:
1293:
1285:
1245:, which turn off their activity when they are no longer needed.
112:
3039:
2574:
2130:
1039:
It was discovered that additional amino acids of the protease,
340:
217:
1679:"hdANM: a new comprehensive dynamics model for protein hinges"
824:, since the same mechanism evolved twice independently during
1062:
can donate backbone hydrogens for hydrogen bonding. When the
835:
1221:
The activation of trypsinogen by trypsin is relatively slow
3035:
1841:"The chemistry of snake venom and its medicinal potential"
994:
Now, water comes into the reaction. Water replaces the
3326:
2093:
online database for peptidases and their inhibitors:
1387:, risk stratification, and post-treatment monitoring.
1248:
Serine proteases are inhibited by a diverse group of
938:, making the nitrogen atom mentioned above much more
3222:
3186:
3155:
3124:
3073:
2935:
2912:
2878:
2828:
2665:
2620:
2549:
2465:
2445:
2388:
2358:
2340:
2293:
2258:
2186:
2173:
1890:
1074:and the overall structure is favored, lowering the
199:
187:
175:
170:
147:
135:
123:
111:
99:
87:
75:
70:
58:
53:
32:
1769:Evnin LB, Vásquez JR, Craik CS (September 1990).
1241:Serine proteases are paired with serine protease
946:The whole reaction can be summarized as follows:
820:. This is the classic example used to illustrate
1184:(self digestion), yielding active chymotrypsin.
1628:Khade PM, Kumar A, Jernigan RL (January 2020).
915:-OH group, thus coordinating the attack of the
318:. The majority belong to the S1 family of the
3051:
2586:
2142:
1199:It is activated by cleavage through trypsin.
1047:, are involved in creating what is called an
1018:carbon moves to attack the hydrogen that the
8:
1934:"Kimball's Biology Pages, Serine Proteases"
860:195 (hence the name "serine protease") and
3058:
3044:
3036:
2593:
2579:
2571:
2183:
2149:
2135:
2127:
1524:- Database of protease evolutionary groups
1205:As can be seen, trypsinogen activation to
1022:just acquired. The now electron-deficient
888:has an -OH group that is able to act as a
329:, P: superfamily, containing a mixture of
167:
38:
2119:at the U.S. National Library of Medicine
2062:
1901:
1864:
1812:
1794:
1710:
1653:
1109:polypeptide cannot bind effectively, and
580:Murein tetrapeptidase LD-carboxypeptidase
294:protease classification system counts 16
1394:, is an important diagnostic marker for
1119:
998:of the cleaved peptide, and attacks the
355:
3333:
1550:
907:nitrogen has the ability to accept the
266:. They are found ubiquitously in both
2606:serine proteases/serine endopeptidases
1082:Regulation of serine protease activity
29:
7:
1834:
1832:
1315:, and may function in the arthropod
1303:serine peptidase inhibitors, called
773:, tissue activating plasminogen and
618:S1, S3, S6, S7, S29, S30, S31, S32,
2519:Amyloid precursor protein secretase
840:serine protease reaction mechanism
298:(as of 2013) each containing many
25:
1506:Protease inhibitor (pharmacology)
700:S48, S62, S68, S71, S72, S79, S81
3336:
2504:Proteasome endopeptidase complex
2004:International Immunopharmacology
1390:Serine protease, as released by
972:carbon, and the nitrogen of the
1972:10.1016/j.peptides.2008.07.026
1736:Trends in Biochemical Sciences
1035:Additional stabilizing effects
1030:of the peptide is now ejected.
900:peptide bond of the substrate.
348:). The hinges predicted using
1:
2818:Urinary plasminogen activator
2368:Serine type carboxypeptidases
2350:Angiotensin-converting enzyme
357:Families of serine proteases
2813:Tissue plasminogen activator
2016:10.1016/j.intimp.2022.108573
1634:Journal of Molecular Biology
1511:Protease inhibitor (biology)
1272:, studied for their role in
1144:When trypsinogen enters the
620:S39, S46, S55, S64, S65, S75
322:(superfamily) of proteases.
302:. Each superfamily uses the
228:, a typical serine protease.
903:A pair of electrons on the
816:, to create a nucleophilic
402:S9, S10, S15, S28, S33, S37
3380:
1912:10.1016/j.bbrc.2009.04.022
1857:10.1038/s41570-022-00393-7
1748:10.1016/j.tibs.2009.08.001
1254:serine protease inhibitors
3214:Michaelis–Menten kinetics
2904:Proteinase 3/Myeloblastin
1845:Nature Reviews. Chemistry
1703:10.1016/j.bpj.2021.10.017
1646:10.1016/j.jmb.2019.11.018
1496:Proteases in angiogenesis
1385:prostate cancer screening
1381:prostate-specific antigen
1307:, has been identified in
1103:postmortem investigations
166:
37:
3106:Diffusion-limited enzyme
2330:Tripeptidyl peptidase II
2121:Medical Subject Headings
1064:tetrahedral intermediate
808:is a serine protease in
278:-like (trypsin-like) or
2499:Threonine endopeptidase
2325:Tripeptidyl peptidase I
1796:10.1073/pnas.87.17.6659
1501:Intramembrane proteases
1396:type 1 hypersensitivity
605:Drosophila melanogaster
462:S21, S73, S77, S78, S80
426:D-Ala-D-Ala peptidase C
306:or dyad in a different
2958:Proprotein convertases
2489:Aspartic acid protease
2313:Dipeptidyl peptidase-4
2111:Saint Louis University
872:catalysis, in which a
841:
797:tend to be preferred.
585:Pseudomonas aeruginosa
391:Bacillus licheniformis
353:
229:
3199:Eadie–Hofstee diagram
3132:Allosteric regulation
2808:Plasminogen activator
2320:Tripeptidyl peptidase
839:
709:Substrate specificity
406:Prolyl oligopeptidase
343:
237:serine endopeptidases
220:
3209:Lineweaver–Burk plot
2948:Prolyl endopeptidase
2494:Metalloendopeptidase
2399:Metalloexopeptidases
2303:Dipeptidyl peptidase
2055:10.1128/JVI.03677-13
1476:Convergent evolution
1415:Antimicrobial effect
1374:chronic pancreatitis
1335:and predisposing to
1333:protein C deficiency
822:convergent evolution
644:Penicillin G acylase
312:convergent evolution
2509:HslU—HslV peptidase
2453:Metalloexopeptidase
2043:Journal of Virology
1787:1990PNAS...87.6659E
1695:2021BpJ...120.4955K
1683:Biophysical Journal
1491:The Proteolysis Map
1402:. More useful than
1270:alpha 1-antitrypsin
1148:from the pancreas,
832:Catalytic mechanism
684:DmpA aminopeptidase
358:
316:catalytic mechanism
3168:Enzyme superfamily
3101:Enzyme promiscuity
2100:2017-04-04 at the
1406:due to the longer
1360:Coagulation factor
1233:There are certain
1099:Acute pancreatitis
1090:Zymogen activation
842:
356:
354:
262:at the (enzyme's)
230:
3324:
3323:
3033:
3032:
2830:Complement system
2622:Digestive enzymes
2568:
2567:
2517:Other/ungrouped:
2484:Cysteine protease
2461:
2460:
2379:
2049:(10): 5608–5616.
1781:(17): 6659–6663.
1689:(22): 4955–4965.
1607:10.1021/cr900368a
1572:10.1021/cr000033x
1566:(12): 4501–4524.
1203:
1202:
1076:activation energy
745:Chymotrypsin-like
706:
705:
689:Brucella anthropi
310:and so represent
222:Crystal structure
215:
214:
211:
210:
130:metabolic pathway
16:(Redirected from
3371:
3341:
3340:
3332:
3204:Hanes–Woolf plot
3147:Enzyme activator
3142:Enzyme inhibitor
3116:Enzyme catalysis
3060:
3053:
3046:
3037:
2595:
2588:
2581:
2572:
2534:Beta-secretase 2
2529:Beta-secretase 1
2405:Carboxypeptidase
2401:
2377:
2184:
2151:
2144:
2137:
2128:
2117:Serine+proteases
2107:Serine Proteases
2095:Serine Peptidase
2077:
2076:
2066:
2034:
2028:
2027:
1998:
1992:
1991:
1955:
1949:
1948:
1946:
1945:
1936:. Archived from
1930:
1924:
1923:
1905:
1885:
1879:
1878:
1868:
1836:
1827:
1826:
1816:
1798:
1766:
1760:
1759:
1731:
1725:
1724:
1714:
1674:
1668:
1667:
1657:
1625:
1619:
1618:
1595:Chemical Reviews
1590:
1584:
1583:
1560:Chemical Reviews
1555:
1439:Serine hydrolase
1398:reactions e.g.,
1167:Chymotrypsinogen
1120:
1072:transition state
968:-OH attacks the
892:, attacking the
669:Escherichia coli
649:Escherichia coli
510:Escherichia coli
490:Escherichia coli
465:Cytomegalovirus
451:Escherichia coli
446:Signal peptidase
431:Escherichia coli
359:
233:Serine proteases
168:
42:
30:
27:Class of enzymes
21:
18:Serine proteases
3379:
3378:
3374:
3373:
3372:
3370:
3369:
3368:
3349:
3348:
3347:
3335:
3327:
3325:
3320:
3232:Oxidoreductases
3218:
3194:Enzyme kinetics
3182:
3178:List of enzymes
3151:
3120:
3091:Catalytic triad
3069:
3064:
3034:
3029:
2931:
2908:
2874:
2824:
2661:
2630:Enteropeptidase
2616:
2599:
2569:
2564:
2545:
2539:Gamma secretase
2524:Alpha secretase
2479:Serine protease
2457:
2446:Other/ungrouped
2441:
2397:
2384:
2380:-Transpeptidase
2354:
2336:
2289:
2254:
2169:
2155:
2102:Wayback Machine
2086:
2081:
2080:
2036:
2035:
2031:
2000:
1999:
1995:
1957:
1956:
1952:
1943:
1941:
1932:
1931:
1927:
1903:10.1.1.150.1086
1887:
1886:
1882:
1838:
1837:
1830:
1768:
1767:
1763:
1733:
1732:
1728:
1676:
1675:
1671:
1627:
1626:
1622:
1592:
1591:
1587:
1557:
1556:
1552:
1547:
1542:
1486:Catalytic triad
1434:
1417:
1370:cystic fibrosis
1353:
1325:
1323:Role in disease
1231:
1178:pi-chymotrypsin
1150:enteropeptidase
1146:small intestine
1092:
1084:
1037:
940:electronegative
834:
814:catalytic triad
803:
801:Subtilisin-like
783:
767:
747:
723:
711:
619:
485:Lon-A peptidase
364:
304:catalytic triad
288:
49:
33:Serine protease
28:
23:
22:
15:
12:
11:
5:
3377:
3375:
3367:
3366:
3361:
3351:
3350:
3346:
3345:
3322:
3321:
3319:
3318:
3305:
3292:
3279:
3266:
3253:
3240:
3226:
3224:
3220:
3219:
3217:
3216:
3211:
3206:
3201:
3196:
3190:
3188:
3184:
3183:
3181:
3180:
3175:
3170:
3165:
3159:
3157:
3156:Classification
3153:
3152:
3150:
3149:
3144:
3139:
3134:
3128:
3126:
3122:
3121:
3119:
3118:
3113:
3108:
3103:
3098:
3093:
3088:
3083:
3077:
3075:
3071:
3070:
3065:
3063:
3062:
3055:
3048:
3040:
3031:
3030:
3028:
3027:
3026:
3025:
3020:
3010:
3005:
2996:
2982:
2977:
2972:
2971:
2970:
2965:
2955:
2950:
2945:
2939:
2937:
2933:
2932:
2930:
2929:
2924:
2918:
2916:
2910:
2909:
2907:
2906:
2901:
2896:
2891:
2885:
2883:
2876:
2875:
2873:
2872:
2867:
2866:
2865:
2860:
2850:
2845:
2840:
2834:
2832:
2826:
2825:
2823:
2822:
2821:
2820:
2815:
2805:
2793:
2792:
2791:
2790:
2785:
2780:
2775:
2770:
2765:
2760:
2755:
2750:
2745:
2740:
2735:
2730:
2725:
2720:
2715:
2705:
2700:
2695:
2690:
2685:
2680:
2671:
2669:
2663:
2662:
2660:
2659:
2658:
2657:
2652:
2642:
2637:
2632:
2626:
2624:
2618:
2617:
2602:Endopeptidases
2600:
2598:
2597:
2590:
2583:
2575:
2566:
2565:
2563:
2562:
2560:Staphylokinase
2556:
2554:
2547:
2546:
2544:
2543:
2542:
2541:
2536:
2531:
2526:
2514:
2513:
2512:
2511:
2506:
2496:
2491:
2486:
2481:
2475:
2473:
2463:
2462:
2459:
2458:
2456:
2455:
2449:
2447:
2443:
2442:
2440:
2439:
2438:
2437:
2432:
2427:
2422:
2417:
2412:
2402:
2394:
2392:
2386:
2385:
2383:
2382:
2374:
2364:
2362:
2356:
2355:
2353:
2352:
2346:
2344:
2338:
2337:
2335:
2334:
2333:
2332:
2327:
2317:
2316:
2315:
2310:
2299:
2297:
2291:
2290:
2288:
2287:
2286:
2285:
2280:
2275:
2264:
2262:
2256:
2255:
2253:
2252:
2251:
2250:
2245:
2244:
2243:
2238:
2228:
2223:
2218:
2213:
2208:
2203:
2196:Aminopeptidase
2192:
2190:
2181:
2171:
2170:
2156:
2154:
2153:
2146:
2139:
2131:
2125:
2124:
2114:
2104:
2085:
2084:External links
2082:
2079:
2078:
2029:
1993:
1966:(3): 622–632.
1950:
1925:
1896:(4): 417–420.
1880:
1851:(7): 451–469.
1828:
1761:
1742:(9): 453–463.
1726:
1669:
1640:(2): 508–522.
1620:
1601:(6): PR1–P31.
1585:
1549:
1548:
1546:
1543:
1541:
1540:
1535:
1530:
1525:
1519:
1513:
1508:
1503:
1498:
1493:
1488:
1483:
1478:
1473:
1468:
1467:
1466:
1461:
1456:
1451:
1441:
1435:
1433:
1430:
1416:
1413:
1412:
1411:
1388:
1377:
1366:Fecal elastase
1363:
1352:
1351:Diagnostic use
1349:
1324:
1321:
1230:
1227:
1226:
1225:
1222:
1201:
1200:
1197:
1192:
1186:
1185:
1174:
1169:
1163:
1162:
1142:
1137:
1131:
1130:
1127:
1124:
1091:
1088:
1083:
1080:
1036:
1033:
1032:
1031:
1008:
992:
981:
962:
944:
943:
932:hydrogen bonds
920:
901:
896:carbon of the
833:
830:
802:
799:
782:
779:
769:These include
766:
763:
746:
743:
722:
719:
710:
707:
704:
703:
701:
698:
694:
693:
681:
678:
674:
673:
661:
658:
654:
653:
641:
638:
634:
633:
621:
616:
610:
609:
597:
594:
590:
589:
577:
574:
570:
569:
557:
554:
550:
549:
537:
534:
530:
529:
522:
519:
515:
514:
502:
499:
495:
494:
482:
479:
475:
474:
463:
460:
456:
455:
443:
440:
436:
435:
423:
420:
416:
415:
403:
400:
396:
395:
383:
380:
376:
375:
372:
367:
287:
286:Classification
284:
255:serves as the
213:
212:
209:
208:
203:
197:
196:
191:
185:
184:
179:
173:
172:
164:
163:
152:
145:
144:
139:
133:
132:
127:
121:
120:
115:
109:
108:
103:
97:
96:
91:
85:
84:
79:
73:
72:
68:
67:
62:
56:
55:
51:
50:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3376:
3365:
3362:
3360:
3357:
3356:
3354:
3344:
3339:
3334:
3330:
3316:
3312:
3311:
3306:
3303:
3299:
3298:
3293:
3290:
3286:
3285:
3280:
3277:
3273:
3272:
3267:
3264:
3260:
3259:
3254:
3251:
3247:
3246:
3241:
3238:
3234:
3233:
3228:
3227:
3225:
3221:
3215:
3212:
3210:
3207:
3205:
3202:
3200:
3197:
3195:
3192:
3191:
3189:
3185:
3179:
3176:
3174:
3173:Enzyme family
3171:
3169:
3166:
3164:
3161:
3160:
3158:
3154:
3148:
3145:
3143:
3140:
3138:
3137:Cooperativity
3135:
3133:
3130:
3129:
3127:
3123:
3117:
3114:
3112:
3109:
3107:
3104:
3102:
3099:
3097:
3096:Oxyanion hole
3094:
3092:
3089:
3087:
3084:
3082:
3079:
3078:
3076:
3072:
3068:
3061:
3056:
3054:
3049:
3047:
3042:
3041:
3038:
3024:
3021:
3019:
3016:
3015:
3014:
3011:
3009:
3008:Streptokinase
3006:
3004:
3000:
2997:
2994:
2990:
2986:
2983:
2981:
2978:
2976:
2973:
2969:
2966:
2964:
2961:
2960:
2959:
2956:
2954:
2951:
2949:
2946:
2944:
2941:
2940:
2938:
2934:
2928:
2925:
2923:
2920:
2919:
2917:
2915:
2911:
2905:
2902:
2900:
2897:
2895:
2892:
2890:
2887:
2886:
2884:
2882:
2881:immune system
2877:
2871:
2870:C3-convertase
2868:
2864:
2861:
2859:
2856:
2855:
2854:
2851:
2849:
2846:
2844:
2841:
2839:
2836:
2835:
2833:
2831:
2827:
2819:
2816:
2814:
2811:
2810:
2809:
2806:
2804:
2801:
2799:
2795:
2794:
2789:
2786:
2784:
2781:
2779:
2776:
2774:
2771:
2769:
2766:
2764:
2761:
2759:
2756:
2754:
2751:
2749:
2746:
2744:
2741:
2739:
2736:
2734:
2731:
2729:
2726:
2724:
2721:
2719:
2716:
2714:
2711:
2710:
2709:
2706:
2704:
2701:
2699:
2696:
2694:
2691:
2689:
2686:
2684:
2681:
2679:
2676:
2673:
2672:
2670:
2668:
2664:
2656:
2653:
2651:
2648:
2647:
2646:
2643:
2641:
2638:
2636:
2633:
2631:
2628:
2627:
2625:
2623:
2619:
2614:
2611:
2607:
2603:
2596:
2591:
2589:
2584:
2582:
2577:
2576:
2573:
2561:
2558:
2557:
2555:
2552:
2548:
2540:
2537:
2535:
2532:
2530:
2527:
2525:
2522:
2521:
2520:
2516:
2515:
2510:
2507:
2505:
2502:
2501:
2500:
2497:
2495:
2492:
2490:
2487:
2485:
2482:
2480:
2477:
2476:
2474:
2472:
2471:Endopeptidase
2468:
2464:
2454:
2451:
2450:
2448:
2444:
2436:
2433:
2431:
2428:
2426:
2423:
2421:
2418:
2416:
2413:
2411:
2408:
2407:
2406:
2403:
2400:
2396:
2395:
2393:
2391:
2387:
2381:
2375:
2373:
2369:
2366:
2365:
2363:
2361:
2357:
2351:
2348:
2347:
2345:
2343:
2339:
2331:
2328:
2326:
2323:
2322:
2321:
2318:
2314:
2311:
2309:
2306:
2305:
2304:
2301:
2300:
2298:
2296:
2292:
2284:
2281:
2279:
2276:
2274:
2271:
2270:
2269:
2266:
2265:
2263:
2261:
2257:
2249:
2246:
2242:
2239:
2237:
2234:
2233:
2232:
2229:
2227:
2224:
2222:
2219:
2217:
2214:
2212:
2209:
2207:
2204:
2202:
2199:
2198:
2197:
2194:
2193:
2191:
2189:
2185:
2182:
2180:
2176:
2172:
2167:
2163:
2159:
2152:
2147:
2145:
2140:
2138:
2133:
2132:
2129:
2122:
2118:
2115:
2112:
2108:
2105:
2103:
2099:
2096:
2092:
2088:
2087:
2083:
2074:
2070:
2065:
2060:
2056:
2052:
2048:
2044:
2040:
2033:
2030:
2025:
2021:
2017:
2013:
2009:
2005:
1997:
1994:
1989:
1985:
1981:
1977:
1973:
1969:
1965:
1961:
1954:
1951:
1940:on 2005-12-13
1939:
1935:
1929:
1926:
1921:
1917:
1913:
1909:
1904:
1899:
1895:
1891:
1884:
1881:
1876:
1872:
1867:
1862:
1858:
1854:
1850:
1846:
1842:
1835:
1833:
1829:
1824:
1820:
1815:
1810:
1806:
1802:
1797:
1792:
1788:
1784:
1780:
1776:
1772:
1765:
1762:
1757:
1753:
1749:
1745:
1741:
1737:
1730:
1727:
1722:
1718:
1713:
1708:
1704:
1700:
1696:
1692:
1688:
1684:
1680:
1673:
1670:
1665:
1661:
1656:
1651:
1647:
1643:
1639:
1635:
1631:
1624:
1621:
1616:
1612:
1608:
1604:
1600:
1596:
1589:
1586:
1581:
1577:
1573:
1569:
1565:
1561:
1554:
1551:
1544:
1539:
1536:
1534:
1531:
1529:
1526:
1523:
1520:
1517:
1514:
1512:
1509:
1507:
1504:
1502:
1499:
1497:
1494:
1492:
1489:
1487:
1484:
1482:
1479:
1477:
1474:
1472:
1469:
1465:
1462:
1460:
1457:
1455:
1452:
1450:
1447:
1446:
1445:
1442:
1440:
1437:
1436:
1431:
1429:
1427:
1423:
1414:
1409:
1405:
1401:
1397:
1393:
1389:
1386:
1382:
1378:
1375:
1371:
1367:
1364:
1361:
1358:
1357:
1356:
1350:
1348:
1346:
1342:
1338:
1334:
1330:
1322:
1320:
1318:
1317:immune system
1314:
1310:
1306:
1302:
1297:
1295:
1291:
1287:
1283:
1279:
1275:
1271:
1267:
1263:
1259:
1256:) can form a
1255:
1251:
1246:
1244:
1239:
1236:
1228:
1223:
1220:
1219:
1218:
1216:
1212:
1208:
1198:
1196:
1193:
1191:
1188:
1187:
1183:
1179:
1175:
1173:
1170:
1168:
1165:
1164:
1160:
1159:autocatalytic
1156:
1151:
1147:
1143:
1141:
1138:
1136:
1133:
1132:
1128:
1125:
1122:
1121:
1118:
1116:
1112:
1106:
1104:
1100:
1096:
1089:
1087:
1081:
1079:
1077:
1073:
1069:
1065:
1061:
1057:
1053:
1051:
1046:
1042:
1034:
1029:
1025:
1021:
1017:
1013:
1009:
1006:
1001:
997:
993:
990:
986:
982:
979:
975:
971:
967:
963:
960:
957:
953:
949:
948:
947:
941:
937:
933:
929:
928:aspartic acid
926:group on the
925:
921:
918:
914:
910:
906:
902:
899:
895:
891:
887:
883:
882:
881:
878:
875:
871:
866:
863:
859:
855:
851:
847:
846:superfamilies
838:
831:
829:
827:
823:
819:
815:
811:
807:
800:
798:
796:
792:
788:
781:Elastase-like
780:
778:
776:
772:
765:Thrombin-like
764:
762:
760:
756:
755:phenylalanine
752:
744:
742:
740:
739:glutamic acid
736:
735:aspartic acid
732:
728:
720:
718:
716:
708:
702:
699:
696:
695:
691:
690:
685:
682:
679:
676:
675:
671:
670:
665:
664:Dipeptidase E
662:
659:
656:
655:
651:
650:
645:
642:
639:
636:
635:
631:
630:
625:
622:
617:
615:
612:
611:
607:
606:
601:
598:
595:
592:
591:
587:
586:
581:
578:
575:
572:
571:
567:
566:
561:
558:
555:
552:
551:
547:
546:
541:
538:
535:
532:
531:
527:
523:
520:
517:
516:
512:
511:
506:
503:
501:S14, S41, S49
500:
497:
496:
492:
491:
486:
483:
481:S16, S50, S69
480:
477:
476:
472:
468:
464:
461:
458:
457:
453:
452:
447:
444:
441:
438:
437:
433:
432:
427:
424:
422:S11, S12, S13
421:
418:
417:
413:
412:
407:
404:
401:
398:
397:
393:
392:
387:
384:
381:
378:
377:
373:
371:
368:
366:
361:
360:
351:
347:
342:
338:
336:
332:
328:
327:superfamilies
323:
321:
317:
313:
309:
305:
301:
297:
296:superfamilies
293:
285:
283:
281:
277:
273:
269:
265:
261:
258:
254:
250:
246:
245:peptide bonds
242:
238:
234:
227:
223:
219:
207:
204:
202:
198:
195:
192:
190:
186:
183:
180:
178:
174:
169:
165:
162:
159:
156:
153:
150:
146:
143:
140:
138:
134:
131:
128:
126:
122:
119:
116:
114:
110:
107:
106:NiceZyme view
104:
102:
98:
95:
92:
90:
86:
83:
80:
78:
74:
69:
66:
63:
61:
57:
52:
47:
41:
36:
31:
19:
3310:Translocases
3307:
3294:
3281:
3268:
3255:
3245:Transferases
3242:
3229:
3086:Binding site
2798:fibrinolysis
2796:
2674:
2640:Chymotrypsin
2605:
2478:
2435:Glutamate II
2230:
2179:Exopeptidase
2046:
2042:
2032:
2007:
2003:
1996:
1963:
1959:
1953:
1942:. Retrieved
1938:the original
1928:
1893:
1889:
1883:
1848:
1844:
1778:
1774:
1764:
1739:
1735:
1729:
1686:
1682:
1672:
1637:
1633:
1623:
1598:
1594:
1588:
1563:
1559:
1553:
1538:Proteinase K
1428:and others.
1418:
1354:
1331:can lead to
1326:
1299:A family of
1298:
1266:antithrombin
1261:
1247:
1240:
1232:
1214:
1211:chymotrypsin
1210:
1206:
1204:
1189:
1177:
1172:chymotrypsin
1166:
1134:
1107:
1093:
1085:
1059:
1055:
1048:
1044:
1040:
1038:
956:nucleophilic
945:
917:peptide bond
879:
867:
843:
804:
784:
768:
748:
724:
721:Trypsin-like
712:
687:
667:
647:
627:
624:Chymotrypsin
603:
583:
565:Homo sapiens
563:
545:Homo sapiens
543:
508:
505:Clp protease
488:
449:
429:
409:
389:
324:
308:protein fold
289:
276:chymotrypsin
257:nucleophilic
243:that cleave
236:
232:
231:
94:BRENDA entry
3081:Active site
2703:Factor XIIa
2683:Factor VIIa
2667:Coagulation
2372:Cathepsin A
2308:Cathepsin C
2268:Dipeptidase
1481:Proteolysis
1400:anaphylaxis
1383:is used in
1274:coagulation
1190:Proelastase
1135:Trypsinogen
1115:proteolysis
1111:proteolysis
952:polypeptide
890:nucleophile
850:amino acids
810:prokaryotes
646:precursor (
560:Lactoferrin
540:Nucleoporin
471:herpesvirus
331:nucleophile
272:prokaryotes
264:active site
82:IntEnz view
54:Identifiers
3353:Categories
3284:Isomerases
3258:Hydrolases
3125:Regulation
2985:Subtilisin
2927:Batroxobin
2708:Kallikrein
2698:Factor XIa
2688:Factor IXa
2655:Pancreatic
2650:Neutrophil
2010:: 108573.
1944:2008-06-02
1545:References
1533:Subtilisin
1528:Keratinase
1454:threonine-
1392:mast cells
1345:SARS-CoV-2
1337:thrombosis
1305:pacifastin
1278:thrombosis
1250:inhibitors
1243:inhibitors
1235:inhibitors
1229:Inhibition
1180:undergoes
1028:C-terminus
996:N-terminus
806:Subtilisin
759:tryptophan
629:Bos taurus
411:Sus scrofa
386:Subtilisin
280:subtilisin
268:eukaryotes
260:amino acid
151:structures
118:KEGG entry
3364:Proteases
3359:EC 3.4.21
3163:EC number
3013:Cathepsin
2999:Sedolisin
2975:Prostasin
2693:Factor Xa
2553:: Unknown
2231:Methionyl
2162:proteases
2158:Hydrolase
1898:CiteSeerX
1459:aspartic-
1449:cysteine-
1422:Influenza
1408:half-life
1404:histamine
1343:activate
1329:protein C
1301:arthropod
1282:emphysema
1182:autolysis
1020:histidine
1005:histidine
985:histidine
974:histidine
936:histidine
934:with the
911:from the
905:histidine
874:substrate
870:ping-pong
826:evolution
526:phage K1F
467:assemblin
374:Examples
71:Databases
3187:Kinetics
3111:Cofactor
3074:Activity
2914:Venombin
2899:Tryptase
2894:Granzyme
2848:Factor I
2843:Factor D
2838:Factor B
2678:Thrombin
2675:factors:
2645:Elastase
2226:Glutamyl
2216:Cystinyl
2211:Aspartyl
2109:site at
2098:Archived
2073:24600012
2024:35183035
1980:18775459
1960:Peptides
1920:19364497
1875:35702592
1756:19726197
1721:34687719
1664:31786268
1615:20377171
1580:12475199
1464:metallo-
1444:Protease
1432:See also
1313:crayfish
1258:covalent
1215:elastase
1195:elastase
1123:Zymogen
1095:Zymogens
1068:carbonyl
1050:oxyanion
1024:carbonyl
1016:carbonyl
1014:and the
1000:carbonyl
989:carbonyl
978:carbonyl
970:carbonyl
930:in turn
924:carboxyl
909:hydrogen
898:scissile
894:carbonyl
771:thrombin
751:tyrosine
731:arginine
640:S45, S63
600:Rhomboid
442:S24, S26
370:Families
335:families
300:families
249:proteins
206:proteins
194:articles
182:articles
155:RCSB PDB
65:3.4.21.-
3343:Biology
3297:Ligases
3067:Enzymes
2953:Pronase
2943:Acrosin
2889:Chymase
2803:Plasmin
2635:Trypsin
2206:Arginyl
2201:Alanine
2064:4019123
1988:8797134
1866:9185726
1823:2204062
1805:2355359
1783:Bibcode
1712:8633836
1691:Bibcode
1655:7029793
1516:TopFIND
1471:PA clan
1341:TMPRSS2
1309:locusts
1262:serpins
1207:trypsin
1140:trypsin
1126:Enzyme
1117:occur.
1060:Ser 195
1056:Gly 193
1054:. Both
1045:Ser 195
1041:Gly 193
791:glycine
787:alanine
775:plasmin
715:enzymes
469:(human
382:S8, S53
350:PACKMAN
320:PA clan
314:of the
282:-like.
241:enzymes
226:Trypsin
142:profile
125:MetaCyc
3329:Portal
3271:Lyases
2980:Reelin
2922:Ancrod
2879:Other
2613:3.4.21
2551:3.4.99
2467:3.4.21
2390:3.4.17
2360:3.4.16
2342:3.4.15
2295:3.4.14
2260:3.4.13
2221:Leucyl
2188:3.4.11
2175:3.4.11
2123:(MeSH)
2091:MEROPS
2071:
2061:
2022:
1986:
1978:
1918:
1900:
1873:
1863:
1821:
1811:
1803:
1754:
1719:
1709:
1662:
1652:
1613:
1578:
1522:MEROPS
1379:Serum
1155:lysine
1129:Notes
1012:serine
966:serine
959:serine
913:serine
886:serine
818:serine
795:valine
727:lysine
365:family
363:Super-
292:MEROPS
253:Serine
239:) are
189:PubMed
171:Search
161:PDBsum
101:ExPASy
89:BRENDA
77:IntEnz
60:EC no.
3223:Types
2989:Furin
2936:Other
2863:MASP2
2858:MASP1
2788:KLK15
2783:KLK14
2778:KLK13
2773:KLK12
2768:KLK11
2763:KLK10
2469:-25:
2177:-19:
2113:(SLU)
1984:S2CID
1814:54596
1801:JSTOR
1290:AEBSF
542:145 (
137:PRIAM
3315:list
3308:EC7
3302:list
3295:EC6
3289:list
3282:EC5
3276:list
3269:EC4
3263:list
3256:EC3
3250:list
3243:EC2
3237:list
3230:EC1
3003:TPP1
2853:MASP
2758:KLK9
2753:KLK8
2748:KLK7
2743:KLK6
2738:KLK5
2733:KLK4
2728:KLK3
2723:KLK2
2718:KLK1
2168:3.4)
2089:The
2069:PMID
2020:PMID
1976:PMID
1916:PMID
1871:PMID
1819:PMID
1752:PMID
1717:PMID
1660:PMID
1611:PMID
1576:PMID
1426:hRSV
1311:and
1294:PMSF
1292:and
1286:A1AT
1280:and
1268:and
1264:are
1213:and
1058:and
1052:hole
1043:and
964:The
950:The
922:The
884:The
856:57,
793:and
757:and
697:None
602:-1 (
346:2PTN
325:For
290:The
270:and
235:(or
201:NCBI
158:PDBe
113:KEGG
46:1CBW
2993:S1P
2713:PSA
2059:PMC
2051:doi
2012:doi
2008:106
1968:doi
1908:doi
1894:383
1861:PMC
1853:doi
1809:PMC
1791:doi
1744:doi
1707:PMC
1699:doi
1687:120
1650:PMC
1642:doi
1638:432
1603:doi
1599:110
1568:doi
1564:102
1372:or
862:Asp
858:Ser
854:His
741:).
737:or
729:or
660:S51
626:A (
596:S54
576:S66
556:S60
536:S59
521:S74
473:5)
448:I (
251:.
247:in
224:of
177:PMC
149:PDB
3355::
2610:EC
2604::
2415:A2
2378:DD
2370::
2166:EC
2160::
2067:.
2057:.
2047:88
2045:.
2041:.
2018:.
2006:.
1982:.
1974:.
1964:30
1962:.
1914:.
1906:.
1892:.
1869:.
1859:.
1847:.
1843:.
1831:^
1817:.
1807:.
1799:.
1789:.
1779:87
1777:.
1773:.
1750:.
1740:34
1738:.
1715:.
1705:.
1697:.
1685:.
1681:.
1658:.
1648:.
1636:.
1632:.
1609:.
1597:.
1574:.
1562:.
1424:,
1319:.
1296:.
1161:.
852::
828:.
789:,
761:.
753:,
692:)
680:P1
677:PE
672:)
657:PC
652:)
637:PB
632:)
614:PA
608:)
593:ST
588:)
573:SS
568:)
553:SR
548:)
533:SP
528:)
518:SO
513:)
498:SK
493:)
478:SJ
459:SH
454:)
439:SF
434:)
419:SE
414:)
399:SC
394:)
379:SB
3331::
3317:)
3313:(
3304:)
3300:(
3291:)
3287:(
3278:)
3274:(
3265:)
3261:(
3252:)
3248:(
3239:)
3235:(
3059:e
3052:t
3045:v
3023:G
3018:A
3001:/
2995:4
2991:/
2987:/
2968:2
2963:1
2800::
2615:)
2608:(
2594:e
2587:t
2580:v
2430:E
2425:C
2420:B
2410:A
2283:3
2278:2
2273:1
2248:O
2241:2
2236:1
2164:(
2150:e
2143:t
2136:v
2075:.
2053::
2026:.
2014::
1990:.
1970::
1947:.
1922:.
1910::
1877:.
1855::
1849:6
1825:.
1793::
1785::
1758:.
1746::
1723:.
1701::
1693::
1666:.
1644::
1617:.
1605::
1582:.
1570::
1376:.
1284:/
1276:/
961:.
942:.
919:.
686:(
666:(
582:(
562:(
507:(
487:(
428:(
408:(
388:(
48:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.